Abstract
Identification of the specific cytogenetic abnormality is one of the critical steps for classification of acute myeloblastic leukemia (AML) which influences the selection of appropriate therapy and provides information about disease prognosis. However at present, the genetic complexity of AML is only partially understood. To obtain a comprehensive, unbiased, quantitative measure, we performed serial analysis of gene expression (SAGE) on CD15+ myeloid progenitor cells from 22 AML patients who had four of the most common translocations, namely t(8;21), t(15;17), t(9;11), and inv(16). The quantitative data provide clear evidence that the major change in all these translocation-carrying leukemias is a decrease in expression of the majority of transcripts compared with normal CD15+ cells. From a total of 1,247,535 SAGE tags, we identified 2,604 transcripts whose expression was significantly altered in these leukemias compared with normal myeloid progenitor cells. The gene ontology of the 1,110 transcripts that matched known genes revealed that each translocation had a uniquely altered profile in various functional categories including regulation of transcription, cell cycle, protein synthesis, and apoptosis. Our global analysis of gene expression of common translocations in AML can focus attention on the function of the genes with altered expression for future biological studies as well as highlight genes/pathways for more specifically targeted therapy.
Keywords: hematopoietic cell differention, diagnostic microarray
The pathogenesis of acute myeloid leukemia (AML) in many patients is linked to oncogenic fusion proteins, generated as a consequence of chromosome translocations or inversions (1). Many different translocations have been described in AML, the most frequent being the t(9;11), t(15;17), t(8;21), and inv(16), which, taken together with their variants, account for ≈20–30% of AML cases (2, 3), although a recent analysis by Mitelman et al. (4) suggests that the proportion may be closer to 10%. These recurring translocations are now the basis for classification of some patients with AML. Despite genetic heterogeneity, there is increasing evidence for some common molecular and biological mechanisms in the genesis of AML. In particular, one of the components of each fusion protein is almost invariably a transcription factor, frequently involved in the regulation of myeloid cell differentiation (5). As a consequence, AML-associated fusion proteins function as aberrant transcriptional regulators with the potential to interfere with the normal processes of myeloid cell differentiation.
Genome-wide gene expression profiling is becoming useful for the classification of many types of cancer (6, 7), including AML and acute lymphoblastic leukemia (8–15). Although AML sub-types can be distinguished by oligonucleotide microarrays, the results of analysis of different translocations between laboratories are not always similar. This lack of consistency has probably resulted from the heterogeneous nature of clinical samples (age, sex, stage of disease, percentage of blasts in the sample, other chromosomal abnormalities, etc.) as well as for technical reasons, such as the various platforms and algorithms used in the analysis. Moreover analysis of the same data set using different algorithms also yields different results (U. Kees, personal communication). However, this question of reproducibility has recently been reviewed by Sherlock (16), who concludes that when very carefully controlled experiments are done in various laboratories, in general the results are comparable. However, when different materials and different platforms are used, the reproducibility is poor.
We used serial analysis of gene expression (SAGE) to obtain quantitative, unbiased gene expression in bone marrow samples from 22 patients with four subtypes of AML, namely de novo AMLM2 with t(8;21), AMLM3 or M3V with t(15;17), AMLM4Eo with inv(16), and AML with t(9;11) or treatment-related t(9;11). The results of this analysis are presented here.
Results
Characterization of the Leukemic Samples. We studied samples obtained from diagnosis of 22 AML cases representing four de novo and one treatment-related subtypes: five each de novo t(8;21), t(15;17), inv(16), four de novo t(9;11), and three treatment-related t(9;11). All samples were verified by cytogenetic analysis showing the balanced abnormalities as the sole karyo-type change (except for no. 10) in >75% of the cells, and reverse-transcriptase PCR showing the presence of the expected fusion transcript (Tables 1 and 2, which are published as supporting information on the PNAS web site).
Distribution of the SAGE Tags and Match of SAGE Tags to Known Expressed Sequences. We collected a total of 1,247,535 SAGE tags from the 22 AML libraries. From these SAGE tags, we identified 209,486 unique SAGE tags. Matching these SAGE tags to the reference database shows that 136,010 SAGE tags matched to known gene transcripts, and 73,476 had no match representing potentially novel transcripts (Table 2). The number of SAGE tags per library ranged from 23,176 to 84,249. Therefore, the libraries were normalized to ≈50,000 tags per library for comparison, as described in Methods. The number of unique transcripts in each translocation varied substantially; however, the number of unique transcripts in the t(8;21) is smaller and the transcripts are more similar between patients compared with other translocations, suggesting that a smaller number of unique transcripts was actively expressed in t(8;21) patients (Table 2). We compared data from our patient samples with our earlier SAGE analysis of CD15+ normal bone marrow samples and selected SAGE tags that showed a difference in expression up or down at least 5-fold and that were significantly different at the 5% level in the individual leukemia samples.
