Abstract
Myotubularins, a large family of catalytically active and inactive proteins, belong to a unique subgroup of protein tyrosine phosphatases that use inositol phospholipids, rather than phosphoproteins, as physiological substrates. Here, by integrating crystallographic and deuterium-exchange mass spectrometry studies of human myotubularin-related protein-2 (MTMR2) in complex with phosphoinositides, we define the molecular basis for this unique substrate specificity. Phosphoinositide substrates bind in a pocket located on a positively charged face of the protein, suggesting an electrostatic mechanism for membrane targeting. A flexible, hydrophobic helix makes extensive interactions with the diacylglycerol moieties of substrates, explaining the specificity for membrane-bound phosphoinositides. An extensive H-bonding network and charge–charge interactions within the active site pocket determine phosphoinositide headgroup specificity. The conservation of these specificity determinants within the active, but not the inactive, myotubularins provides insight into the functional differences between the active and inactive members.
Keywords: x-ray structure
The myotubularin gene (MTM1) was identified as a gene mutated in X-linked myotubular myopathy, a congenital muscle disorder in which muscle cell differentiation is impaired (1). Since the identification of MTM1, 13 related genes, named MTM1-related (MTMR) 1–13, have been identified in the human genome (2). Recently, it was discovered that mutations in MTMR2 or MTMR13 (also known as SBF2) cause Charcot–Marie–Tooth disease Type 4B, a neuropathy characterized by abnormal myelination of peripheral nerves (3, 4).
Myotubularin and related genes encode proteins with sequence similarity to protein tyrosine phosphatases (PTPs) (1). PTPs are a large and diverse family of enzymes characterized by a C(X)5R active site motif within an ≈250-aa catalytic domain (5). Despite this similarity, myotubularin phosphatases have poor activity toward phosphoprotein substrates in vitro (6, 7). Numerous studies have now shown that the myotubularins use phosphoinositide lipids (PIs), rather than phosphoproteins, as physiological substrates (6–12). Myotubularin phosphatases specifically dephosphorylate the D-3 position of phosphatidylinositol 3-phosphate [PI(3)P] and phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2], generating phosphatidylinositol and phosphatidylinositol 5-phosphate [PI(5)P], respectively. PI(3)P and PI(3,5)P2 regulate endosomal trafficking events through the recruitment of effector proteins containing specific binding modules such as FYVE, PH, and ENTH domains (13, 14).
Interestingly, a subset of myotubularin family proteins contain substitutions of residues within the C(X)5R active site motif and are catalytically inactive (2). Six of the 14 human proteins (MTMR5 and MTMR9–13) are catalytically inactive, including MTMR13, which is mutated in Charcot–Marie–Tooth disease Type 4B (4). It was originally proposed that the inactive myotubularins might bind to the substrates of the active members and protect them from dephosphorylation, thereby acting as antagonists of endogenous phosphatase activity (15, 16). Recent evidence indicates that the inactive myotubularins function as adaptors for the active members, altering their localization and/or activity (17–20).
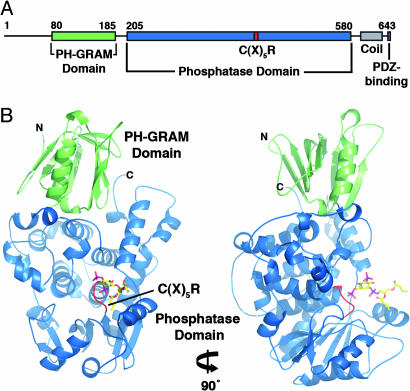
The myotubularins are multidomain proteins that share a common structural core comprised of a PH-GRAM domain, a PTP-like catalytic domain, and a coiled-coil motif (Fig. 1A). Several family members also contain FYVE, PH, or DENN domains, as well as PDZ-binding sites (2). The crystal structure of the PH-GRAM and PTP domains of a representative family member, MTMR2, was recently reported (21). The structure revealed that the PH-GRAM domain (previously known as the GRAM) has the PH domain β-sandwich fold (Fig. 1B). Subsequently, PH-GRAM domains from myotubularin family proteins have been reported to bind PIs, with specificity for PI(3,5)P2 and PI(5)P (22, 23). The catalytic domain is structurally similar to other PTPs, consisting of a central β-sheet sandwiched by α-helices, but it is much larger (≈400 residues) (Fig. 1B). The coiled-coil motif, which was missing from the MTMR2 structure, has, in several cases, been reported to mediate the interactions between active and inactive myotubularins (17, 19, 20).
Fig. 1.
MTMR2 structure. (A) Domain organization of MTMR2. (B) Ribbon diagram of MTMR2 [PI(3,5)P2 complex] in two orientations. Bound substrate is shown in stick form. Figure was created using pymol (DeLano Scientific, South San Francisco, CA; http://pymol.sourceforge.net).
Here we describe crystal structures of MTMR2 in complex with PI(3)P and PI(3,5)P2. In addition, we investigated the solution structure of MTMR2 in the absence or presence of PI(3)P or PI(3,5)P2 using deuterium-exchange mass spectrometry (DXMS). The results reveal how myotubularin family phosphatases bind and dephosphorylate membrane-embedded PIs and provide insight into the functional differences between the active and inactive members.
