Abstract
The nosocomial pathogen Enterococcus faecalis has a unique pheromone-inducible conjugative mating system. Conjugative transfer of the E. faecalis plasmid pCF10 is specifically induced by the cCF10 peptide pheromone (LVTLVFV). Genomic sequence information has recently allowed the identification of putative structural genes coding for the various enterococcal pheromones (D. B. Clewell et al., Mol. Microbiol. 35:246-247, 2000). The cCF10 pheromone sequence LVTLVFV was found within an open reading frame designated ccfA, encoding a putative lipoprotein precursor. Several other pheromone sequences were found in similar locations within other predicted lipoproteins. CcfA shows significant sequence relatedness to the Escherichia coli protein YidC, an inner membrane protein translocase, as well as to a large number of homologs identified in gram-positive and in gram-negative bacteria. Analysis of the deduced CcfA amino acid sequence suggested that mature cCF10 peptide could be formed from the proteolytic degradation of its signal peptide. Expression of the cloned ccfA gene with an inducible expression vector dramatically increased cCF10 production by E. faecalis and also resulted in cCF10 production by Lactococcus lactis, a non-pheromone producer. Site-directed mutagenesis of the ccfA sequence encoding the cCF10 peptide confirmed that ccfA was a functional genetic determinant for cCF10.
The medical importance of the nosocomial pathogen Enterococcus faecalis is in large part due to its remarkable ability to efficiently transfer genetic determinants (31). One extensively studied enterococcal mobile genetic element is the tetracycline resistance plasmid pCF10 (15). Comparative studies of enterococcal clinical isolates collected over 15 years from the same hospital suggest a possible scenario in which endemic pCF10-related plasmids selectively acquired different resistance determinants as they transferred horizontally among the enterococci in the hospital environment (15, 21).
pCF10 is one of numerous conjugative plasmids whose transfer is mediated by peptide pheromones excreted by recipient cells lacking the plasmid. Pheromone-induced conjugation is a highly specific process, dependent not only on the specialized plasmid-encoded transfer machinery, but also upon the distinct peptide sequence of the cognate hepta- or octapeptide pheromone (12, 17). These hydrophobic peptides are excreted into the culture supernatants in small amounts and can display biological activity at concentrations as low as <5 × 10−12 M (29).
One to five molecules of exogenously supplied cCF10 (LVTLVFV) are sufficient to specifically induce the conjugative transfer of pCF10 from a donor cell (29). The genetic determinant of cCF10 is chromosomal, and donor cells containing the pCF10 plasmid possess an intricate regulatory system composed of plasmid-encoded proteins to prevent self-induction of conjugation, since they are still capable of pheromone production (10).
It has been over 15 years since the amino acid sequences of the first enterococcal pheromones were reported (30, 33), yet their biosynthesis has remained a mystery. An obvious possibility is that the mature pheromones are derived from proteolytic processing of larger polypeptides. Support for this notion comes from physiological and genetic analysis of cCF10 production and control of endogenous pheromone by cells carrying pCF10 (10). In addition, An et al. (3) have identified a genetic determinant, eep, encoding a putative membrane peptidase which seems to enhance production of several enterococcal peptide pheromones.
Previous attempts to identify the pheromone genetic determinants have been unsuccessful due to the excessive probe degeneracy resulting from the high sequence homologies among the various peptides. Recently, the E. faecalis genome sequencing projects allowed the identification of putative genetic determinants of the peptide pheromones (13). In this study, we describe experimental evidence indicating that ccfA is the functional genetic determinant of peptide pheromone cCF10.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown in Luria-Bertani (LB) broth (Gibco-BRL). E. faecalis strains were grown in Todd-Hewitt broth (THB; Difco Laboratories, Detroit, Mich.) or in M9-YE medium (18). Lactococcus lactis strains were grown in GM17-glucose medium (34).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Enterococcus faecalis OG1RF | Wild type, Rifr Fusr | 15 |
| Lactococcus lactis LM2301 | Nonproducer of cCF10 | 28 |
| Escherichia coli DH5α | Gibco-BRL | |
| Plasmids | ||
| pGEM-T-Easy | AmprlacZ, cloning vector | Promega |
| PMSP3535 | ErmrnisA nisR nisK | 9 |
| pGEMmha | AmprlacZ, contains 880-bp ccfA fragment produced from PCR in pGEM-T-Easy vector | This study |
| pGEMLAT | AmprlacZ, contains 880-bp ccfA mutated fragment produced by PCR-based site-directed mutagenesis in pGEM-T-Easy vector | This study |
| pMHAnis | Ermr, contains PstI/SalI ccfA fragment in pMSP3535 | This study |
| pLAT | Ermr, contains PstI/SalI ccfA mutated fragment in pMSP3535 | This study |
| pCF10 | Tetr, conjugative plasmid | 15 |
Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; erythromycin, 200 μg/ml for E. coli and 20 μg/ml for E. faecalis and L. lactis; fusidic acid, 25 μg/ml; rifampin, 25 μg/ml; and tetracycline, 10 μg/ml. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Gibco-BRL, Inc., Rockville, Md.) was used at a concentration of 100 μg/ml. Nisin (Sigma Chemical Co., St. Louis, Mo.) was used at a concentration of 25 ng/ml. Restriction enzymes used were purchased from Promega (Madison, Wis.) and Gibco-BRL (Rockville, Md.). Reactions were done as recommended by the manufacturer. Synthetic cCF10 (LVTLVFV) and its peptide analog (LATLVFV) were prepared by the microchemical facility at the University of Minnesota.