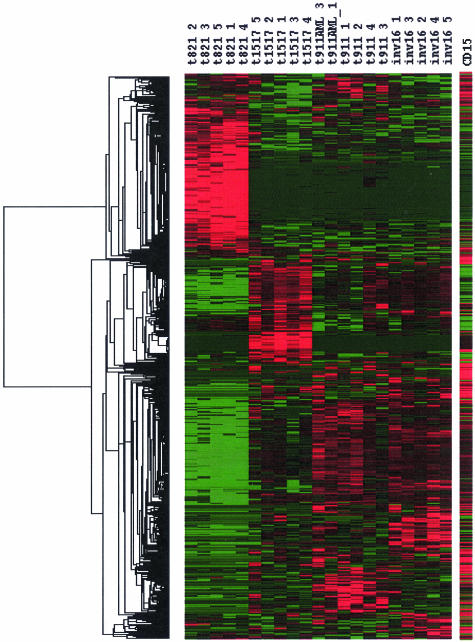
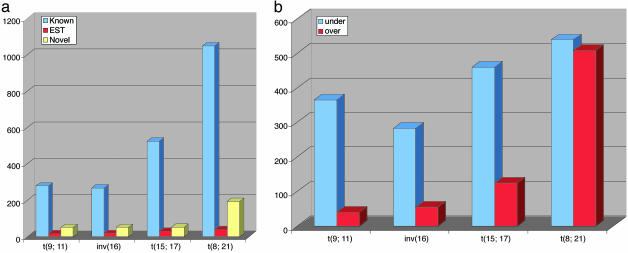
We have identified 2,604 transcripts that were significantly different between the four translocations, except for 56 common to all. A total of 1,882 of the transcripts showed a decrease, and 722 of those showed an increase in expression level. To provide a graphical representation of these selected SAGE tags in a manner comparable to that used for microarray expression data, we converted our quantitative data into a “heat” map (Fig. 1). It is clear that the selected SAGE tags can discriminate between the four translocations. Fig. 1 also clearly shows that t(8;21) has the largest numbers of overexpressed transcripts, and t(9;11) and inv(16) have the fewest relative to the t(8;21) and t(15;17). Among the 2,604 SAGE tags, 2,248 SAGE tags were known genes or ESTs, and 356 were novel. The identity of 378 of the multiple matched and novel transcripts was resolved by using glgi. The novel SAGE tags detected in each translocation were: 195 in t(8;21), 53 in t(15;17), 54 in inv(16), and 51 in t(9;11), and the known genes and ESTs were 1,072 in t(8;21), 546 in t(15;17), 284 in inv(16) and 293 in t(9;11); the remainder were common to all translocations (Fig. 2a). The number of up-and down-regulated transcripts in each translocation is summarized in Fig. 2b. The expression pattern was relatively uniform between patients with the same translocation compared with other translocations, as illustrated in Table 3, which is published as supporting information on the PNAS web site, showing data from 1,110 SAGE tags for all four translocations. The importance of quantitative SAGE data is illustrated in Table 3, which allows for direct comparison of expression levels with no manipulation of the primary information except for normalization of all leukemia samples to ≈50,000 tags. We also identified the transcripts whose expression pattern, either increased or decreased, was common in all four translocations (Table 4, which is published as supporting information on the PNAS web site). t(8;21) had the largest number of transcripts that showed a statistically significant difference from the normal CD15+ cells and had the highest expression level, whereas inv(16) and t(9;11) had the smallest number of transcripts with altered expression (Fig. 1). As a consequence, a smaller number of transcripts were specific discriminators for t(9;11), inv(16), and t(15;17) than for t(8;21).
Fig. 1.
The expression level of the 2,604 SAGE tags whose expression was statistically significantly different from CD15+ control cells has been converted to a “heat” map with red representing overexpression and green underexpression of the transcript. Patient samples sorted by translocation are in vertical columns and the individual SAGE tags are in the horizontal columns.
Fig. 2.
Classification of SAGE tags whose expression is significantly different from normal CD15+ cells. (a) Classification of the 2,604 SAGE tags by whether they represent a known gene, EST, or novel transcript. (b) Distribution of SAGE tags by whether their transcription is up or down in each translocation compared with normal CD15+ cells.
Abnormally Expressed Genes in Each Type of Translocation Related to Cellular Function. To determine the nature of the highly expressed genes in each translocation, we selected the top 20 genes that were significantly highly expressed in each translocation (Table 5, which is published as supporting information on the PNAS web site). Interestingly, nine of the 20 genes highly expressed in t(8;21) were related to ribosomal proteins. This finding is not unexpected, because these are cells involved in very active protein synthesis. However, it is perplexing that only some of them are overexpressed and they are overexpressed only in the t(8;21). The list of those SAGE tags that are underexpressed also includes ribosomal proteins, most of which (eight of 20) are in the t(8;21), but two and three are in the inv(16) and t(15;17), respectively. Note that hemoglobin alpha was the top gene and hemoglobin gamma 2 was the third nonribosomal protein gene in the list. Expression of hemoglobin genes was not expected. Prothymosin alpha (PTMA) is a histone H1-binding protein that interacts with the transcription coactivator CREB-binding protein and potentiates transcription (17). PTMA, a regulator of estrogen receptor transcriptional activity (18) and a negative regulator of caspase-9 activation by inhibiting apoptosome formation, was highly expressed in inv(16) as compared with normal CD15+ cells. CCNB1IP1, which interacts with cyclin B1, the E2 ubiquitin-conjugating enzyme UBCH7, and PFKL were each highly expressed in the inv(16), t(15;17) and t(9;11), respectively.