Results
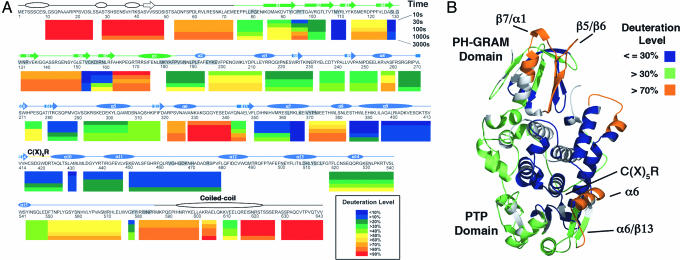
Deuterium Incorporation into Apo-MTMR2. DXMS was used to investigate the structural properties of MTMR2 in solution and provide a reference for subsequent experiments with PI(3)P and PI(3,5)P2. This approach measures the solvent accessibility of main-chain amides in defined segments of a protein through a combination of time-dependent deuterium exchange, limited proteolysis, and mass spectrometry (24). The DXMS profile of MTMR2 is shown in Fig. 2A and mapped onto the existing crystallographic model (residues 74–586) in Fig. 2B (21).
Fig. 2.
DXMS results for apo-MTMR2. (A) Percent deuteration of peptides derived from MTMR2. Each colored bar below the primary sequence represents the percent deuteration of one or more peptides at five time points. Secondary structural elements are shown above the primary sequence. Predicted secondary structure (PSI-PRED) is shown in white, and secondary structure based on the crystal structure of MTMR2 is shown in green (PH-GRAM domain) and blue (phosphatase domain) (21). Residues that form the PH-GRAM/phosphatase domain interface are shaded gray. (B) The average deuterium exchange of each peptide was mapped onto the MTMR2 model (residues 74–586). For clarity, peptides were grouped into three classes: ≤30%, >30%, or >70% deuterated. Parts of the model for which exchange was not measured are shown in gray.
Based on the results, the protein can be divided into three distinct regions. The first region, consisting of the N terminus (residues 1–74), is highly solvent accessible, with all peptides covering this region heavily deuterated at the earliest time points (Fig. 2A). Consistent with this result, truncation of the N terminus, which contains no predicted structural domains and little predicted secondary structure, is required for crystallization (discussed below).
The second region (residues 75–579), consisting of the PH-GRAM and phosphatase domains, is less deuterated overall (Fig. 2A). Among the least solvent accessible areas are segments that form the PH-GRAM/phosphatase domain interface in the crystal structure, indicating that a similar interface exists in solution (Fig. 2). The PH-GRAM and phosphatase domains also contain several solvent-accessible segments that are likely to be functionally important. The PH-GRAM domain contains two highly solvent-accessible loops: the β5/β6 (residues 132–152) and β7/α1 (residues 164–176) connecting loops (Fig. 2). These loops are located on the same face of the protein as the active site and could therefore participate in PI binding (Fig. 2B). The phosphatase domain contains a highly solvent-accessible segment (residues 322–342) that includes helix α6 and the α6/β13 connecting loop (Fig. 2). This region forms one side of the active site (Fig. 2B), and its accessibility is likely to be important for substrate binding (discussed below).
The third region (residues 582–643) includes the C-terminal coiled-coil motif (Fig. 2A). The MTMR2 coiled-coil mediates its interaction with two inactive family members, MTMR5 and MTMR13 (17, 20). In MTMR2, this region is highly solvent accessible, indicating that the coiled-coil is part of an extended region of the protein, accessible to binding partners, and distinct from the slower-exchanging PH-GRAM and phosphatase domains (Fig. 2A).
Generation of MTMR2–PI Complexes. The protein used in crystallization has an inactivating mutation (C417S) in the C(X)5R active site motif and consists of residues 73–586, encompassing the PH-GRAM and phosphatase domains. As found previously, truncation of the N and C termini (residues 1–72 and 587–643) was essential for obtaining crystals (21). Crystals of the truncated protein were soaked with 2 mM concentrations of short-chain versions of PI(3)P (Km = 78 μM), PI(3,5)P2 (Km = 71 μM), or PI(5)P (see Fig. 6, which is published as supporting information on the PNAS web site). This approach generated complexes with PI(3)P and PI(3,5)P2 (Table 1); PI(5)P binding was not observed. The crystals are isomorphous with the previously reported structure of MTMR2 (Protein Data Bank ID code 1LW3), and bound substrates were visualized by difference Fourier methods (Fig. 6). Least-squares comparisons of the MTMR2–PI(3)P and MTMR2–PI(3,5)P2 complexes with the previously reported structure showed that they are essentially identical (rms deviation of 0.5 and 0.4 Å, respectively) (21).