Detection of cCF10 activity.
Clumping assays for the detection and determination of ccfA-related pheromone activity, including culture supernatants, were done as described previously (10). cCF10 activity in culture filtrates was confirmed by high-pressure liquid chromatography (HPLC) analysis as described previously (10). The elution time of the active fractions was compared with that of synthetic peptide.
General DNA techniques.
Plasmid DNA from E. coli was prepared with a miniprep kit (Qiagen). Plasmid DNA from E. faecalis and L. lactis was prepared as described earlier (23, 26). Plasmid DNA was analyzed by restriction enzyme digestion on 0.9% Tris-borate-EDTA-agarose gels. Plasmid DNA was introduced into cells by electroporation (16). For Southern hybridization, E. faecalis OG1RF chromosomal DNA was digested with EcoRI, electrophoresed through agarose gels, and transferred to a positively charged nylon membrane (Boehringer, Mannheim, Germany) with a Genie apparatus (Idea Scientific Co., Minneapolis, Minn.). Digoxigenin-labeled probes were prepared and hybridized DNA was detected as specified in the user's guide (Boehringer).
PCR was done with a Perkin-Elmer apparatus under the conditions recommended by the manufacturer. Oligonucleotide primers were synthesized by the microchemical facility at the University of Minnesota.
To amplify the ccfA gene, the primers used were 6221 EcoF (5′-GCTAGAATTCTAGATGTAAGAGAGGGA-3′; incorporated EcoRI site underlined) and 6221 XhoR (5′-GTAGCTCGAGCATAAAATCGGCATCTGA-3′; incorporated XhoI site underlined). E. faecalis OG1RF chromosomal DNA served as the template for PCR amplification of ccfA. The DNA polymerase used in the amplification reaction was Bio-X-Act (Midwest Scientific, St. Louis, Mo.), which allowed the incorporation of A overhangs, facilitating the subsequent cloning of the PCR product in the pGEM-T-Easy cloning vector (Promega, Madison, Wis.); this resulted in pGEMmha.
Site-directed mutagenesis was done using the megaprimer method described by Barik (5), incorporating the following primers: 5′-TTAGGTCGCTAACCCAGCCCA-3′ (LATR), 5′-GTTTTCCCAGTCACGAC-3′ (M4), and 5′-CAGGAAACAGCTATGAC-3′ (RV). Using pGEMmha as the template and PfuTurbo (Stratagene, La Jolla, Calif.) DNA polymerase, the first round of PCR was done using primers M4 and LATR, yielding the LAT megaprimer. This megaprimer was purified according to the methods of Barik (5) and then used in the second round of PCR along with the RV primer to yield the final PCR product containing the desired V14A mutation. Since PfuTurbo DNA polymerase lacks terminal deoxynucleotidyltransferase activity, a blunt-ended PCR product resulted.
In order to produce the A overhangs, the PCR product was subjected to an A-tailing protocol described by the manufacturer (Stratagene, La Jolla, Calif.). The reaction mixture contained purified PCR product, 25 mM MgCl2, 0.2 mM dATP, 10× Taq buffer, and 5 U of Taq. The reaction was done at 70°C for 25 min. Subsequently the product was cloned into the vector pGEM-T-Easy; this resulted in pGEMLAT.
Cloning of ccfA.
A nisin-inducible vector, pMSP3535 (9), was chosen as the expression vector for ccfA. An 880-bp SpeI/XhoI fragment containing ccfA from pGEMmha was ligated into pMSP3535 to yield pMHAnis. Similarly, the 880-bp SpeI/XhoI fragment from pGEMLAT was ligated into pMSP3535 to yield pLAT. Ligated products were transformed into E. coli DH5α competent cells by electroporation and then plated on LB plates supplemented with X-Gal and ampicillin to allow blue-white screening of transformants.
Potential white colonies were picked, and the isolated plasmid DNA was subjected to restriction enzyme digestion analysis to determine whether it contained the proper insert. The confirmed cloned plasmid DNAs were then transformed into E. faecalis OG1RF competent cells, resulting in strains MSPmha001 (with pMHAnis) and MSPmha002 (with pLAT). Resulting plasmid DNA isolated from E. faecalis was used to verify the nucleotide sequence of ccfA. Sequencing was performed by the microchemical facility at the University of Minnesota. pMHAnis and pLAT were introduced into E. faecalis OG1RF(pCF10) by conjugation with MSPmha001 and MSPmha002, respectively, as described previously (36). Plasmid pMHAnis was also electroporated into L. lactis competent cells (27).
Expression of ccfA.
In order to check cCF10 production from either MSPmha001 or MSPmha002, a time course experiment was carried out. Parallel cultures (10 ml of THB, one tube for each time point) were inoculated with the appropriate strain and then induced with 25 ng of nisin per ml. Pheromone activity was determined at each time point as described above. Time course experiments were repeated thrice. L. lactis (pMHAnis) was also induced using 25 ng of nisin per ml .
