Abstract
Repair of mature skeletal muscle is mediated by adult muscle progenitors. Satellite cells have long been recognized as playing a major role in muscle repair, whereas side population (SP) cells have more recently been identified as contributing to this process. The developmental source of these two progenitor populations has been considerably debated. We explicitly tested and quantified the contribution of embryonic somitic cells to these progenitor populations. Chick somitic cells were labeled by using replication-defective retroviruses or quail/chick chimeras, and mouse cells were labeled by crossing somite-specific, Pax3-derived Cre driver lines with a Cre-dependent reporter line. We show that the majority of, if not all, limb muscle satellite cells arise from cells expressing Pax3 specifically in the hypaxial somite and their migratory derivatives. We also find that a significant number of, but not all, limb muscle SP cells are derived from the hypaxial somite. Notably, the heterogeneity in the developmental origin of SP cells is reflected in their functional heterogeneity; somitically derived SP cells are intrinsically more myogenic than nonsomitically derived ones. Thus, we show that the somites, which supply embryonic and fetal myoblasts, are also an important source of highly myogenic adult muscle progenitors.
Keywords: regeneration, satellite cells, somite
In the adult vertebrate, muscle is composed primarily of terminally differentiated, multinucleate myofibers. Muscle repair therefore depends on adult mononuclear muscle progenitors (e.g., ref. 1). In recent years, several populations of potential adult muscle progenitors have been identified.
The progenitors that appear to be responsible for most of the repair of adult myofibers are satellite cells residing within muscle (1). Satellite cells are mononuclear cells lying between the plasmalemma and the basal lamina of adult myofibers within all skeletal muscles (2). These cells are normally mitotically quiescent, and in response to stimuli such as injury or exercise, they are activated to reenter the cell cycle whereupon they self-renew, express myogenic markers, and fuse into myotubes (3). In general, satellite cells have been considered to be a committed muscle progenitor population (although see refs. 4 and 5). The transcription factor Pax7 has been identified as a good molecular marker of and functionally essential for satellite cell renewal and maintenance (6, 7).
Recently, multipotential progenitors cells residing outside of or within adult muscle have been identified that can differentiate into myofibers. One of the progenitor populations resident within muscle is the side population (SP) of cells. SP cells were first identified in bone marrow, via staining with the vital dye Hoechst 33342 and FACS analysis, as a population that actively excludes Hoechst and is enriched in hematopoietic stem cells (8). Subsequently, SP cells have been identified in several other tissues, including muscle (9, 10). Muscle SP cells are multipotential progenitors that initially express no muscle markers (9, 10). However, when injected intravenously into lethally irradiated dystrophic mice, the cells can both reconstitute the hematopoietic system and fuse into myofibers (10). Also, when injected into injured muscle, the cells can differentiate into satellite cells and contribute to myofibers (9). In addition, Pax7–muscle SP cells in coculture with primary myoblasts or C2C12 cells differentiate into Pax7+ cells and myotubes (9–11). The presence of muscle SP cells in Pax7–/– mice suggests that SP cells arise independently from Pax7+ satellite cells (7). These experiments suggest that muscle SP cells are endogenous multipotential progenitors able to give rise to satellite cells and myofibers. Furthermore, the ability of muscle SP cells to be delivered systemically through the circulation and to repair dystrophic muscle has generated significant interest in their use for cell-based therapies for muscular dystrophy (11).
The developmental origin of satellite cells and muscle SP cells has been the subject of considerable debate. Several lines of evidence suggest that satellite cells derive developmentally from the somites. Quail/chick chimeras (12, 13) and retroviral labeling experiments in chick (14, 15) have firmly established that somitic cells give rise to embryonic and fetal myoblasts and differentiated myofibers. In particular, the dorsomedial dermomyotome supplies epaxial (back) muscles, and the ventrolateral dermomyotome supplies hypaxial (ventral body wall and limb) muscles. Similarly, quail/chick experiments show that limb and epaxial satellite cells are somitic in origin (16, 17), although it is unknown whether they are derived from the same region of the somite. In mouse, evidence for a somitic origin of adult muscle progenitors derives from analysis of mice expressing various Pax3 alleles. Pax3 is a transcription factor initially expressed in the presomitic mesoderm (psm), newly formed somites, and in older somites restricted to the dermomyotome, then concentrated in the dorsomedial and ventrolateral dermomyotome and the somitic myogenic precursors migrating into the limbs, and finally down-regulated upon myogenic differentiation (18–20). Lineage analysis, through crossing Pax3CreKI mice with Cre-dependent reporter mice, reveals that epaxial and hypaxial skeletal muscles are derived from Pax3+ cells (21). The persistent presence of Pax3/7+ muscle progenitor cells in embryonic, fetal, and adult muscle has suggested that satellite cells are somitic in origin (22). Furthermore, recent analysis of Pax3 GFP knock-in mice indicates that Pax3+ cells give rise to hypaxial trunk satellite cells (23). However, whether Pax3+ cells give rise to limb satellite cells has not been explicitly tested. Overall, these studies strongly suggest that satellite cells are derived from Pax3+ somitic cells, but other sources, such as bone marrow or endothelial cells, have also been proposed (e.g., refs. 24 and 25).