Recent studies of AML have indicated how disruption of transcription-factor function can disrupt normal cellular differentiation and lead to malignancy (19). We searched our database to identify those genes related to cellular differentiation by focusing on the genes that were related to cell proliferation, cell cycle, and cell death. Different genes related to cell proliferation were abnormally expressed in all four translocations. The examples of the genes specific in each translocation are described below.
t(9;11). Cell survival is associated with defects in either the extrinsic or the intrinsic pathways of apoptosis. Expression of ROCK1, STK17B, and CASP8 genes was down-regulated. CASP8 triggers apoptosis. Down-regulation of CASP8 could suppress apoptosis. AIF1, which can arrest cell cycle, was also down-regulated. Expression of the GSTP1, TYMS, NUP210, C5ORF13, MYB, and WDR1 genes was up-regulated. The MYB gene encodes for proteins that are critical for hematopoietic cell proliferation and development. Previous experiments showed that when human leukemia (K562)-SCID chimeric mice were exposed to antisense MYB RNA, they survived 3.5 times longer than untreated mice (20). These data support earlier observations that overexpression of MYB contributes to leukemogenesis in AML.
inv(16). Contrary to the previously reported results, MYH11 was not a predictor for AML M4eo with inv(16) in our data. Kohlmann et al. (10) suggested that increased expression of MYH11 in inv(16) compared to other translocations likely is due to hybridization of the MYH11-oligonucleotides on the microarray to the M4eo-specific fusion transcript CBFB-MYH11. However, SAGE detects the 3′ part of the normal transcripts as well as the fusion transcripts. We had searched SAGEmap database for the alternatively spliced transcripts of MYH11 to identify the expressed transcript(s) in inv(16) samples. We detected one copy for CAGACCACAA, and no copies for ATCTCGGATC and GCGCAGAAGG of MYH11 in inv(16) samples. None of these SAGE tags was detected in CD15+ normal cells. In our study, expression of MAPK3, FOSL2, RASSF5, and CUL2 were down-regulated, whereas expression of SOX4, PAK1, and RAB13 were up-regulated in inv(16) cells. CUL2 is expressed in proliferating cells and is required at two distinct points in the cell cycle, the G1-to-S-phase transition and in mitosis. CUL2 mutant cells undergo a G1-phase arrest that correlates with accumulation of CKI1, a member of the CIP/KIP family of cyclin-dependent-kinase inhibitors (21), which should lead to decreased cell growth. In contrast, PAK1 is essential for RAS-induced up-regulation of CCND1 during the G1-to-S transition (22). CCND1 (also known as BCL1 or PRAD1) is a proto-oncogene that encodes a regulatory subunit of the cyclin-dependent kinase holoenzyme. Activation of the holoenzyme leads to phosphorylation and inactivation of the RB tumor suppressor protein and thereby promotes entry into S phase (23). Increased expression of PAK1 could contribute to the induced expression of CCND1 and thus increased cell growth. Thus, changes in expression of these genes would appear to result in opposite effects.
t(15;17). MCL1, S100A6, GNAI2, OGFR were underexpressed, and TNFSF10, MPO, FBXL10 were overexpressed. OGFR is an inhibitory peptide that modulates cell proliferation and tissue organization during development, cellular renewal, wound healing, angiogenesis, and cancer. The down-regulation of OGFR resulting in decreased growth inhibition could contribute to cell proliferation. MPO is present in azurophilic granules that appear in the promyelocyte stage of differentiation, and is the most common functional protein of myeloid cells. TNFSF10 (TRAIL) can induce apoptosis in a wide variety of transformed cell lines of diverse lineages, but its expression does not appear to kill normal cells even though it is expressed at significant levels in most normal tissues (24). Because the t(15;17) results in the PML/RARA fusion gene, we investigated the expression of the both genes. We obtained two SAGE tags (AGCACAGGGA and TGGCAGGAAA) for PML and two SAGE tags (TGACCCCGCA and CGCGTGCGCA) for RARA from SAGEmap and compared their expression between the normal and t(15;17) patient's samples. Only AGCACAGGGA of PML showed one copy in both normal and patient's sample. The other SAGE tags were not detected in our analysis.
t(8;21). Like CBFB-MYH11 in the inv(16), the increased expression of CBFA2T1 (formerly ETO) in AML with t(8;21) may be due to the hybridization effect of the subtype-specific AML1-ETO fusion transcript (25) in microarray experiments. However, we detected no expression of AML1 or ETO in our SAGE analysis. MCL1, PNUTL1, FOSB, and DAP were underexpressed, and TRAF4, BCL2L11, and TPDP1 were overexpressed genes. Induction of BCL2L11 causes apoptosis, whereas MCL1 is an antiapoptotic protein that opposes the effect of p53. Because down-regulation of MCL1 and up-regulation of BCL2L11 should increase apoptosis, the excess proliferation of t(8;21) cells likely occurs by other mechanisms.