Culture supernatant was collected by centrifugation at 8,000 rpm (Beckman J2-21 centrifuge, JA20 rotor) and subjected to a clumping assay (10) to detect pheromone activity. E. faecalis OG1RF/pMHAnis and E. faecalis OG1RF/pLAT in 20 ml of THB were induced with nisin at a concentration of 25 ng/ml, incubated at 37°C with shaking for 4 h, and then observed for any visible clumps formed at the bottom of the tubes.
Computer sequence analysis.
Preliminary sequence data were obtained from the Institute for Genomic Research (TIGR) website (http://www.tigr.org). tBLASTn (1) searches were done at the TIGR Comprehensive Microbial Resource website at http://tigrblast.tigr.org/cmr-blast. Most of the sequence analyses were done using the Wisconsin package (version 10.2) of the Genetics Computer Group (Madison, Wis.). Multiple sequence alignment using ClustalW 1.81 (35) was done through the Biology Workbench (version 3.2; San Diego Supercomputer Center, University of California at San Diego) website at http://workbench.sdsc.edu.
RESULTS
Molecular characterization of ccfA gene of E. faecalis OG1RF.
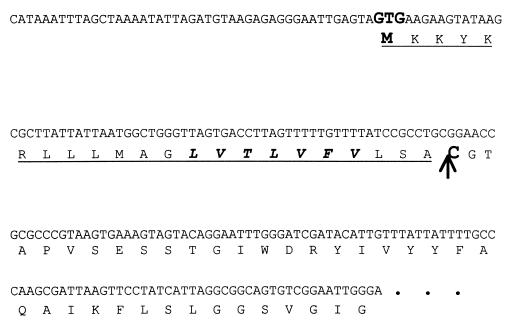
The amino acid sequence of the cCF10 pheromone, LVTLVFV (29), was used to perform a tBLASTn (4) search of the E. faecalis V583 genomic sequence, available from the TIGR database website at http://www.tigr.org. Initial results matched to two noncontiguous TIGR loci, EF2045 and EF3331. The former match showed the LVTLVFV sequence at amino acid positions 169 to 175 of a 348-amino-acid putative type II secretion system protein. In contrast, the EF3331 match mapped the pheromone sequence within an open reading frame (ORF) predicted to encode a prolipoprotein, with the LVTLVFV amino acid sequence encoded within the signal peptide of the lipoprotein precursor (Fig. 1). This gene organization seemed more likely to result in cCF10 expression, since sequence analysis predicts that the cAD1, cPD1, cOB1, and cAM373 peptide pheromones correspond to the carboxy-terminal residues of the signal sequences of putative prolipoproteins (13).
FIG. 1.
Partial nucleotide sequence of ccfA. Underlined is the hypothesized lipoprotein signal sequence. The amino acid sequence of the pheromone cCF10 is italicized. Sequencing of this particular locus in E. faecalis OG1RF verified its identity to that of strain E. faecalis V583 as determined by TIGR (http://www.tigr.org).
Unlike the tBLASTn search results for cCF10, the other pheromones only matched to a single locus in the genome. For lipoprotein synthesis, signal peptidase II cleaves the signal peptide at the conserved cysteine residue within the lipo-box processing site (14). In the case of cCF10, the LVTLVFV amino acid sequence is three residues upstream of this processing site; the other pheromone sequences are found at the C termini of the respective cleaved signal peptides (13).
A Blast (1) search of the microbial genome database revealed that CcfA is most closely related to the Bacillus subtilis gene SpoIIIJ (19). Sequence alignment analysis showed 65% similarity and 39% identity. The cCF10 LVTLVFV amino acid sequence, however, is not present within the spoIIIJ signal peptide. Furthermore, culture filtrates from B. subtilis do not exhibit any cCF10 pheromone activity (results not shown).
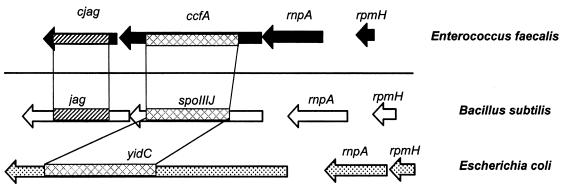
spoIIIJ appears to be in an operon coexpressed with a downstream gene, jag, of unknown function (Fig. 2). Examination of the surrounding nucleotide sequence around ccfA suggests a similar organization, whereby a jag homolog, cjag, is also present downstream. The function of SpoIIIJ is still unknown; however, it appears to be essential for sigma G activity in stage III of sporulation (19). The jag gene appears to be essential for SpoIIIJ activity (19).
FIG. 2.
Genetic map of the ccfA cluster in E. faecalis, in comparison to homologous loci in B. subtilis and E. coli. Regions of significant homology are shaded. ccfA is homologous to spoIIIJ and yidC. SpoIIIJ has an unknown function, whereas YidC is an inner membrane translocase. cjag is a homolog of jag, a spoIIIJ-associated gene of unknown function. RnpA is an RNase P protein involved in tRNA processing. rpmH encodes ribosomal protein component L34. Genomic sequence information and annotation were obtained from the NCBI Entrez Genomes website at http://www.ncbi.nlm.nih.gov.
CcfA also appears to be homologous to a protein of gram-negative bacteria, YidC. Comparative sequence alignment with E. coli YidC analysis shows 37% identity and 55% similarity. YidC is an inner membrane translocase which is essential for normal growth (32). The YidC protein is considerably larger than the homologs from gram-positive organisms, with the former proteins showing strong similarity to the carboxy-terminal segment of YidC (Fig. 2). It is interesting that although YidC does not appear to have a jag homolog, all three genes have a common genetic organization, having rnpA and rpmH genes directly upstream from them (Fig. 2). Although it can be speculated that the jag homologs might have the same function as the amino-terminal half of YidC, we did not find any amino acid sequence conservation that would support this notion.