To date, no studies have explicitly tested the developmental origin of muscle SP cells. The efficacy of transplanted bone marrow cells in contributing myofibers to damaged muscle (10, 26, 27) and the capacity of transplanted or cultured muscle SP cells to give rise to hematopoietic as well as myogenic cells (9, 10) have suggested that muscle SP cells are of mixed origin. One possible source is bone marrow, as suggested by the expression of CD45, a pan marker of nucleated hematopoietic stem cells, found by some in muscle SP cells (9, 28). However, the findings of others that muscle SP cells are CD45– (10, 29) and not derived from labeled bone marrow transplants (30) argue against a bone marrow origin of muscle SP cells. Alternatively, it is reasonable to hypothesize, but not yet tested, that muscle SP cells may be derived from somites.
In the present study, we explicitly test the contribution of somitic cells to limb muscle satellite and SP cells. Using three labeling techniques in birds and mice, we quantify that nearly all, if not all, limb satellite cells are derived from Pax3-expressing cells in the hypaxial dermomyotome and their migratory derivatives. We also demonstrate that somitic cells give rise to a substantial portion, but not all, limb muscle SP cells, and these somitically derived SP cells are highly myogenic.
Results
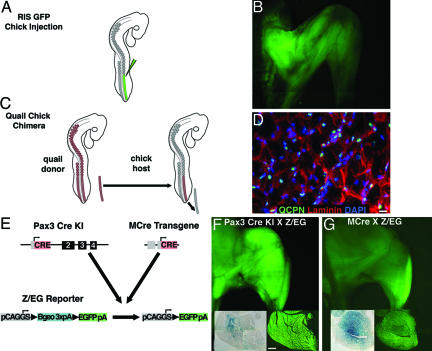
Labeling of Somitic Cells in Chick and Mouse. To determine whether satellite and SP cells are derived from somites, we labeled somitic cells and their progeny using three different methods. In chick, somitic cells were labeled via injection of a replication-defective GFP retrovirus, RIS-GFP (replication-incompetent splice acceptor GFP). Right hindlimb-level psm was infected at stage 15 before the migration of myogenic precursors into the limb (Fig. 1A). Because the virus cannot spread, GFP expression is limited to the somitic cells and their progeny. In infected chicks, right hindlimb muscles expressed GFP (Fig. 1B). The efficiency of labeling ranged from 34% to 47% of myonuclei (DAPI+, Pax7–nuclei under the laminin+ basal lamina of myofibers) in tissue sections (Table 1, which is published as supporting information on the PNAS web site), with no cells labeled in control, contralateral limbs (data not shown). Because retroviruses can only infect mitotic cells (31), the overall rate of infection is limited.
Fig. 1.
Methods for labeling somitic cells in chick and mouse. (A) Labeling of chick psm through injection of RIS-GFP. (B) GFP labeling of chick hindlimb muscle. (C) Somitic labeling through replacement of chick psm with quail psm. (D) QCPN+ myonuclei in chimeric hindlimb muscle. (Scale bar: 5 μm.) (E) Labeling of mouse Pax3+ cells and their progeny through Pax3CreKI or MCre;Z/EG mice. (F and G) GFP labeling of mouse hindlimb muscle in both crosses. Sections through quadriceps muscles were simultaneously stained for LacZ and visualized for GFP. (Scale bar: 0.5 mm for all sections.)