Abnormal Genes Common in Four Types of AMLs. We have identified 56 genes that were abnormally expressed in all four translocations, 52 were underexpressed, and four genes were up-regulated. For example, NUBPL, TRAM2, and PTRF were up-regulated, and Ficolin1, Lipocalin2, and FASN were down-regulated (Table 4). PTRF is known to interact selectively with ribosomal protein. FASN is involved in various cellular processes such as apoptosis and proliferation. RNA interference-mediated silencing of FASN attenuates growth and induces morphological changes and apoptosis of prostate cancer cells (26), but it might have different role in AML. Some genes show altered expression in only some translocations. For example, TNFSF10, which plays an important role in IFN-induced apoptosis (27) was overexpressed in t(15;17), t(9;11), and inv(16), but not in t(8;21) samples.
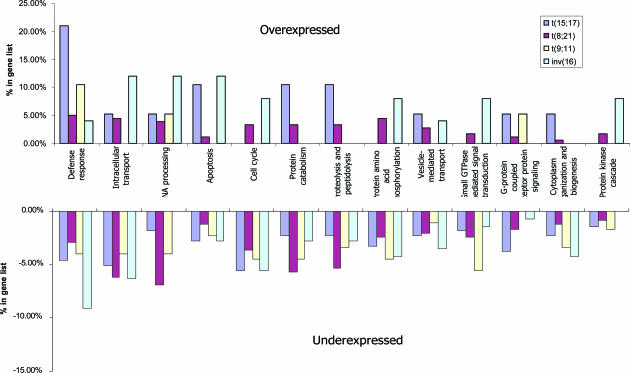
Functional Classification of the Identified Genes in Each Translocation. To gain further insight into the biological importance of these 2,604 differentially expressed SAGE tags, we analyzed the functional categories of known genes by using gene ontology. The function of the 1,110 known genes was classified by gene ontology, including 179 in inv(16), 254 in t(15;17), 468 in t(8;21), and 209 in t(9;11). These genes were grouped into 12 functional categories, including defense response, intracellular transport, cell cycle, apoptosis, signal transduction, and protein biosynthesis. More than half of the genes were underexpressed in patient's sample compared to normal cells. For example, the majority of genes related to cell cycle were underexpressed and a small number of genes were overexpressed only in t(8;21) and inv(16) samples. The majority of t(9;11) specific genes in almost all of the categories showed underexpression (Fig. 3). The detailed information is presented in Table 3.
Fig. 3.
Representation of expression levels of 1,110 known genes by functional categories and by translocation.
Comparison of the Identified AML Genes with Previously Published Results. It is of great importance to validate published candidate genes intended for diagnostic purposes. We reviewed five published reports describing gene expression in AML and we selected 48 genes that were said to be important in distinguishing patients with the individual translocations in AML in at least one of these five published microarray analysis of leukemia samples; more than half of the genes were identified in two independent reports (Table 6, which is published as supporting information on the PNAS web site) (8, 10–13). As mentioned previously, the published data showed some agreement among the results, but generally the analysis indicated that the results varied between different reports, likely due to the use of different patient samples (16), selection bias (28) and the analysis algorithm used (29). Only six of 48 genes, CST7, LGALS9, CLECSF2, RUNX3, SELL, and STAB1 were also appropriately differentially expressed in our SAGE data set; this raises the issue of the sensitivity of SAGE compared with microarrays. It is estimated that microarrays can detect a minimum of five transcripts per cell; we used only 50,000 tags or ≈1/8 of the expected ≈400,000 transcripts per cell, so on average we would detect one transcript if eight transcripts were present. CST7, SELL, and STAB1 were underexpressed in our t(15;17) samples as well as in published reports. However, CLECSF2 was an underexpressed discriminator of t(15;17) in published reports (8, 11) but it was an underexpressed discriminator of t(8;21) in our samples. LGALS9 was underexpressed in t(15;17) in Valk et al. (13), but it was overexpressed in our t(8;21) samples. RUNX3 was an overexpressed discriminator in t(15;17) of Debernardi et al. (11) and underexpressed in inv(16) of Valk et al. (13), and it was underexpressed in our t(8;21) samples. A number of these genes are not included among our 2,604 discriminatory genes.