Cloning and expression of ccfA.
In order to confirm the existence of ccfA in E. faecalis OG1RF, primers 6221 EcoF and 6221 XhoR (Fig. 1) were used to amplify an 880-bp PCR product from genomic DNA. The PCR product was cloned into the nisin-inducible expression vector pMSP3535 (9). The sequence of the cloned PCR product and flanking vector sequences were determined (not shown) and found to be identical to the sequence from strain V543 in the TIGR database, confirming the identity of the two alleles of the gene.
A digoxigenin-labeled version of the same PCR product was used as a probe on Southern blots of EcoRI-digested genomic DNA. Analysis showed hybridization to a 10-kb fragment, confirming the presence of ccfA in E. faecalis OG1RF (results not shown).
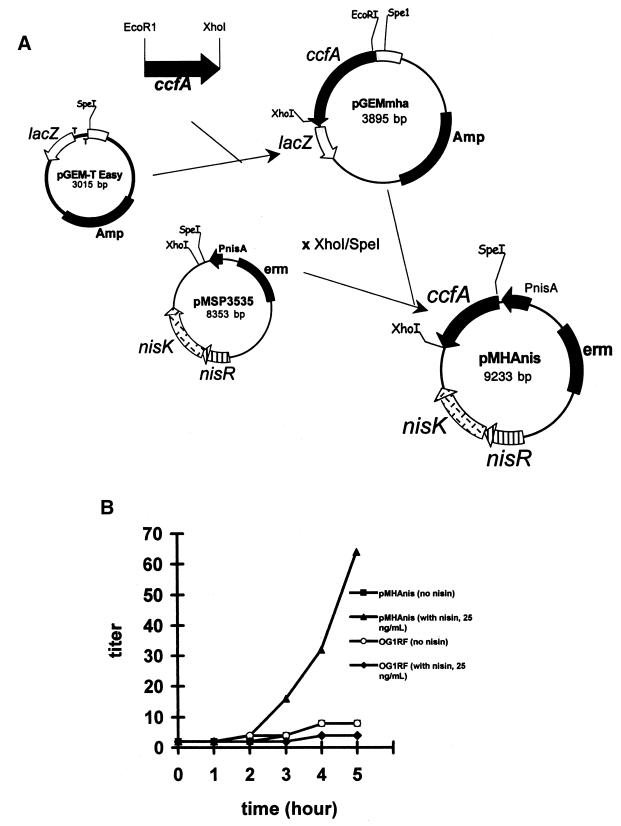
E. faecalis secretes cCF10 into the medium at a low concentration (10−10 to 10−12 M), and pheromone activity can be detected in late logarithmic phase (29). Plasmid pMHAnis contains the ccfA gene cloned from OG1RF under control of a nisin-inducible promoter, PNisA (Fig. 3A). To determine whether ccfA is involved in cCF10 synthesis, the titer of pheromone activity was monitored from an E. faecalis OG1RF(pMHAnis) culture and compared to that from the wild type. The hypothesis tested is that increased expression of ccfA would result in pheromone activity released into the growth medium.
FIG. 3.
(A) Construction of plasmid pMHAnis. (B) Detection of pheromone activity in culture supernatants. Cultures of E. faecalis OG1RF and E. faecalis OG1RF(pMHAnis) were grown overnight in the presence (25 ng/ml) or absence of nisin. Titer of cCF10 in the corresponding culture supernatants was determined.
Culture supernatants were sampled at different time points, precipitated with trichloroacetic acid (TCA), and then subjected to a clumping assay to determine the titer of pheromone activity. In the absence of nisin induction, there was no significant difference in cCF10 titer compared to wild-type levels (Fig. 3B). However, upon nisin induction, pheromone activity was significantly increased, as much as eightfold. In our hands, expression of several other cloned enterococcal proteins under similar conditions resulted in a much higher level of induction (9, 22). It is thus possible that a posttranslational processing step involved in generation of mature cCF10 from the CcfA polypeptide may be rate limiting for cCF10 production.
Physiological effects of ccfA expression.
E. faecalis OG1RF(pCF10) continues to produce peptide pheromone, and the plasmid encodes a complicated control system to prevent self-induction. Control of self-induction in donor cells involves several cell-associated regulators, as well as a precise stoichiometry between excreted inhibitor peptide (iCF10) and excreted cCF10 of endogenous origin (10). In the presence of recipient cells, exogenous cCF10 contributes to upsetting the “balance” in the iCF10-cCF10 ratio in the growth medium, leading to induction.
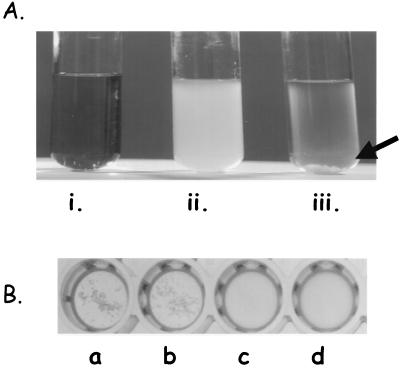
To test whether ccfA expression could result in higher production of endogenous cCF10, plasmid pMHAnis was transformed into E. faecalis OG1RF(pCF10). The resulting strain containing both plasmids was subjected to nisin induction (25 ng/ml). In the presence of nisin, after 2 h of growth with shaking at 37°C, the culture exhibited self-clumping (Fig. 4A). However, without nisin induction, no clumping was observed, and the culture appeared similar to the wild-type uninduced state.