To more efficiently label chick somitic cells, we produced quail/chick chimeras in which right hindlimb-level psm of the chick was replaced with isochronic quail psm (Fig. 1C). Quail cells were identified with a perinuclear antibody, QCPN, that labels the majority of most quail cell types (94% of quail myonuclei, satellite, and muscle mononuclear cells and 82% of bone marrow cells) but does not label any chick cells. Using QCPN on chimeras, we found that 41–71% of myonuclei were labeled in right-limb sections (Fig. 1D and Table 1), with no labeling of cells in control, contralateral limbs (data not shown). Unlabeled cells on the experimental side derive from host chick somites as well as unlabeled quail cells.
Somitic cells and their progeny were also labeled in mouse through two Pax3Cre driver lines. We used a Pax3CreKI mouse in which Cre is faithfully expressed in all regions where Pax3 is normally expressed (Fig. 1E; ref. 21), and when crossed with Cre-dependent Z/EG reporter mice (in which GFP is activated in response to Cre recombinase), Pax3+ cells and their progeny express GFP (Fig. 1E). Muscle development is normal in these Pax3 heterozygous mice, and all back, body wall, and limb muscles are GFP+ (Fig. 1F; ref. 21). Lineage and mutational studies (18–21) suggest that all limb myofibers should initially be derived from Pax3+ somitic cells. However, in our experiments, labeling of limb muscles was not 100%. In sections simultaneously stained for LacZ activity (indicating cells in which GFP has not been Cre activated) and visualized for GFP (Fig. 1F), 95% of multinucleate myotubes were GFP+ and 11% were LacZ+ (8% were GFP+/LacZ+ and 1% were GFP–/LacZ–). Because the GFP is cytoplasmic and myotubes are a multinucleate syncytium, it is likely that <95% of the myonuclei are GFP+. Furthermore, the presence of LacZ+ myotubes is likely due to the inefficiency of the Pax3CreKI, but it also may indicate that not all myotubes are derived from Pax3+ cells. In tissue sections prepared for antigen retrieval for simultaneous Pax7 labeling, only 58–67% of limb myonuclei were GFP-labeled (Table 1), and this decreased labeling is likely due to poor preservation of GFP under these conditions. Because Pax3 is expressed in lineages other than muscle (notably neural crest cells) (32), in these mice labeled satellite and SP cells may potentially be derived from nonsomitic (although not bone marrow) sources.
To specifically label Pax3+ cells only in the hypaxial somitic lineage, we used a transgenic line, MCre, in which a transgene containing a Pax3 enhancer element and the proximal promoter drives Cre expression only in the hypaxial dermomyotome and the body wall and migrating limb myogenic cells between embryonic day 9 (E9) and E14.5 (ref. 33; data not shown). As expected, when MCre mice were crossed with Z/EG reporter mice, limb and body wall muscle (but not back muscle) expressed GFP (Fig. 1G). The efficiency of labeling limb muscle cells was considerably lower (37–57% of myonuclei) and more variable in these mice (Table 1 and Fig. 1G) compared with the Pax3CreKI;Z/EG mice perhaps because not all of the elements necessary for Pax3 expression in hypaxial somitic cells are present.
Because Pax3 has been reported to be expressed in adult muscle (6, 34), we also specifically tested whether MCre was active in adult muscle. MlacZ mice (transgenic consisting of Pax3 enhancer element, proximal promoter, and lacZ) did not express lacZ in adult muscle, and in MCre mice, Cre was not detected by RT-PCR in adult whole limb muscle or muscle mononuclear preparations (Fig. 4, which is published as supporting information on the PNAS web site).
Somitic Cells Give Rise to Satellite Cells in both Chick and Mouse Limb Muscle. To assess the contribution of somitic cells to satellite cells in chick and mouse limb muscles, we labeled somitic cells by the methods described above and harvested and sectioned mature hindlimb muscles. Satellite cells begin to appear in limb muscle by E16 (16, 35–37) and E18 (38) in the chicks and mice, respectively. In our study, we identified satellite cells in E16 to postnatal day 4 (P4) chicks and 4- to 7-week-old mice on the basis of their expression of Pax7 (7, 39) and their location under the laminin+ basal lamina of myofibers (2).