Table 1. Data collection and refinement statistics.
| Statistic | diC4Ptdlns(3)P | diC4Ptdlns(3,5)P2 |
|---|---|---|
| Data collection | ||
| Space group | P41212 | P41212 |
| Unit cell, Å | a = b = 66.23; c = 261.68 | a = b = 66.18; c = 262.37 |
| Resolution, Å | 50.0-1.82 | 50.0-1.98 |
| Completeness, %* | 98.0 (85.0) | 96.7 (79.4) |
| I/σI* | 21.9 (3.5) | 32.3 (2.1) |
| Rsym*† | 10.2 (45.2) | 6.4 (38.9) |
| Refinement | ||
| Resolution, Å | 50.0-1.82 | 50.0-1.98 |
| No. of reflections (cryst/free) | 43,539/4,858 | 33,411/3,716 |
| Rcryst/Rfree‡ | 0.218/0.243 | 0.228/0.252 |
| No. of protein atoms | 4,187 | 4,198 |
| No. of ligand atoms | 35 | 39 |
| No. of water atoms | 399 | 398 |
| rms deviation | ||
| Bond lengths, Å | 0.010 | 0.008 |
| Bond angles, ° | 1.7 | 1.5 |
| Average B-factors, Å2 | ||
| Protein | 42.6 | 47.7 |
| Ligand | 44.5 | 62.5 |
| Solvent | 51.1 | 50.6 |
Highest-resolution shell in parentheses.
Rsym = Σ|I — 〈I 〉|/ΣI, where I is the observed intensity and 〈I 〉 is the average intensity of multiple observations of symmetry-related reflections.
Rcryst = Σ∥Fo| — |Fc∥/Σ|Fo|, where Fo and Fc are observed and calculated structure factor amplitudes, respectively. Rfree is Rcryst for 10% of the reflections excluded from the refinement.
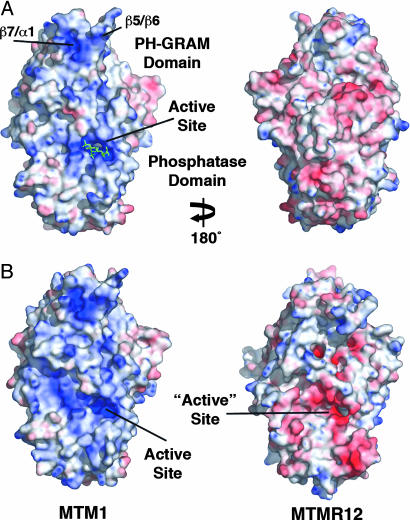
Surface of MTMR2 Is Electrostatically Polarized. The active-site pocket and surrounding surface of MTMR2 are strongly electropositive, whereas the remaining surface is predominantly electronegative (Fig. 3A). This polarization includes the PH-GRAM domain where positive charge coincides with the solvent-accessible β5/β6 and β7/α1 loops (Fig. 2B and 3A). The positively charged, membrane-proximal face of MTMR2 would create electrostatic interactions with negatively charged, PI-containing membranes, contributing to substrate-binding affinity.
Fig. 3.
Surface electrostatic potentials of human myotubularin proteins. (A) The surface of MTMR2 colored by electrostatic potential. Saturating blue and red are 10 and –10 kT/e, respectively. Bound PI(3,5)P2 is shown in green. (B) The membrane-proximal surfaces of representative myotubularins, colored by electrostatic potential.
To assess whether electrostatic polarization is characteristic of the PH-GRAM and phosphatase domains of myotubularin proteins, homology models were generated using MTMR2 (74–586) as a template. Models were created for every active member (36–74% sequence identity to the PH-GRAM and phosphatase domains of MTMR2) and four of the six inactive members (MTMR9–MTMR12; 24–39% identity). As expected, models of the PH-GRAM and phosphatase domains of every active myotubularin were electrostatically polarized, with positive charge localized to the membrane-proximal surface (Fig. 3B; data not shown). In contrast, electrostatic polarization is not characteristic of the PH-GRAM and phosphatase domains of the inactive members (Fig. 3B; data not shown). The pocket and surface equivalent to the substrate-binding pocket and membrane-proximal surface of MTMR2 are often electronegative in the inactive members. For example, MTMR12, which associates with MTM1, relocalizing MTM1 from the plasma membrane to the cytosol, has both an acidic pocket and acidic surrounding surface (18) (Fig. 3B). Based on the MTMR2 structure and homology modeling, electrostatic polarization of the PH-GRAM and phosphatase domains is likely to be characteristic of the active, but not the inactive, myotubularins, suggesting that the two subgroups may have different affinities for PI-containing membranes.
PI Specificity. The myotubularin phosphatases are unique among PTPs because of their specificity for membrane-embedded PI substrates. MTMR2 specifically hydrolyzes the D-3 position of PI(3)P and PI(3,5)P2; significant activity toward other PIs has not been reported (11). In addition, MTMR2 strongly prefers lipid substrates to the isolated inositol headgroups of PI(3)P and PI(3,5)P2 [Ins(1,3)P2 and Ins(1,3,5)P3, respectively] (7). The structures of the MTMR2–PI(3)P and MTMR2–PI(3,5)P2 complexes reveal the basis for this specificity.