FIG. 4.
(A) Overriding self-clumping control mechanism with nisin induction. (i) Uninoculated medium (THB); (ii) E. faecalis(pCF10/pMHAnis) 4-h culture; and (iii) E. faecalis(pCF10/pMHAnis) 4-h culture induced with nisin (25 ng/ml). The arrow points to the characteristic clumpy phenotype that results upon pheromone induction. (B) Heterologous expression of cCF10. Results of a clumping assay for pheromone activity in culture filtrates from the following cultures: L. lactis(pMHAnis) induced with nisin (25 ng/ml) (b) and L. lactis(pMHAnis) uninduced (c) in comparison with E. faecalis(pCF10/pMHAnis) induced with nisin (25 ng/ml) (a) and E. faecalis(pCF10/pMHAnis) uninduced (d). E. faecalis OG1RF(pCF10) was used as a responder strain for this assay.
In order to prove that CcfA was not simply playing a regulatory role in cCF10 synthesis, plasmid pMHAnis was transformed into a heterologous host. L. lactis, which does not normally produce the peptide pheromone. L. lactis carrying pMHAnis was grown in the presence and absence of nisin. The culture supernatant was collected and subjected to a clumping assay and HPLC analysis. Samples from nisin-induced cultures exhibited pheromone activity, whereas those from uninduced cultures and wild-type L. lactis did not (Fig. 4B). In C18 reverse-phase HPLC analysis of concentrated culture filtrates using the method described in detail by Buttaro et al. (10), the active fraction exhibited a retention time of 27 min, which was identical to that of synthetic cCF10.
Genetic evidence that the LVTLVFV sequence encoded by ccfA is responsible for pheromone activity.
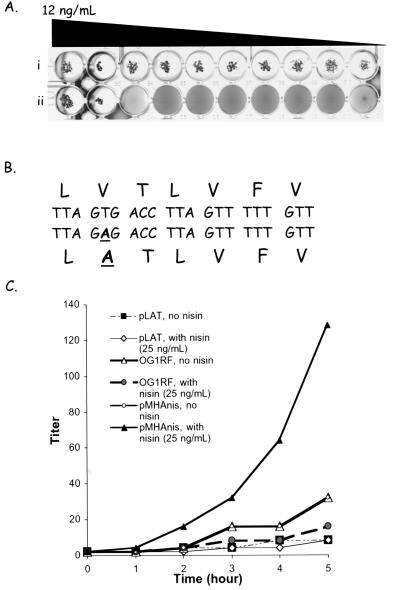
A synthetic peptide analog of cCF10, LATLVFV, was used in a clumping assay to determine whether this peptide could induce clumping in E. faecalis OG1RF(pCF10) cells. At physiological concentrations, this analog was incapable of inducing a pheromone response (Fig. 5A). To prove that mature peptide pheromone is processed proteolytically from the CcfA signal peptide, a single Τ→A mutation was introduced at nucleotide 40 of the ccfA gene, so that LATLVFV was the peptide produced in vivo from processing of the signal peptide (Fig. 5B). Site-directed mutagenesis of pGEMmha was done using the megaprimer method (5). The mutated gene was then cloned into the nisin-inducible expression vector to yield the construct pLAT.
FIG. 5.
(A) Clumping assay using synthetic (i) cCF10 peptide LVTLVFV and (ii) LATLVFV peptide. The starting concentration (12 ng/ml) was serially diluted (twofold) before it was inoculated with the indicator strain E. faecalis OG1RF(pCF10). At this starting concentration, cCF10 had a clumping titer of 210 (i), whereas LATLVFV had a titer of only 22 (ii). (B) Nucleotide modification generated by site-directed mutagenesis to yield the appropriate V14A change with the prolipoprotein signal peptide. (C) Pheromone activity in culture supernatants. Cultures of E. faecalis OG1RF, E. faecalis OG1RF(pMHAnis), and E. faecalis OG1RF(pLAT) were grown for 5 h in the presence (25 ng/ml) or absence of nisin. Titer of cCF10 in the corresponding culture supernatants was determined.
Experiments were performed on batch cultures of E. faecalis OGIRF(pLAT), and the titer of secreted pheromone activity was determined at different time points. Unlike E. faecalis OG1RF(pMHAnis), the pheromone titer was comparable to wild-type levels whether or not E. faecalis OG1RF(pLAT) was induced with nisin. RNA dot blot analysis (not shown) indicated no significant difference in the levels of expression induced by nisin in cells carrying pLAT. Furthermore, pLAT was also transformed into E. faecalis OG1RF(pCF10). In contrast to the results obtained previously with E. faecalis(pCF10/pMHAnis), nisin induction of E. faecalis(pCF10/pLAT) did not result in self-clumping (data not shown).