To determine the percentage of satellite cells derived from the somites in the avian experiments, we first assessed the overall efficiency of labeling somitic cells in each experiment. Because all myonuclei (DAPI+, Pax7–nuclei under the myofiber basal lamina) are initially derived from the somites (12, 13), all myonuclei should be labeled if the methods of labeling somitic cells were 100% efficient. For each set of muscles, we calculated the percentage of myonuclei that were labeled (GFP+ or QCPN+). In regions with efficient myonuclei labeling, we calculated (one to three sections of two different muscles) the percentage of satellite cells labeled. To calculate the percentage of satellite cells derived from the somite, we normalized the percentage of satellite cells labeled by the percentage of myonuclei labeled.
A similar calculation was done to determine the percentage of satellite cells derived from Pax3+ cells in the mouse experiments. As described above, not all myonuclei were labeled in either Pax3CreKI or MCre;Z/EG experiments. Therefore, because labeling was not 100% efficient, we determined the percentage of satellite cells derived from Pax3+ cells as the percentage of satellite cells labeled normalized by the percentage of myonuclei labeled.
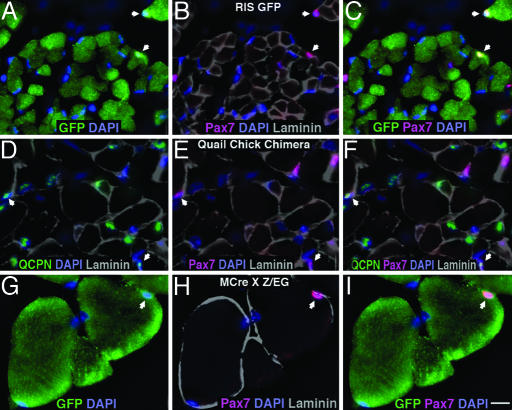
In all experiments, a large number of myonuclei and satellite cells in chick and mouse limbs were labeled (Fig. 2 and Table 1). In three of four chimera experiments and in one of four RIS-GFP chick experiments, the percentage of satellite cells labeled exceeded the percentage of myonuclei labeled (103–146%). In the remaining experiments, the percentage of labeled satellite cells normalized by labeled myonuclei ranged from 79% to 90% (with one RIS-GFP experiment at 39%). These experiments show that avian limb satellite cells are largely derived from the somites. In addition, in the Pax3CreKI;Z/EG mice, 57–62% of the limb satellite cells when normalized were labeled and therefore derived from Pax3+ cells. Furthermore, in the MCre;Z/EG mice, at least 79% of satellite cells after normalization were labeled, demonstrating that a large portion of limb satellite cells were derived from Pax3+ cells, specifically in the hypaxial dermomyotome and its derivatives. After pooling all 12 experiments and using a Sign test to compare satellite cells and myonuclei, we found no significant difference in the labeling of these two populations. Given that all myonuclei are initially derived from the somites, these findings suggest that all satellite cells are derived from the somites, specifically from Pax3+ cells in the hypaxial dermomyotome.
Fig. 2.
Somitic cells give rise to limb satellite cells. (A–C) Section through P2–P3, RIS-GFP-labeled chick iliofibularis muscle with two GFP+, somitically derived Pax7+ satellite cells (arrows). (D–F) Section through E18 quail/chick chimeric iliofibularis muscle with two QCPN+, somitically derived Pax7+ satellite cells (arrows). (G–I) Section through 6-week-old mouse tibialis anterior muscle with one GFP+ Pax7+ satellite cell (arrow) derived from a hypaxial Pax3+ cell. DAPI+ nuclei are shown in blue (A–I); Pax7+ satellite cells are shown in red (B, C, E, F, H, and I); basal lamina is highlighted with laminin shown in gray (B, D–F, and H); GFP+ cells are shown in green (A, C, G, and I); and QCPN+ nuclei are shown in green (D and F). (Scale bar: 5 μm for all sections.)