Discussion
The current approach to the diagnosis of AML in addition to the standard clinical features and laboratory analyses requires additional extensive procedures including pathology, immunophenotyping, cytogenetics, and molecular diagnostics. Molecular classification based on expression profiling offers a powerful means of distinguishing distinct AML subclasses if it is based on reliable data. Using gene expression profiling based on SAGE, we have demonstrated that distinct features of gene expression were identified in 22 AML samples with the t(9;11), t(8;21), t(15;17), and inv(16). Moreover, the major observation of our study was the remarkable underexpression of the majority of transcripts in all leukemia except t(8;21). These data suggest that the expression of many genes related to cellular differentiation is suppressed or not activated.
Genome-wide analysis of gene expression in AML has been reported by several groups using microarray analysis (8, 11–13, 30, 31). Each study described genes that were identified as being reliable in distinguishing the common translocations. Unfortunately, the data from various groups often failed to agree; as noted by Sherlock (16), some of this variability may be due to use of different materials and platforms. Our use of SAGE was a unique strategy to acquire complete, unbiased, quantitative data from >106 individual SAGE tags. Compared with microarray, SAGE has many advantages: (i) it requires no prior genetic information about the samples; (ii) the output of the data are quantitative and therefore does not require any conversion; (iii) it is very sensitive, being able to detect lower abundant transcripts; and (iv) once the data are generated, one can use them continually for comparison between different samples.
Our study identified 2,604 unique transcripts whose expression varied significantly in different translocations. Contrary to a previously published report (12), our study reveals that t(8;21) has a highly correlated pattern of expression among different patients, followed by the t(15;17). The t(9;11) and inv(16) have more variable patterns between samples. The t(8;21) also has the lowest number of unique transcripts, but they are expressed at the highest level. Interestingly, >2/3 of the transcripts were down-regulated in t(9;11), inv(16), and t(15;17) compared with only 1/2 in the t(8;21) (Fig. 2b). It appears that the level of certain transcripts is specific for only one translocation; however, the expression level of other transcripts may be altered in two or more translocations. We also identified 52 genes that were underexpressed in all four translocations, including TRRAP, YWHAQ, CAPN10, C1QTNF6, CFL1, and CAST. TRRAP is known to be an essential cofactor for both the MYC and E1A/E2F oncogenic transcription factor pathways. It has been shown recently that MYC regulates E2F by activating micro-RNAs (32). These common genes could potentially be used as universal markers for leukemia diagnosis.
We had compared our SAGE data with 48 translocation specific genes mentioned in at least one of five published papers including the two just discussed (Table 6). Six genes, CLECSF2, CST7, LGALS9, RUNX3, SELL, and STAB1, matched with our SAGE data. We saw no expression at all in our SAGE data for the two genes that were mentioned in all five reports, namely CBFA2T1 and MYH11. One possible explanation is that SAGE detected the 3′ part of the UTR and did not necessarily detect the same region of the transcripts recognized by the microarray. For example, Schoch et al. (8) identified 36 genes that could differentiate the three AML subtypes including t(8;21), inv(16), and t(15;17). From these genes, they identified 13 genes as a minimal set as the discriminators including PRKAR1B that is down-regulated and MYH11 overexpression and HOXB2 that are overexpressed in inv(16). Our study confirmed the overexpression of HOXB2 and the underexpression of PRKAR1B in inv(16), but we did not detect MYH11 overexpression in inv(16). We also confirmed that GNAI2 is down-regulated in t(15;17). In an extension of their study, Kohlmann et al. (10) analyzed expression patterns in eight different types of acute leukemia (AML and acute lymphoblastic leukemia), and identified 25 genes that were sufficient for classifying AML subtypes. Of these 25 genes, MYH11 was a specific discriminator for inv(16) and showed increased expression in inv(16) samples. Increased expression of CBFA2T1 and POU4F1 was observed in t(8;21) samples; POU4F1 has been shown to confer an oncogenic potential when cotransfected with HRAS. ARGHGAP4 is pre-dominantly expressed in hematopoietic cells but showed a lower expression level in AML with t(15;17) (33). We did not observe any differences in expression levels for these 25 genes in our SAGE data.
Gene ontology (GO) provides a tool for functional interpretation of abnormally expressed genes in leukemia. We have classified the functional category of 1,110 AML genes by GO. A total of 1,110 genes were classified within 12 major functional categories, whereas for the remaining 1,271 transcripts, their gene equivalent could not be determined (Table 3). This analysis demonstrates that SAGE could be used as a tool not only for distinguishing subclasses of AML but also to identify new transcripts whose function has yet to be defined.
In addition, we have identified 73,476 unique transcripts in this study; however, the expression level of most transcripts was relatively low being less than three copies of each transcript in 1,247,535 total transcripts. As a result, the majority of these tags were not included in the final 2,604 tags after the selection process. However, a low level of expression does not mean that a transcript has no importance. The critical importance of previously unidentified noncoding RNAs is becoming increasingly recognized. The detection by Cheng et al. (34) of a very large number of “intronic” transcripts provides further support for the existence of noncoding RNAs, which include small interfering RNAs (35), sense–antisense pairs (36), and microRNAs (32, 37–39), some of which have been shown to play a critical role in cancer (32) and leukemia (37, 38). The ground-breaking paper from Golub and colleagues (38) on the use of beads to measure microRNAs revealed that the analysis was more efficient in distinguishing specific acute lymphoblastic leukemia subtypes than the Affymetrix microarray. Further studies are needed to reveal the importance of the novel transcripts, which might have future applications, including the identification of markers for early diagnosis, targets for drug design, and indicators for treatment responsiveness and prognosis.