DISCUSSION
E. faecalis has evolved numerous mechanisms of genetic exchange (11). Pheromone-induced mobilization of conjugative plasmids carrying various genetic determinants such as antibiotic resistance or virulence factors is perhaps one of the most widely studied systems (12, 17). Biochemical studies by Suzuki and coworkers and Clewell et al. led to the isolation and identification of several peptide pheromones which induce conjugative transfer of plasmids with exquisite specificity (reviewed in reference 12).
The E. faecalis genome sequencing projects shed light on the origin of pheromone biosynthesis. As reported by Clewell et al. (13), computer sequence search and analysis identified various potential ORFs encoding prolipoproteins, each containing a specific pheromone amino acid sequence. In each case, the pheromone sequence was contained within the signal sequence of the prolipoprotein. This suggests that the prolipoprotein signal peptide is potentially a pheromone precursor, which could subsequently be proteolytically modified to yield the mature peptide pheromone. This model is substantiated by results from experiments reported by Buttaro et al. (10), which showed that mature cCF10 is predominantly cell wall associated, and pheromone activity generated by incubation of subcellular fractions can be significantly decreased by the addition of protease inhibitors. In addition, An et al. (3) identified Eep, believed to be involved in the processing of pheromone. The Eep protein has structural features characteristic of a membrane-associated zinc metalloprotease. Eep belongs to the family of RIP (regulated intramembrane proteolysis) proteins involved in intramembrane processing of nascent proteins, a mechanism which appears to be conserved from bacteria to humans (8).
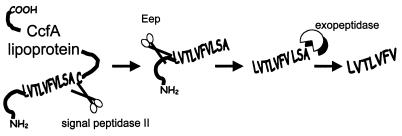
The present study was undertaken to confirm the identity of the structural gene from which cCF10 pheromone is biosynthesized. Sequence analysis showed that the cCF10 LVTLVFV amino acid sequence, like that of other pheromones such as cAD1, cOB1, cPD1, and cAM373, corresponded to the carboxy-terminal end of a prolipoprotein signal peptide (13). Lipoprotein signal peptides are liberated upon signal peptidase II cleavage at the cysteine residue contained within the conserved lipobox processing site (7, 14). The fate of signal peptides released during protein biosynthesis is further degradation by signal peptide peptidases (14). In the case of cCF10, however, the LVTLVFV sequence is three bases upstream of the cysteine cleavage site. Assuming that Eep is involved in an endoproteolytic cleavage of the carboxy-terminal portion of the signal peptide, this suggests that another protease could be required during processing of the CcfA signal peptide to generate mature cCF10. An et al. showed that although the absence of the eep product significantly reduces cCF10 production, elevated expression of eep does not drastically increase cCF10 production, in contrast to the results observed with cAD1 (3). This suggests that exoproteolytic processing of the decapeptide pro-cCF10 molecule produced by Eep could be a rate-limiting step. A model for cCF10 production is shown in Fig. 6.
FIG. 6.
Model for cCF10 biosynthesis. Signal peptidase II cleaves off the signal peptide of nascent prolipoprotein CcfA. The signal peptide is processed further by Eep, which cleaves at the amino-terminal end of the cCF10 peptide sequence. The decapeptide cCF10 precursor is further processed by an exopeptidase, resulting in mature cCF10. The scissors represent an endopeptidase; whereas the pie figure represents an exopeptidase.
Increased expression of CcfA resulted in a significant increase in pheromone production by E. faecalis OG1RF(pMHAnis). Furthermore, when this protein was overexpressed in pCF10-containing cells, the strains were no longer capable of regulating self-induction. This observation concurs with the current model, in which regulation of self-induction by donor cells is based on maintenance of a proper ratio between inhibitor peptide and endogenous pheromone (10). In the presence of excess pheromone provided by neighboring recipient cells, or in this case when CcfA is overexpressed, the balance is tipped and leads to induction. When ccfA was expressed in a heterologous host, L. lactis, it conferred pheromone production in the otherwise cCF10-nonproducing organism.
L. lactis is a fastidious organism with a wide array of proteases needed for proteolytic digestion of milk proteins to obtain amino acids essential for growth (24). Database search of the L. lactis genomic sequence at http://www.tigr.org identified Yvjb, a putative zinc metalloprotease highly homologous to Eep. Therefore, it is not surprising that L. lactis carrying pMHAnis was capable of properly processing the CcfA signal peptide. The introduction of a single base change in the ccfA coding region (pLAT) of the cloned gene, which would result in substitution of an alanine for a valine within the hydrophobic portion of the signal peptide region of the protein, would be unlikely to affect transcription from the nisin promoter or processing and secretion of CcfA. However, this mutation abolished the enhancement of cCF10 production that was observed when the cloned wild-type gene was overexpressed. This result argues strongly that the observed biological effects are due to the change in amino acid sequence of the processed peptide.
Numerous efforts have been made to genetically inactivate the ccfA determinant in strain OG1RF. The strategies employed for these experiments included efforts to make ccfA “knockouts” by integration of replication-deficient plasmids carrying internal fragments of the gene by a single crossover. We also attempted to carry out replacement (via double crossover) of the wild-type gene with the “pLAT” allele (Fig. 5C), encoding the A→V substitution in the second position of the mature pheromone. In spite of the fact that the same methods have been used successfully in our laboratory to generate mutations in other E. faecalis genes, we have been unable to generate a stable clone carrying any type of ccfA mutation.