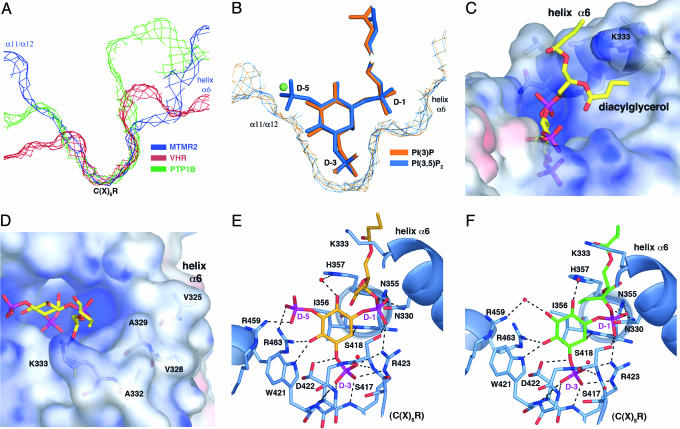
In general, the depth of the active-site pocket is a key determinant of substrate specificity within the PTP family (25). The shallow pocket of dual-specificity PTPs, such as VHR, allows for the hydrolysis of phosphorylated serine, threonine, or tyrosine residues. The deeper pocket of tyrosine-specific PTPs, such as PTP1B, limits hydrolysis to longer phosphotyrosine residues. The MTMR2 active-site pocket is similar in depth to PTP1B but is significantly wider than PTP1B or VHR (Fig. 4A), providing selectivity for the large, multiply phosphorylated inositol headgroups of its PI substrates. The larger width results from two pocket extensions, formed from the α11/α12 connecting loop on one side and helix α6 on the other (Fig. 4B). In the PI(3,5)P2 complex, the D-5 phosphate fills the extension formed by the α11/α12 loop (Fig. 4B). The terminal oxygens of the D-5 phosphate make a salt bridge with Arg-459 and a long hydrogen bond (H-bond) with Arg-463 (Fig. 4E). In contrast, in the PI(3)P complex there are no direct interactions between the protein and 5-hydroxyl. An ordered water molecule fills the α11/α12 extension and mediates a H-bond between the 5-hydroxyl and Arg-459 (Fig. 4 B and F).
Fig. 4.
PI specificity. (A) Slices of active-site surfaces showing the MTMR2 pocket in comparison with VHR and PTP1B. (B) Slices of the active-site surfaces of superimposed MTMR2–PI(3)P and MTMR2–PI(3,5)P2 models. Substrates are shown as sticks, and a water molecule seen in the MTMR2-PI(3)P structure is shown as a green sphere. (C and D) Active-site surface colored by electrostatic potential. Saturating blue and red are 10 and –10 kT/e, respectively. Bound PI(3,5)P2 is shown as a stick. The interaction between the diacylglycerol moiety and helix α6(C) and solvent-exposed hydrophobic residues on helix α6(D) are shown. (E and F) The PI(3,5)P2 (E) and PI(3)P (F) active sites. The phosphatase domain is shown in blue, side chains interacting with the ligands are shown as sticks, and water molecules are red spheres. H-bonds and salt bridges are shown as dashed lines. Several H-bonds between the substrates and water molecules have been omitted for clarity.
The remaining contacts between PI(3)P and PI(3,5)P2 and the protein are identical. The three terminal phosphate oxygens of the D-3 phosphate make seven H-bonds and one salt bridge with the backbone amides of the C(X)5R motif and the guanidium side chain of Arg-423 (Fig. 4 E and F). This bonding pattern is characteristic of PTPs. The D-1 phosphoryl group fills the pocket extension created by helix α6 (Fig. 4B). The two nonbridging oxygens of the D-1 phosphoryl make H-bonds with the side chains of Asn-355, Arg-423, and Ser-418 (Fig. 4 E and F). Asn-330 on helix α6 makes a H-bond with the bridging oxygen between the D-1 phosphoryl and the diacylglycerol moiety (Fig. 4 E and F).
One face of the inositol ring is solvent exposed, whereas the other face makes van der Waals contacts with Ile-356 (Fig. 4 E and F). This interaction is similar to phosphotyrosine binding by PTP1B in which a conserved isoleucine on the P-loop forms part of the recognition site for the phenyl ring (26). The 2-hydroxyl of the inositol ring makes H-bonds with the main-chain amide of Ile-356 and an ordered water molecule (Fig. 4 E and F). The 4-hydroxyl makes H-bonds with the side chains of Arg-463 and Trp-421 of the C(X)5R motif (Fig. 4 E and F). Modeling a phosphate in the position of the 4-hydroxyl of PI(3)P or PI(3,5)P2 creates a steric clash with Arg-463 and Trp-421, accounting for the selectivity of MTMR2 against PI(3,4)P2 and PI(3,4,5)P3. Similarly, hydrolysis of PI(4)P, PI(4,5)P2, or PI(5)P would require a different headgroup orientation as compared with PI(3)P or PI(3,5)P2, creating steric clashes in the active site. The region surrounding the 6-hydroxyl is solvent exposed, and a water-mediated H-bond is observed between the 6-hydroxyl and His-357 (Fig. 4 E and F).
The diacylglycerol group makes extensive interactions with the aliphatic moieties of main- and side-chain groups from helix α6, particularly the side chain of Lys-333 (Fig. 4C). These nonspecific, hydrophobic interactions are likely to make a significant contribution to substrate-binding affinity. This interaction would create significant contact between helix α6, which projects above the substrate-binding pocket (Fig. 4 E and F) and the lipid bilayer. Helix α6 is the most solvent-accessible region of the phosphatase domain (Fig. 2), and this flexibility may allow it to slide past adjacent lipid headgroups and partially insert into the membrane to facilitate entry of the substrate headgroup into the active site. Helix α6 contains a number of surface-exposed, hydrophobic residues (Val-325, Val-328, Ala-329, and Ala-332), consistent with partial membrane insertion of this region upon substrate binding (Fig. 4D).