Somitic Cells Give Rise to SP Cells in Chick and Mouse Limb Muscle. We hypothesized that limb muscle SP cells may also be derived from somites. We labeled somitic cells using the methods described above, harvested and dissociated mature limb muscles, and determined the appropriate concentrations of Hoechst 33342 to identify SP cells (see Methods and ref. 29). Consistent with previous studies (29), we found that SP cells constitute 0.05–2.07% of the total mononuclear cells derived from avian and mouse limb muscle and were enriched in Sca1 (Tables 2 and 3, which are published as supporting information on the PNAS web site). To assess the contribution of somitic cells to limb muscle SP cells, we had to establish a method for determining the efficiency of somitic cell labeling. The main population (MP) cells are the cells centrally located in a FACS Hoechst/propidium iodide profile (Fig. 5, which is published as supporting information on the PNAS web site) and enriched in satellite cells and myoblasts (30). Because satellite cells and myoblasts are derived from Pax3+ somitic cells, we wanted to use the percentage of MP-cell labeling as our measure of the efficiency of somitic labeling. However, MP cells are a mixed population containing nonsomitically derived cell types, such as fibroblasts and blood-derived cells. We estimated that 70% of the MP cells are somitic in origin (because in the Pax3CreKI;Z/EG mice with consistent high levels of labeling, 67.6–70.6% of MP cells are labeled) and used this to calculate the percentage of labeling of somitically derived MP cells. Therefore, for each experiment we determined the contribution of somitic Pax3+ cells to muscle SP cells as the percentage of labeled SP cells normalized by the percentage of labeled somitically derived MP cells.
In all avian and mouse experiments, we found labeled muscle SP cells (Fig. 5 and Table 2). In RIS-GFP chicks and chimeras, the percentage of SPs labeled normalized by the percentage of MPs was variable, ranging from 8% to 52%. The relatively low percentage of labeled SP cells (8–11%) in the E16 RIS-GFP chicks may reflect the relative quiescence of the E16 SP progenitors, and consequent lower infectivity, at the time of retroviral injection. In the Pax3CreKI and MCre;Z/EG mice, we also consistently found labeled SPs in all experiments. However, in the MCre crosses, the percentage of labeled SP cells was highly variable (from 39% to >100%), reflecting the variability in the MCre transgene expression. In the Pax3CreKI mice, we found consistent high labeling of MP cells and 41% labeling of SP cells after normalization. Overall, these experiments indicate that Pax3+ cells in the hypaxial somites give rise to a substantial portion of limb muscle SP cells. Nevertheless, after pooling all 19 experiments and using a Sign test to pairwise compare SP and MP cells, we found a significant difference (P < 0.001) in the labeling of these two populations. These findings indicate that although somitic cells do substantially contribute to the limb muscle SP cells, other nonsomitic cell types also are likely to give rise to some SP cells.
Marker Characterization of Somitically Derived Muscle SP Cells. To examine the relationship between somitically derived muscle SP cells and bone marrow-derived hematopoietic cells, we examined the expression of the protein tyrosine phosphatase CD45. CD45 is a pan marker of nucleated hematopoietic cells (40) and is expressed at high levels in bone marrow-derived SPs and MPs. In our samples, 49–64% of chick and 73–89% mouse bone marrow mononuclear cells are CD45+. CD45 is also expressed in other tissues and interpreted as an indicator of hematopoietic origin. We found only a low percentage of chick (0–1.1%) and mouse (0–5.6%, with one small sample at 11%) muscle SP cells expressing CD45 (Table 3). Moreover, in GFP+ SPs and MPs, we found background levels of CD45 expression. The low percentage of CD45 in SPs suggests that most SPs are unrelated to hematopoietic cells. The lack of CD45 expression in somitically derived GFP+ SPs also suggests that these cells did not go through a bone marrow intermediate. Further supporting this inference, we never found QCPN+ or GFP+ cells in bone marrow cells in any of our experiments (data not shown).
We also examined the relationship between muscle SP cells and Pax7+ cells. In muscle mononuclear cell preparations, Pax7 labels satellite cells and also, in less mature muscle, satellite cell precursors (20, 23). As expected, some MP cells expressed Pax7: 6–28% in avian muscle and 3–9% in mouse muscle. In both P5–P6 chick limb muscle and 6-week-old mouse limb muscle, we found no SP cells that expressed Pax7. In E16–E17 chick muscle, we did find that 1–13% of SP cells expressed Pax7, but in any particular sample, SP levels of Pax7 were considerably lower than MP levels of Pax7.