In conclusion, our data illustrate and further confirm the applicability of gene expression profiling by using SAGE for the stratification of leukemia subtypes, as a means to identify some targets that could then be used in a smaller format for diagnosis. By combining these analyses with molecular biological methods, this approach may provide a more valid basis for the accurate diagnosis of subtypes of AML than current methods.
Methods
Isolation of Myeloid Progenitor Cells from Patient's Samples. Samples from four patients with de novo t(9;11), three patients with treatment-related t(9;11), and five patients each each with inv(16), t(15;17), and t(8;21) were selected based on the cytogenetic examination of metaphase bone marrow cells. The samples were obtained at diagnosis with informed consent at The University of Chicago or other hospitals and contained at least 75% translocation-positive cells that, with one exception, had no other chromosome abnormalities. Mononuclear cells were purified by Nycoprep 1.077A (Axis-Shield, Oslo) according to the manufacturer's recommendation. Myeloid progenitor cells were isolated from these mononuclear cells by using immunomagnetic anti-CD15 beads (Dynal Biotech).
SAGE Analysis. Poly(A)+ RNA isolation, cDNA synthesis, and SAGE analysis were carried out according to Lee et al. (40). For each SAGE library, ≈3,000 sequencing reactions were performed. Tags were extracted from the raw sequence data with sage2000 analysis software kindly provided by Kenneth W. Kinzler (The Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore). A total of 1,247,535 SAGE tags was collected from 22 SAGE libraries. SAGE tags were matched to the SAGE references database (SAGEmap reliable) for gene identification. For statistical analysis, the SAGE tags from each SAGE library were extracted to yield a total tag copy close to 50,000 per library. Each library had ≈3,000 sequence files, each of which had ≈15–30 SAGE tags. We randomly selected sequence files and extracted the files until the total tag number reached 50,000.
Bioinformatics and Statistical Analyses. Individual libraries from each of the four types of leukemia were compared to the normal control CD15+ library pooled from three normal samples (40). Differentially expressed transcripts were identified by using two criteria: first, >5-fold difference between the average tag count in each type of leukemia library and the control, and then the unadjusted P value <0.05 for the modified t test (41). The modified t test, based on a β-binomial sampling model, appropriately accounts for the between-library variability, and assigns different weights to each library according to its size. Because no between-library variability could be assessed for the pooled library, a one-sample version of the t test was used and the normal tag count was treated as a constant. This test could not be applied to transcripts that were not detected in all leukemic samples (i.e., all 0 counts); therefore, undetected transcripts were selected if the corresponding control count was greater than 5. These analyses resulted in the selection of 2,604 tags that were uniquely over- or underexpressed in a single leukemia type relative to each other and to normal CD15+ cells. Additional analyses were carried out comparing the level of expression of these 2,604 tags with data we had previously obtained from our SAGE analysis of normal CD34+ bone marrow cells (42).
Gene Confirmation Using the GLGI Technique. For the selected unique and multimatched SAGE tags, GLGI analysis (43) was performed to obtain longer 3′ ESTs corresponding to each SAGE tag.
Clustering of SAGE Data. cluster and treeview software were used for visualization of the commonly deregulated transcripts in AML (44). The average clustering of the SAGE data were based on the fold change of tag counts for each transcript comparing AML cells to normal CD15+ cells. Two-way (by gene and AML sample) hierarchical clustering was used to examine the relationships among the AML libraries.
Functional Classification of SAGE Data. For functional classification of the identified genes in AML, we used ease (version 2.0) software (http://david.niaid.nih.gov/david/ease.htm) for gene ontology analysis. ease performs a statistical analysis of gene categories in the gene list to find those that are the most overrepresented either because of under- or overexpression. This allows us to define the “biological process” for the analyzed genes.
Supplementary Material
Acknowledgments
We appreciate the thoughtful comments and criticisms of Professor Felix Mitelman and Dr. Nancy Zeleznik-Le. This research has been supported by National Institutes of Health Grants CA40046 and CA84405, Babe and Marvin Conney, and the University of Chicago (J.D.R.). This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (E.D.G., J.T., and P.P.L.).
Conflict of interest statement: No conflicts declared.
Abbreviations: AML, acute myeloid leukemia; SAGE, serial analysis of gene expression.