Comparison with Other Myotubularins. Only one residue in MTMR2, Arg-459, that specifically coordinates PI(3,5)P2 and PI(3)P, is not absolutely conserved among the active human myotubularin phosphatases (see Fig. 7, which is published as supporting information on the PNAS web site). Arg-459, which forms a salt bridge with the D-5 phosphate of PI(3,5)P2 and a water-mediated H-bond with the 5-hydroxyl of PI(3)P, is a lysine in most of the active human myotubularins. In addition, the hydrophobic character of helix α6 is a conserved feature of the active members. The high level of conservation suggests that all human myotubularin phosphatases have the same substrate specificity and a similar mode of substrate binding. In contrast, the residues in MTMR2 that coordinate PI(3)P and PI(3,5)P2 are not conserved in the inactive human members (Fig. 7). Moreover, the region equivalent to helix α6 of MTMR2 is highly variable, in sequence and predicted secondary structure, in the inactive members. The lack of conservation suggests that the inactive myotubularins not only cannot catalyze bond cleavage but are unlikely to bind PI(3)P or PI(3,5)P2 via their substrate-binding pockets.
Two residues that coordinate PI(3,5)P2 and PI(3)P in MTMR2, Ser-418 and Arg-463, are sites of missense mutations in MTM1 that cause X-linked myotubular myopathy (27). Ser-418 (Ser-376 in MTM1) makes a H-bond with the D-1 phosphoryl group of each substrate, whereas Arg-463 (Arg-421 in MTM1) H-bonds the 4-hydroxyl of PI(3)P and the 4-hydroxyl and D-5 phosphate of PI(3,5)P2 (Fig. 4 and F). Based on the structures, mutation of either of these residues would be expected to disrupt substrate binding. Consistent with this finding, the Ser-376 → Asn disease mutation eliminates MTM1 activity in vitro (6).
Implications for Catalysis. In the PTP catalytic mechanism, the C(X)5R cysteine functions as a nucleophile, attacking the phosphorous atom of the substrate. A conserved aspartic acid located on a loop near the top of the active-site pocket (termed the WPD-loop) acts as a general acid/base (5). In the MTMR2-PI(3)P and -PI(3,5)P2 complexes, Asp-422 of the C(X)5R motif is H-bond distance to the scissile oxygens, indicating that it functions as the general acid/base (Fig. 4 E and F). Consistent with this result, mutation of Asp-422 → Ala renders the enzyme inactive (21). The location of the general acid in MTMR2 in the C(X)5R motif, rather than on the WPD-loop, distinguishes it from most other PTPs. The WPD-loop of MTMR2, which contains Asn-355, Ile-356, and His-357, plays an important role in determining substrate specificity.
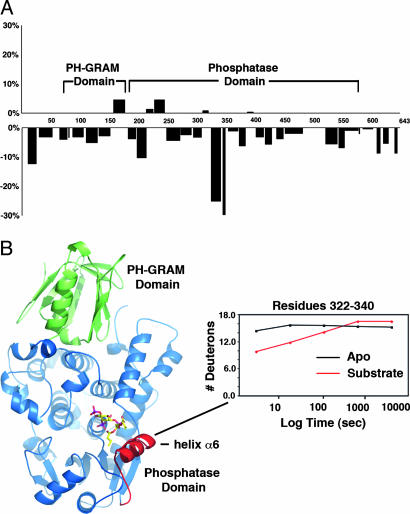
Deuterium Incorporation into PI-Bound MTMR2. DXMS was used to assess changes in the structure of MTMR2 induced by PI(3)P or PI(3,5)P2 binding. Exchange was performed in the presence of 1 mM PI and compared with apo-MTMR2 (Fig. 2). Significant substrate-induced changes were localized to a single segment of the phosphatase domain (residues 320–345) centered on helix α6 (Fig. 5). Helix α6 forms an extension to the active-site pocket and makes significant interactions with the D-1 phosphate and diacylglycerol moiety of each substrate (Fig. 4). In the absence of substrate, this segment is the most heavily exchanged in the phosphatase domain (Fig. 2). Upon substrate binding, the degree of exchange decreases dramatically, indicating an extensive interaction between this segment and substrate in solution (Fig. 5B). As expected, longer durations of deuterium exchange gave rise to increased deuteration throughout the protein and masked differences evident shortly after substrate binding. None of the other changes detected after substrate binding are as significant (Fig. 5). No changes are evident in the PH-GRAM domain, including the highly solvent-accessible β5/β6 and β7/α1 connecting loops. Similarly, there are no significant changes at the PH-GRAM/phosphatase domain interface, indicating that the extensive interaction between the two domains is a fixed property of the protein in solution. Finally, there are no additional changes upon substrate binding within the phosphatase domain, suggesting that the structural changes centered on helix α6 at the active site do not propagate throughout the domain.
Fig. 5.