Somitically Derived Muscle SP Cells Are Highly Myogenic in Culture. To test the myogenic capacity of somitically derived SP cells, we cultured SP cells in vitro. Similar to others (9), we found that mouse muscle SP cells isolated from uninjured muscle are nonmyogenic when cultured alone but myogenic when cocultured with myogenic cells. Therefore, we cocultured RIS-GFP SPs with quail primary myoblasts, chimeric SPs with chick primary myoblasts, and mouse MCre;Z/EG SPs with C2C12 cells. Because of the relatively small number of labeled SPs, we cultured labeled and unlabeled SPs in these experiments. Cells were initially cultured in growth medium, upon confluency, switched to differentiation medium, and then assayed for the presence of mononuclear Pax7+ cells and α-actinin+ or dystrophin+ multinucleate myofibers.
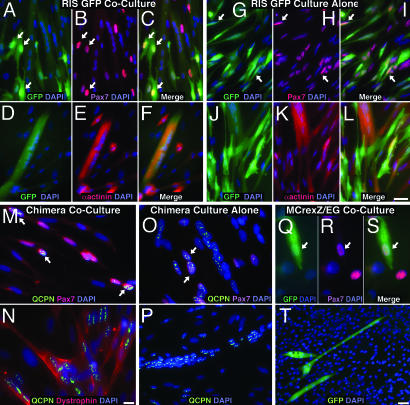
In all experiments, labeled, somitically derived SP cells differentiated into myogenic cells when cocultured (Fig. 3 and Table 4, which is published as supporting information on the PNAS web site). In avian coculture experiments, 81–97% of the GFP+ SPs from RIS-GFP muscle and 94–99% of QCPN+ SPs from chimeric muscle differentiated into myogenic cells (Table 4), either mononuclear Pax7+ cells (Fig. 3 A–C and M) or α-actinin+ or dystrophin+ myofibers (Fig. 3 D–F and N). In cocultures of SPs derived from MCre;Z/EG mouse muscle, we isolated small initial numbers of GFP+ SPs, and so at the end of the coculture experiments, we found few adherent GFP+ cells. Of the five coculture experiments in which we found GFP+ cells, all contained cells that were myogenic, either as Pax7+ mononuclear cells (Fig. 3 Q–S) or as fused myofibers (Fig. 3T). The presence in chick and mouse cultures of QCPN+ or GFP+ Pax7+ mononuclear cells indicates that somitically derived muscle SP cells did not become myogenic simply by fusion with cocultured primary myoblasts.
Fig. 3.
Somitically derived limb muscle SP cells are myogenic in culture. (A–F) GFP+ SP cells from hindlimb muscle of RIS-GFP chick in coculture differentiate into Pax7+ mononuclear cells (A–C, arrows) or multinucleate, α-actinin+ myofibers (D–F). (G–L) GFP+ SP cells from hindlimb muscle of RIS-GFP chick cultured alone differentiate into Pax7+ mononuclear cells (G–I, arrows) or multinucleate, α-actinin+ myofibers (J–L). (M and N) QCPN+ SP cells from quail/chick chimeric hindlimb muscle in coculture differentiate into Pax7+ mononuclear cells (M, arrows) or multinucleate, dystrophin+ myofibers (N). (O and P) QCPN+ SP cells from quail/chick chimeric hindlimb muscle cultured alone differentiate into Pax7+ mononuclear cells (O, arrows) or multinucleate myofibers (P). (Q–T) GFP+ SP cells derived from MCre;Z/EG mice in coculture differentiate into Pax7+ mononuclear cells (Q–S, arrows) or multinucleate myofibers (T). DAPI+ nuclei are shown in blue; Pax7+ cells are shown in red (B, H, M, O, R, and S); α-actinin+ myofibers are shown in red (E and K); dystrophin+ myofiber is shown in red (N); GFP+ cells are shown in green (A, C, D, F, G, I, J, L, Q, S, and T); and QCPN+ cells are shown in green (M–P). (Scale bars: A–L, 0.025 μm; M–O and Q–S, 0.0125 μm; P and T, 0.05 μm.)
In the course of determining appropriate avian culture conditions, we discovered that unlike mouse, avian muscle SP cells are able to differentiate into myogenic cells when cultured alone in growth and then differentiation medium. The greater myogenic capacity of the avian SPs may be due to their younger age or to intrinsic differences between mouse and avian SP cells. In these culture-alone experiments (which include labeled and unlabeled SP cells), 73–99% of the GFP+ SPs from RIS-GFP muscle and 79–87% of QCPN+ SPs from chimeric muscle differentiated into myogenic cells (Table 4) either as Pax7+ mononuclear cells (Fig. 3 G–I and O) or multinucleate α-actinin+ myofibers (Fig. 3 J–L and P). Because we found a small percentage of E16–E17 avian Pax7+ muscle SPs, we tested whether myogenic cells in the culture-alone experiments may simply result from an expansion of small numbers of initially Pax7+ SPs. During a 2-week time period of culturing RIS-GFP SP cells, we found that myogenic cells were not derived from clonal expansion of initially Pax7+ SPs. Daily monitoring of the cultures revealed that Pax7+ cells only appeared after 2 days in growth medium (data not shown). Thus, these experiments demonstrate that even when cultured alone, somitically derived avian SP cells are highly myogenic.