References
- 1.Rowley, J. D. (1999) Semin. Hematol. 36, 59–72. [PubMed] [Google Scholar]
- 2.Smith, M., Barnett, M., Bassan, R., Gatta, G., Tondini, C. & Kern, W. (2004) Crit. Rev. Oncol. Hematol. 50, 197–222. [DOI] [PubMed] [Google Scholar]
- 3.Jaffe, E. S., Harris, N. L., Diebold, J. & Muller-Hermelink, H. K. (1999) Am. J. Clin. Pathol. 111, S8–S12. [PubMed] [Google Scholar]
- 4.Mitelman, F., Mertens, F. & Johansson, B. (2005) Genes Chromosomes Cancer 43, 350–366. [DOI] [PubMed] [Google Scholar]
- 5.Sjin, R. M., Krishnaraju, K., Hoffman, B. & Liebermann, D. A. (2002) Blood 100, 80–88. [DOI] [PubMed] [Google Scholar]
- 6.Watson, M. A., Perry, A., Tihan, T., Prayson, R. A., Guha, A., Bridge, J., Ferner, R. & Gutmann, D. H. (2004) Brain Pathol. 14, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haven, C. J., Howell, V. M., Eilers, P. H., Dunne, R., Takahashi, M., van Puijenbroek, M., Furge, K., Kievit, J., Tan, M. H., Fleuren, G. J., et al. (2004) Cancer Res. 64, 7405–7411. [DOI] [PubMed] [Google Scholar]
- 8.Schoch, C., Kohlmann, A., Schnittger, S., Brors, B., Dugas, M., Mergenthaler, S., Kern, W., Hiddemann, W., Eils, R. & Haferlach, T. (2002) Proc. Natl. Acad. Sci. USA 99, 10008–10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeoh, E. J., Ross, M. E., Shurtleff, S. A., Williams, W. K., Patel, D., Mahfouz, R., Behm, F. G., Raimondi, S. C., Relling, M. V., Patel, A., et al. (2002) Cancer Cell 1, 133–143. [DOI] [PubMed] [Google Scholar]
- 10.Kohlmann, A., Schoch, C., Schnittger, S., Dugas, M., Hiddemann, W., Kern, W. & Haferlach, T. (2003) Genes Chromosomes Cancer 37, 396–405. [DOI] [PubMed] [Google Scholar]
- 11.Debernardi, S., Lillington, D. M., Chaplin, T., Tomlinson, S., Amess, J., Rohatiner, A., Lister, T. A. & Young, B. D. (2003) Genes Chromosomes Cancer 37, 149–158. [DOI] [PubMed] [Google Scholar]
- 12.Bullinger, L., Dohner, K., Bair, E., Frohling, S., Schlenk, R. F., Tibshirani, R., Dohner, H. & Pollack, J. R. (2004) N. Engl. J. Med. 350, 1605–1616. [DOI] [PubMed] [Google Scholar]
- 13.Valk, P. J., Verhaak, R. G., Beijen, M. A., Erpelinck, C. A., Barjesteh van Waalwijk van Doorn-Khosrovani, S., Boer, J. M., Beverloo, H. B., Moorhouse, M. J., van der Spek, P. J., Lowenberg, B. & Delwel, R. (2004) N. Engl. J. Med. 350, 1617–1628. [DOI] [PubMed] [Google Scholar]
- 14.Ross, M. E., Mahfouz, R., Onciu, M., Liu, H. C., Zhou, X., Song, G., Shurtleff, S. A., Pounds, S., Cheng, C., Ma, J., et al. (2004) Blood 104, 3679–3687. [DOI] [PubMed] [Google Scholar]
- 15.Haferlach, T., Kohlmann, A., Schnittger, S., Dugas, M., Hiddemann, W., Kern, W. & Schoch, C. (2005) Blood 106, 1189–1198. [DOI] [PubMed] [Google Scholar]
- 16.Sherlock, G. (2005) Nat. Methods 2, 329–330. [DOI] [PubMed] [Google Scholar]
- 17.Karetsou, Z., Martic, G., Tavoulari, S., Christoforidis, S., Wilm, M., Gruss, C. & Papamarcaki, T. (2004) FEBS Lett. 577, 496–500. [DOI] [PubMed] [Google Scholar]
- 18.Joosten, S. A., Smit van Dixhoorn, M. G., Borrias, M. C., Ham, V., Groot Koerkamp, M. J., Savolainen-Peltonen, H. M., Hayry, P., Daha, M. R., Kooten, C. & Paul, L. C. (2005) Transpl. Int. 18, 1010–1015. [DOI] [PubMed] [Google Scholar]
- 19.Yan, M., Burel, S. A., Peterson, L. F., Kanbe, E., Iwasaki, H., Boyapati, A., Hines, R., Akashi, K. & Zhang, D. E. (2004) Proc. Natl. Acad. Sci. USA 101, 17186–17191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratajczak, M. Z., Kant, J. A., Luger, S. M., Hijiya, N., Zhang, J., Zon, G. & Gewirtz, A. M. (1992) Proc. Natl. Acad. Sci. USA 89, 11823–11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng, H., Zhong, W., Punkosdy, G., Gu, S., Zhou, L., Seabolt, E. K. & Kipreos, E. T. (1999) Nat. Cell Biol. 1, 486–492. [DOI] [PubMed] [Google Scholar]