In one experiment (data not shown), in which allelic replacement was attempted, we obtained several very slow-growing clones which produced no cCF10 and which were confirmed (by pulsed-field genomic DNA analysis; not shown) to be strain OG1RF. Although these isolates did not maintain viability beyond one or two serial transfers, sequence analysis of amplified ccfA region DNA taken from the initial colonies indicated that several unintended point mutations of the DNA upstream of the ccfA coding region had apparently been generated during the recombination process, but the protein coding sequence had not been altered. These mutations may have abolished expression of the gene and generated the pheromone-negative, growth-defective phenotype, although it is also possible that these strains contained additional unlinked point mutations. Therefore, we cannot conclude with certainty that this gene is essential in E. faecalis. However, the available information suggests that either the mature lipoprotein, the processed cCF10 peptide, or both might be required for normal growth of strain OG1RF. To examine this question further, attempts are under way to construct a derivative of OG1RF in which ccfA expression from an ectopic locus can be controlled experimentally to allow inactivation of the wild-type determinant even if the gene is essential.
The function of SpoIIIJ is still unknown, yet much work has been done on the other CcfA homolog, YidC. YidC is a homolog of the eukaryotic Oxa1p protein found in mitochondria and chloroplasts (32). Like Oxa1p, YidC is believed to be a translocase required for the proper insertion of nascent proteins into the inner membrane of bacteria (32). YidC was shown to be required for membrane insertion of Sec-independent proteins and also appears to be involved to a lesser extent in the membrane insertion of some Sec-dependent proteins. More interestingly, Samuelson et al. (32) showed that YidC is essential for cell viability. The growth defect of a YidC deletion mutant was rescued by arabinose-controlled expression of yidC in trans. When arabinose-grown overnight cultures were reinoculated into minimal medium or arabinose-free rich medium, the growth defect was restored.
The work presented here shows that the peptide pheromone cCF10 can be produced from proteolytic processing of the signal sequence of the polypeptide encoded by the ccfA gene. Although the cCF10 sequence was also found within an internal sequence in the C-terminal portion of another polypeptide, our results indicate that the expression of the ccfA gene is responsible for the vast majority, if not all, of the cCF10 produced by E. faecalis OG1RF. Previous analysis of enterococcal genomic sequence data (13), along with the identification of the eep determinant encoding a putative membrane protease activity which enhances production of several pheromones, and recent analysis of production of pheromone cAD1 and the cognate inhibitor iAD1 (F. Y. An and D. B. Clewell, Abstracts, ASM Conference on Cell-Cell Communication in Bacteria, abstr. 66, 2001) suggest a common processing pathway for maturation of enterococccal pheromone and inhibitor peptides.
Since Eep-mediated cleavage of the signal peptides is believed to occur in the membrane (2, 8), and mature cCF10 is actually more hydrophobic than the signal peptide precursor, it seems likely that the cleavage process could be accompanied by a simultaneous active export of the peptide from the membrane to the outside environment. A similar process is believed to occur in the secretion of bacteriocin and bacteriocin-inducing factor peptides produced by numerous gram-positive bacteria (20).
It is interesting that the pheromone plasmids have apparently evolved to allow their host cells to use a by-product of normal metabolism which might be considered cellular waste as an indicator of the proximity of potential recipient cells. There are an extremely large number of additional functional peptides that could be generated in these organisms by similar mechanisms, as well as peptides that could be produced from expression of ORFs in the genome whose lengths are too short to be considered functional genes in standard computerized genomic analyses. Thus, it would not be surprising to find many more peptide-regulated physiological processes than those currently recognized.
The plasmid-determined detection system for the cCF10 molecule needs to be exquisitely sensitive because of the low levels of active signal produced. At the same time, the low levels of cCF10 production could make it easier to control the response to endogenous pheromone in donor cells. Interestingly, the levels of induction of pCF10 transfer genes observed in donor cells at physiologically relevant cCF10 concentrations are relatively modest (5- to 50-fold) and probably transient (6, 25, 29), but these are obviously sufficient to promote frequent transfer of the plasmid under favorable conditions, i.e., availability of recipient cells. When considered as autonomous (“selfish”) DNA elements, the pheromone plasmids seem to have adapted an economical evolutionary strategy to optimize their dissemination while minimizing their metabolic burden on the host cell.
Acknowledgments
We acknowledge Dawn Manias, Helmut Hirt, Ed Bryan, Don Clewell, and Don Morrison for technical assistance and helpful discussion of unpublished results and Tim Leonard for technical assistance on the photographs and figures. We thank Larry McKay for providing us with the L. lactis strains used in this study. We also thank Dave Boxrud for performing pulsed-field gel analysis of the mutant strains. We also acknowledge Linda Banerjee for valuable assistance with the TIGR database.
Preliminary sequence data were obtained from the TIGR website at http://www.tigr.org, where sequencing of E. faecalis V583 was accomplished with support from NIAID. This work was supported by PHS grant GM49530 to G.M.D. from the NIH. M.H.A. was a recipient of the Dennis Watson graduate fellowship from the Dept. of Microbiology, Univ. of Minnesota.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.An, F. Y., and D. B. Clewell. 1994. Characterization of the determinant (traB) encoding sex pheromone shutdown by the hemolysin/bacteriocin plasmid, pAD1 in Enterococcus faecalis. Plasmid 31:215-221. [DOI] [PubMed] [Google Scholar]
- 3.An, F. Y., M. C. Sulavik, and D. B. Clewell. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 181:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 2000. TBLASTN, 2.0MP-WashU ed. Washington University, St. Louis, Mo.