Changes in deuteration after PI binding. (A) Difference in deuteration (expressed as percentage of total amides) between PI-bound [PI(3)P] and apo-MTMR2 after a 10-s incubation (the results were nearly identical for each substrate; data not shown). A positive value represents increased deuteration, and a negative value represents decreased deuteration after binding. The x axis is the primary sequence of MTMR2. Domain boundaries are indicated. (B) Ribbon diagram of MTMR2 highlighting the region around helix α6 (red) that showed decreased deuteration after PI binding. A plot of deuteron incorporation as a function of time for residues 322–340 is shown.
Discussion
The results of our crystallographic and DXMS analyses of MTMR2 in complex with PI(3)P and PI(3,5)P2 reveal several mechanisms through which specificity for membrane-embedded PI substrates is achieved. First, MTMR2 is electrostatically polarized, with positive charge localized to the membrane-proximal face. The positive charge would provide nonspecific, electrostatic interactions with negatively charged PI headgroups, contributing to the protein's affinity for PI-containing membranes. Second, the surface surrounding the active-site pocket contains several highly solvent accessible loops. The accessibility of these regions would facilitate interactions with lipid head-groups in the interfacial region of the bilayer. The accessibility of helix α6, which makes extensive interactions with the diacylglycerol moieties of the substrates, would allow it to slide past the headgroups of adjacent lipids, facilitating insertion of the substrate headgroup into the active site. Helix α6 also contains several solvent-exposed, hydrophobic side chains. Partial penetration of these residues into the hydrophobic portion of the membrane during catalysis would provide additional affinity for PI(3)P- or PI(3,5)P2-containing membranes. Insertion of hydrophobic side chains into the apolar interior of membranes has been reported for FYVE and PX domains, as well as for phosphoinositide-specific phospholipase c (PLCδ1), which also has a hydrophobic ridge adjacent to its active site (28–31). Finally, the substrate-binding pocket of MTMR2 is wider and deeper than that of most other PTPs, which provides selectivity for the large, phosphorylated headgroup of its PI substrates. Within the pocket, a network of interactions specifically coordinate the headgroups of PI(3)P or PI(3,5)P2 while sterically prohibiting the binding of other PIs.
Based on homology modeling and primary sequence conservation, the specificity determinants identified in MTMR2 are expected to be general determinants of substrate specificity for the human myotubularin family phosphatases. In contrast, the specificity determinants are not well conserved among the inactive members. It is therefore unlikely that the inactive proteins bind phosphorylated substrates via their substrate-binding pockets as a mechanism to oppose endogenous phosphatase activity, as originally proposed (15, 16). Recent results suggest that the inactive myotubularin family members function as adaptors for the active forms, altering their localization and/or activity (17–20).
The PH-GRAM domain of MTMR2, based on its position relative to the active site, would be expected to make considerable contact with PI-containing membranes. The size and rigidity of the PH-GRAM/phosphatase domain interface places an important constraint on the orientation of the PH-GRAM domain relative to the bilayer. Based on the structural data, the β5/β6 and β7/α1 loops would be the most likely location of a PI-binding site in the MTMR2 PH-GRAM domain. In support of this hypothesis, the PH-GRAM domain is electrostatically polarized, with positive charge approximately coinciding with these loops. However, we did not detect PI binding to the PH-GRAM domain in our crystal structures or DXMS experiments. Our inability to observe binding may indicate that the PH-GRAM domain of MTMR2 binds PIs with low affinity, a characteristic of the majority of PH domains (32). PI-binding at the β5/β6 and β7/α1 loops would be most similar to the binding mode of the PH domain of β-spectrin, which binds inositides between the β1/β2 and β5/β6 loops (33).
We have described the results of structural analyses of a PI-specific PTP, human MTMR2, in complex with substrates. The results reveal the structural elements that contribute to specificity for membrane-embedded, PI substrates and provide important insight into the functions of myotubularin family proteins.
Methods
Protein Expression and Purification. MTMR2 (C417S) was expressed in Escherichia coli as a C-terminal His-tagged fusion protein and purified as described in ref. 21. MTMR2 (C417S)73–586 was cloned by PCR using MTMR2 (C417S) as a template, expressed in E. coli as a C-terminal His-tagged fusion protein, and purified as described in ref. 21.
Crystallization. Purified MTMR2 (C417S)73–586 was concentrated to 7 mg/ml in 10 mM Tris (pH 8.0), 150 mM NaCl, and 2 mM DTT. Crystals were obtained at 20°C by vapor diffusion vs. 0.1 M Tris (pH 7.5), 2 mM TCEP, and either 3–6% poly(ethylene glycol) (PEG) 10,000 or 1–5% PEG 35,000. To generate complexes, single crystals were soaked 16 h in reservoir buffer in which the PEG concentration was increased 2–3%, and 2 mM diC4PtdIns(3)P, diC4PtdIns(3,5)P2, or diC4PtdIns(5)P was added. For data collection, crystals were transferred to cryoprotectant consisting of the same overnight soak buffer supplemented with 25% ethylene glycol and flash frozen.