- 22.Nheu, T., He, H., Hirokawa, Y., Walker, F., Wood, J. & Maruta, H. (2004) Cell Cycle 3, 71–74. [PubMed] [Google Scholar]
- 23.Sherr, C. J. & Roberts, J. M. (1999) Genes Dev. 13, 1501–1512. [DOI] [PubMed] [Google Scholar]
- 24.Nakata, S., Yoshida, T., Horinaka, M., Shiraishi, T., Wakada, M. & Sakai, T. (2004) Oncogene 23, 6261–6271. [DOI] [PubMed] [Google Scholar]
- 25.Downing, J. R., Head, D. R., Curcio-Brint, A. M., Hulshof, M. G., Motroni, T. A., Raimondi, S. C., Carroll, A. J., Drabkin, H. A., Willman, C., Theil, K. S., et al. (1993) Blood 81, 2860–2865. [PubMed] [Google Scholar]
- 26.De Schrijver, E., Brusselmans, K., Heyns, W., Verhoeven, G. & Swinnen, J. V. (2003) Cancer Res. 63, 3799–3804. [PubMed] [Google Scholar]
- 27.Crowder, C., Dahle, O., Davis, R. E., Gabrielsen, O. S. & Rudikoff, S. (2005) Blood 105, 1280–1287. [DOI] [PubMed] [Google Scholar]
- 28.Ambroise, C. & McLachlan, G. J. (2002) Proc. Natl. Acad. Sci. USA 99, 6562–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dallas, P. B., Gottardo, N. G., Firth, M. J., Beesley, A. H., Hoffmann, K., Terry, P. A., Freitas, J. R., Boag, J. M., Cummings, A. J. & Kees, U. R. (2005) BMC Genomics 6, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armstrong, S. A., Staunton, J. E., Silverman, L. B., Pieters, R., den Boer, M. L., Minden, M. D., Sallan, S. E., Lander, E. S., Golub, T. R. & Korsmeyer, S. J. (2002) Nat. Genet. 30, 41–47. [DOI] [PubMed] [Google Scholar]
- 31.Rozovskaia, T., Ravid-Amir, O., Tillib, S., Getz, G., Feinstein, E., Agrawal, H., Nagler, A., Rappaport, E. F., Issaeva, I., Matsuo, Y., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 7853–7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V. & Mendell, J. T. (2005) Nature 435, 839–843. [DOI] [PubMed] [Google Scholar]
- 33.Liu, W., Khare, S. L., Liang, X., Peters, M. A., Liu, X., Cepko, C. L. & Xiang, M. (2000) Development (Cambridge, U.K.) 127, 3237–3247. [DOI] [PubMed] [Google Scholar]
- 34.Cheng, J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., Long, J., Stern, D., Tammana, H., Helt, G., et al. (2005) Science 308, 1149–1154. [DOI] [PubMed] [Google Scholar]
- 35.Hemann, M. T., Fridman, J. S., Zilfou, J. T., Hernando, E., Paddison, P. J., Cordon-Cardo, C., Hannon, G. J. & Lowe, S. W. (2003) Nat. Genet. 33, 396–400. [DOI] [PubMed] [Google Scholar]
- 36.Chen, J., Sun, M., Hurst, L. D., Carmichael, G. G. & Rowley, J. D. (2005) Trends Genet. 21, 326–329. [DOI] [PubMed] [Google Scholar]
- 37.Calin, G. A., Liu, C. G., Sevignani, C., Ferracin, M., Felli, N., Dumitru, C. D., Shimizu, M., Cimmino, A., Zupo, S., Dono, M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 11755–11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, J., Getz, G., Miska, E. A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B. L., Mak, R. H., Ferrando, A. A., et al. (2005) Nature 435, 834–838. [DOI] [PubMed] [Google Scholar]
- 39.He, L., Thomson, J. M., Hemann, M. T., Hernando-Monge, E., Mu, D., Goodson, S., Powers, S., Cordon-Cardo, C., Lowe, S. W., Hannon, G. J. & Hammond, S. M. (2005) Nature 435, 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, S., Zhou, G., Clark, T., Chen, J., Rowley, J. D. & Wang, S. M. (2001) Proc. Natl. Acad. Sci. USA 98, 3340–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baggerly, K. A., Deng, L., Morris, J. S. & Aldaz, C. M. (2003) Bioinformatics 19, 1477–1483. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, G., Chen, J., Lee, S., Clark, T., Rowley, J. D. & Wang, S. M. (2001) Proc. Natl. Acad. Sci. USA 98, 13966–13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen, J., Lee, S., Zhou, G. & Wang, S. M. (2002) Genes Chromosomes Cancer 33, 252–261. [DOI] [PubMed] [Google Scholar]
- 44.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.