Our FACS analysis of limb muscle SP cells indicates that although a substantial portion of SP cells are derived from the somites, not all are. Potentially, differences in the developmental origin of muscle SPs could affect their subsequent myogenic capacity. Therefore, in RIS-GFP and chimera culture-alone experiments, we explicitly compared the myogenic capacity of labeled, somitically derived versus unlabeled muscle SP cells. It should be noted that in these experiments, not all somitically derived SPs are either GFP+ or QCPN+ because of the labeling inefficiencies of retroviral labeling and quail/chick psm transplantations. Therefore, unlabeled SP cells also include some somite-derived, yet unlabeled cells and could potentially mask differences in the myogenic capacity of somitically derived versus nonsomitically derived SPs. Nevertheless, pairwise comparisons of the myogenic capacity of GFP+ versus GFP–SPs in four culture-alone experiments and QCPN+ versus QCPN–SPs in one culture-alone experiment revealed significant differences in myogenic capacity of labeled versus unlabeled SPs (Table 4). The myogenic capacity of GFP+ SPs ranged from 73% to 99% whereas that of GFP–SPs ranged from 60% to 86%, and the myogenic capacity of QCPN+ SPs was 87% whereas that of QCPN–SPs was 33%. In all five pairwise comparisons, a χ2 test of homogeneity confirmed that labeled SPs were significantly more myogenic than unlabeled SPs (P < 0.001 in all cases). These findings demonstrate that the developmental origin of SP cells has a significant impact on their subsequent myogenicity.
Discussion
Understanding the developmental origin of adult muscle progenitors is critical for establishing when, where, and how adult progenitors are determined and maintained for the repair of adult muscle and may provide important insights into their potential therapeutic value for cell-based therapy of muscle diseases. We have conducted a lineage analysis, using three different types of labeling techniques in chick and mouse, to determine whether somitic cells give rise to two classes of adult limb muscle progenitors, satellite and muscle SP cells.
In avian somite labeling experiments, we demonstrate that nearly all limb satellite cells are derived from the somites. Our data confirm and extend the classic quail/chick studies of Armand et al. (16), which first identified somites as a source of satellite cells. Using specific antibodies, we have improved the efficiency of identifying both quail and satellite cells and find that nearly all limb satellite cells are of somitic origin. Gros et al. (17) have also found, through quail/chick grafts, that nearly all epaxial satellite cells are derived from the somite. Thus, somites are the major source of satellite cells in all body muscles.
We further characterized the genetic origin of limb satellite cells with two Pax3Cre mouse lines. Pax3 is expressed in the somitic hypaxial dermomyotome and somitic cells migrating into the limb and is required for formation of limb muscles (18–20). Recent analysis (23) of a Pax3GFP knock-in mouse in which GFP perdures after endogenous Pax3 expression has revealed that Pax3+ cells give rise to cells lying in the satellite position within hypaxial trunk myofibers. Now, using Pax3CreKI and MCre mice, we explicitly demonstrate and quantify that all, or almost all, limb satellite cells arise from Pax3+ cells in the hypaxial, ventrolateral dermomyotome and their migratory derivatives in the limb, unlike epaxial satellite cells, which arise from the central region of the dermomyotome (17, 23).
In our lineage experiments, we also found that substantial numbers of limb muscle SP cells were labeled, explicitly demonstrating that a significant portion of muscle SP cells are derived from Pax3+ hypaxial somitic cells. Consistent with their non-bone marrow origin, we found that these labeled SP cells were CD45–. The absence of CD45+ SP cells agrees with the findings of others (10, 29, 30) who similarly identified SPs using a relatively high Hoechst concentration and a narrowly defined SP gate. Nevertheless, in contrast to our finding that nearly all satellite cells are derived from the somites, clearly not all muscle SP cells are somitically derived, and therefore they are heterogeneous with respect to their developmental origin. Potentially, some endogenous muscle SPs are derived in development from other sources, such as bone marrow, hematopoietic stem cells, or endothelial cells. The overall heterogeneity of marker expression within muscle SP cells (9, 10) may be indicative of their heterogeneous developmental origin.