Data Collection and Refinement. Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory and processed by using hkl2000 (34). All crystals were isomorphous with the previously reported MTMR2 crystals; therefore, these coordinates (Protein Data Bank ID code 1LW3) were used in the initial refinement of the models described here (21). Refinement was done in cns by using torsion-angle dynamics, energy minimization, and B-factor refinement (35). Manual model building was done in xtalview (36). cns parameter and topology files were generated for diC4PtdIns(3)P, and diC4PtdIns(3,5)P2 by using the HIC-Up server. Refinement was monitored by using the free R factor calculated with 10% of observed reflections. Four patches of density at the surface of the protein in the diC4PtdIns(3)P structure and three patches in the diC4PtdIns(3,5)P2 structure were interpreted as ethylene glycol molecules based on the cryoprotectant and the shape of the density. All residues are within allowed regions of the Ramachandran plot. Statistics of the structure determination are presented in Table 1.
DXMS. Before studying experimental samples, proteolytic digestion conditions that generated peptides of MTMR2 of optimal size and distribution for deuterium-exchange analyses were determined as described in ref. 37. Experimental samples were prepared by mixing 107 μM full-length MTMR2 (C417S) with 1 mM diC4PtdIns(3)P or 1 mM diC4PtdIns(3,5)P2. An apo sample was prepared by mixing 107 μM MTMR2 with an equivalent volume of buffer. All samples were equilibrated overnight at 4°C. Deuterium exchange was initiated by diluting 10 μl of the apo sample with 30 μl of deuterated buffer (8.3 mM Tris, pH 7.0/50 mM NaCl in D2O) or 10 μl of the experimental samples with 30 μl of deuterated buffer containing 1 mM diC4PtdIns(3)P or diC4PtdIns(3,5)P2. Samples were deuterated for varying times (10–3,000 s) and processed, along with control samples of nondeuterated and fully deuterated MTMR2, as described in ref. 37.
Sequence Alignment and Homology Modeling. Protein sequence alignments were calculated by using the algorithm clustalw (macvector 7.2.2; Accelrys, Inc., San Diego). Molecular modeling was done with swiss-model (38). Human MTMR2 (Protein Data Bank ID code 1LW3; residues 74–586) was the template. MTM1 (33–544), MTMR1 (96–607), MTMR3 (27–582), MTMR4 (32–576), MTMR6 (2–509), MTMR7 (2–510), MTMR8 (2–506), MTMR9 (2–504), MTMR10 (49–667), MTMR11 (62–594), and MTMR12 (44–649) were the target sequences. Initial models and structure-based alignments were iteratively improved based on the structure of MTMR2 and primary sequence conservation.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant 2R01 DK018024-31 (to J.E.D.) and National Institutes of Health National Cancer Institute Innovative Molecular Analysis Technologies Grants CA099835 and CA118595 (to V.L.W.). M.J.B. is supported by a National Cancer Institute Cancer Cell Biology Training Grant.
Conflict of interest statement: No conflicts declared.
Abbreviations: DXMS, deuterium exchange MS; PI, phosphoinositide lipid; PI(3)P, phosphatidylinositol 3-phosphate; PI(3,5)P2, phosphatidylinositol 3,5-bisphosphate; PI(5)P, phosphatidylinositol 5-phosphate; PTP, protein tyrosine phosphatase.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1ZSQ and 1ZVR).
References
- 1.Laporte, J., Hu, L. J., Kretz, C., Mandel, J.-L., Kioschis, P., Coy, J. F., Kluack, S. M., Poustka, A. & Dahl, N. (1996) Nat. Genet. 13, 175–182. [DOI] [PubMed] [Google Scholar]
- 2.Laporte, J., Bedez, F., Bolino, A. & Mandel, J.-L. (2003) Hum. Mol. Genet. 12, R285–R292. [DOI] [PubMed] [Google Scholar]
- 3.Bolino, A., Muglia, M., Conforti, F. L., LeGuern, E., Salih, M. A. M., Georgiou, D.-M., Christodoulou, K., Hausmanowa-Petrusewicz, I., Mandich, P., Schenone, A., et al. (2000) Nat. Genet. 25, 17–19. [DOI] [PubMed] [Google Scholar]
- 4.Senderek, J., Bergmann, C., Weber, S., Ketelsen, U., Schorle, H., Rudnik-Shoneborn, S., Buttner, R., Buchheim, E. & Zerres, K. (2003) Hum. Mol. Genet. 12, 349–356. [DOI] [PubMed] [Google Scholar]
- 5.Denu, J. & Dixon, J. E. (1998) Curr. Opin. Chem. Biol. 2, 633–641. [DOI] [PubMed] [Google Scholar]