- 5.Barik, S. 1996. Site-directed mutagenesis in vitro by megaprimer PCR. Methods Mol. Biol. 57:203-215. [DOI] [PubMed] [Google Scholar]
- 6.Bensing, B. A., D. A. Manias, and G. M. Dunny. 1997. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act posttranscriptionally to activate expression of downstream conjugation functions. Mol. Microbiol. 24:295-308. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V. A. W., and H. C. Wu. 1994. Lipoproteins: structure, function, biosynthesis, and model for protein export, p. 319-341. In J.-M. Ghuysen and R. Hackenbeck (ed.), Bacterial cell wall, vol. 1. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 8.Brown, M. S., J. Ye, R. Rawson, and J. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 11:391-398. [DOI] [PubMed] [Google Scholar]
- 9.Bryan, E., T. Bae, M. Kleerebezem, and G. Dunny. 2000. Improved vectors for nisin-controlled gene expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 10.Buttaro (Leonard), B. A., M. H. Antiporta, and G. M. Dunny. 2000. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J. Bacteriol. 182:4926-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clewell, D. B. 1990. Movable genetic elements and antibiotic resistance in enterococci. Eur. J. Clin. Microbiol. Infect. Dis. 9:90-102. [DOI] [PubMed] [Google Scholar]
- 12.Clewell, D. B. 1999. Sex pheromone systems in enterococci, p. 10-20. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 13.Clewell, D. B., F. Y. An, S. E. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35:246-248. [DOI] [PubMed] [Google Scholar]
- 14.Dev, I. K., and P. H. Ray. 1990. Signal peptidases and signal peptide hydrolases. J. Bioenerg. Biomembr. 22:271-290. [DOI] [PubMed] [Google Scholar]
- 15.Dunny, G., C. Funk, and J. Adsit. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6:270-278. [DOI] [PubMed] [Google Scholar]
- 16.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunny, G. M., and B. A. B. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 18.Dunny, G. M., D. L. Zimmerman, and M. L. Tortorello. 1985. Induction of surface exclusion (entry exclusion) by Streptococcus faecalis sex pheromones: use of monoclonal antibodies to identify an inducible surface antigen involved in the exclusion process. Proc. Natl. Acad. Sci. 82:8582-8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Errington, J., L. Appleby, R. A. Daniel, H. Goodfellow, S. R. Partridge, and M. D. Yudkin. 1992. Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for sG activity at an intermediate stage of sporulation. J. Gen. Microbiol. 138:2609-2618. [DOI] [PubMed] [Google Scholar]
- 20.Havarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 21.Heaton, M. P., L. F. Discotto, M. J. Pucci, and S. Handwerger. 1996. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-response plasmid. Gene 171:9-17. [DOI] [PubMed] [Google Scholar]
- 22.Hirt, H., S. L. Erlandsen, and G. M. Dunny. 2000. Heterologous inducible expression of Enterococcus faecalis pCF10 aggregation substance Asc10 in Lactococcus lactis and Streptococcus gordonii demonstrates contribution to cell hydrophobicity and adhesion to fibrin. J. Bacteriol. 182:2299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao, S.-M., S. B. Olmsted, A. S. Viksnins, J. C. Gallo, and G. M. Dunny. 1991. Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis. J. Bacteriol. 173:7650-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunji, E. R., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 25.Leonard, B. A. B., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills, D. A., C. K. Choi, G. M. Dunny, and L. L. McKay. 1994. Genetic analysis of regions of the Lactococcus lactis subsp. lactis plasmid pRS01 involved in conjugative transfer. Appl. Environ. Microbiol. 60:4413-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mills, D. A., D. A. Manias, L. L. McKay, and G. M. Dunny. 1997. Homing of a Group II Intron from Lactococcus lactis subsp. lactis ML3. J. Bacteriol. 179:6107-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills, D. A., L. L. McKay, and G. M. Dunny. 1996. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 178:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori, M., Y. Sakagami, Y. Ishii, A. Isogai, C. Kitada, M. Fujino, J. C. Adsit, G. M. Dunny, and A. Suzuki. 1988. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J. Biol. Chem. 263:14574-14578. [PubMed] [Google Scholar]
- 30.Mori, M., Y. Sakagami, M. Narita, A. Isogai, M. Fujino, C. Kitada, R. A. Craig, D. B. Clewell, and A. Suzuki. 1984. Isolation and structure of the bacterial sex pheromone, cAD1, that induces plasmid transfer in Streptococcus faecalis. FEBS Lett. 178:97-100. [DOI] [PubMed] [Google Scholar]
- 31.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, A., M. Mori, Y. Sakagami, A. Isogai, M. Fujino, C. Kitaga, R. A. Craig, and D. B. Clewell. 1984. Isolation and structure of bacterial sex pheromone, cPD1. Science 226:849-850. [DOI] [PubMed] [Google Scholar]
- 34.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trotter, K. M., and G. M. Dunny. 1990. Mutants of Enterococcus faecalis deficient as recipients in mating with donors carrying pheromone-inducible plasmids. Plasmid 24:57-67. [DOI] [PubMed] [Google Scholar]