- 6.Taylor, G. S., Maehama, T. & Dixon, J. E. (2000) Proc. Natl. Acad. Sci. USA 97, 8910–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, S.-A., Taylor, G. S., Torgersen, K. M. & Dixon, J. E. (2002) J. Biol. Chem. 277, 4526–4531. [DOI] [PubMed] [Google Scholar]
- 8.Blondeau, F., Laporte, J., Bodin, S., Superti-Furga, G., Payrastre, B. & Mandel, J.-L. (2000) Hum. Mol. Genet. 9, 2223–2229. [DOI] [PubMed] [Google Scholar]
- 9.Zhao, R., Qi, Y., Chen, J. & Zhao, Z. J. (2001) Exp. Cell Res. 265, 329–338. [DOI] [PubMed] [Google Scholar]
- 10.Walker, D. M., Urbe, S., Dove, S. K., Tenza, D., Raposo, G. & Clague, M. J. (2001) Curr. Biol. 11, 1600–1605. [DOI] [PubMed] [Google Scholar]
- 11.Berger, P., Bonneick, S., Willi, S., Wymann, M. & Suter, U. (2002) Hum. Mol. Genet. 11, 1569–1579. [DOI] [PubMed] [Google Scholar]
- 12.Schaletzky, J., Dove, S. K., Short, B., Lorenzo, O., Clague, M. J. & Barr, F. A. (2003) Curr. Biol. 13, 504–509. [DOI] [PubMed] [Google Scholar]
- 13.Lemmon, M. A. (2003) Traffic 4, 201–213. [DOI] [PubMed] [Google Scholar]
- 14.Friant, S., Pecheur, E. I., Eugster, A., Michel, F., Lefkir, Y., Nourrisson, D. & Letourneur, F. (2003) Dev. Cell 3, 499–511. [DOI] [PubMed] [Google Scholar]
- 15.Cui, X., De Vivo, I., Slany, R., Miyamoto, A., Firestein, R. & Cleary, M. L. (1998) Nat. Genet. 18, 331–337. [DOI] [PubMed] [Google Scholar]
- 16.Hunter, T. (1998) Nat. Genet. 18, 303–305. [DOI] [PubMed] [Google Scholar]
- 17.Kim, S.-A., Vacratsis, P. O., Firestein, R., Cleary, M. L. & Dixon, J. E. (2003) Proc. Natl. Acad. Sci. USA 100, 4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nandurkar, H. H., Layton, M., Laporte, J., Selan, C., Corcoran, L., Caldwell, K. K., Mochizuki, Y., Majerus, P. W. & Mitchell, C. A. (2003) Proc. Natl. Acad. Sci. USA 100, 8660–8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mochizuki, Y. & Majerus, P. W. (2003) Proc. Natl. Acad. Sci. USA 100, 9768–9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson, F. L. & Dixon, J. E. (2005) J. Biol. Chem. 280, 31699–31707. [DOI] [PubMed] [Google Scholar]
- 21.Begley, M. J., Taylor, G. S., Kim, S.-A., Veine, D. M., Dixon, J. E. & Stuckey, J. A. (2003) Mol. Cell 12, 1391–1402. [DOI] [PubMed] [Google Scholar]
- 22.Berger, P., Schaffitzel, C., Berger, I., Ban, N. & Suter, U. (2003) Proc. Natl. Acad. Sci. USA 100, 12177–12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenzo, O., Urbe, S. & Clague, M. J. (2005) J. Cell Sci. 118, 2005–2012. [DOI] [PubMed] [Google Scholar]
- 24.Woods, V. L. & Hamuro, Y. (2001) J. Cell. Biochem. S37, 89–98. [DOI] [PubMed] [Google Scholar]
- 25.Yuvaniyama, J., Denu, J. M., Dixon, J. E. & Saper, M. A. (1996) Science 272, 1328–1331. [DOI] [PubMed] [Google Scholar]
- 26.Jia, Z., Barford, D., Flint, A. J. & Tonks, N. K. (1995) Science 268, 1754–1758. [DOI] [PubMed] [Google Scholar]
- 27.Laporte, J., Biancalana, V., Tanner, S. M., Kress, W., Schneider, V., Wallgren-Pettersson, C., Herger, F., Buj-Bello, A., Blondeau, F., Liechti-Gallati, S. & Mandel, J.-L. (2000) Hum. Mutat. 15, 393–409. [DOI] [PubMed] [Google Scholar]
- 28.Kutateladze, T. & Overduin, M. (2001) Science 291, 1793–1796. [DOI] [PubMed] [Google Scholar]
- 29.Stahelin, R. V., Long, F., Diraviyam, K., Bruzik, K. S., Murray, D. & Cho, W. (2002) J. Biol. Chem. 277, 26379–26388. [DOI] [PubMed] [Google Scholar]
- 30.Cheever, M. L., Sato, T. K., de Beer, T., Kutateladze, T. G., Emr, S. D. & Overduin, M. (2001) Nat. Cell Biol. 3, 613–618. [DOI] [PubMed] [Google Scholar]
- 31.Essen, L-O., Perisic, O., Cheung, R., Katan, M. & Williams, R. L. (1996) Nature 380, 595–602. [DOI] [PubMed] [Google Scholar]
- 32.Lemmon, M. A. & Ferguson, K. M. (2000) Biochem. J. 350, 1–18. [PMC free article] [PubMed] [Google Scholar]
- 33.Hyvonen, M., Macias, M. J., Nilges, M., Oschkinat, H., Saraste, M. & Wilmanns, M. (1995) EMBO J. 14, 4676–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 461–472. [DOI] [PubMed] [Google Scholar]
- 35.Brunger, A. T. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 36.McRee, D. E. (1999) J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- 37.Hamuro, Y., Anand, G. S., Kim, J. S., Juliano, C., Stranz, D. D., Taylor, S. S. & Woods, V. L. (2004) J. Mol. Biol. 340, 1185–1196. [DOI] [PubMed] [Google Scholar]
- 38.Schwede, T., Kopp, J., Guex, N. & Peitsch, M. C. (2003) Nucleic Acids Res. 31, 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.