Our experiments culturing somitically derived chick and mouse muscle SP cells in vitro confirm that these cells are myogenic. We found that SPs differentiated into mononuclear Pax7+ cells and fused into multinucleate dystrophin+ or α-actinin+ myofibers. The presence of Pax7+ mononuclear cells indicates that SPs do not require fusion with primary myoblasts to become myogenic. The up-regulation of Pax7 by SP cells (also seen in refs. 9 and 11) suggests that these cells go through a Pax7+, satellite cell intermediate on their way to forming myofibers. However, it is unclear whether expression of Pax7 is required for muscle SP cell differentiation because muscle SPs in the absence of Pax7 can undergo myogenic differentiation (7, 9).
Most importantly, experiments culturing chick or quail muscle SP cells alone revealed that they are heterogeneous with respect to their myogenicity: labeled, somitically derived muscle SPs are significantly more myogenic than unlabeled SPs. These findings demonstrate that developmental origin is important for determining the intrinsic myogenic capacity of these adult muscle progenitors. Recently, Sherwood et al. (41) have identified that marker expression, cellular history, and muscle condition (whether the muscle is injured or not) all affect the myogenicity of adult muscle progenitors. In addition, the systemic environment (42) and physiological differences between different anatomical muscles (43) also significantly impact the myogenic capacity of adult muscle progenitors. Our findings implicate another important influence on myogenic capacity: developmental origin.
Methods
Chick Surgeries and Retroviral Injections. Chicken and Japanese quail eggs were incubated until the embryos had a total of 25–26 somites. To retrovirally label chick psm, right-hindlimb-level psm was injected with a replication-defective retrovirus containing GFP, RIS-GFP (gift of E. Laufer, Columbia University, New York). Retrovirally infected chicks were harvested at E16–P6. RIS-GFP was created by cloning EGFP into RIS (44), in which the pol gene had been frameshifted. Virus was produced by cotransfecting RIS-GFP with a vesicular stomatitis virus glycoprotein-encoding plasmid into chicken fibroblast cell line DF1 cells as described in ref. 45. To produce quail/chick chimeras, quail donor hindlimb-level psm was isolated via dispase digestion. Chick psm was removed from the right side with tungsten needles and replaced with quail donor tissue. Left-side psm remained as chick and served as a negative control. Chimeras were harvested at E15–E18.
Mouse Cre and Reporter Lines. To identify derivatives of Pax3+ cells, two mouse Cre driver lines were used. The targeted Pax3CreKI line drives Cre expression in both somites and dorsal neural tube where Pax3 is expressed (21). Hypaxial muscle derivatives of the somitic Pax3+ cells were specifically identified through a transgenic line, MCre (33). To identify derivatives of Pax3CreKI- or MCre-expressing cells, these mice were crossed with Z/EG reporter mice (46). Z/EG mice constitutively express lacZ throughout embryonic development and as adults, but in tissues expressing Cre recombinase, lacZ is removed and cytoplasmic EGFP expression is activated. Pax3CreKI or MCre;Z/EG mice were harvested at 4–7 weeks.
Isolation of SP Cells. Muscles were isolated from RIS-GFP and quail/chick chimera hindlimbs and from Pax3CreKI and MCre;Z/EG mouse fore- and hindlimbs. For RIS-GFP chicks and MCre;Z/EG mice, only the most GFP+ muscles were selected. Muscle mononuclear cells were isolated from muscle and bone marrow mononuclear cells extracted from femurs and tibias as described in ref. 29.
Hoechst titration curves and antibody stains were performed as described in ref. 29. Hoeschst 33342 at concentrations of 1.5 μg/ml, 4.0 μg/ml, and 7.5 μg/ml were used to sort E15–E18 chick and quail muscle cells, post-hatch chick muscle cells, and mouse muscle cells, respectively. Propidium iodide (2 μg/ml) was added to all samples to exclude dead cells. For each experiment, 106 avian cells per ml were incubated with Hoechst and 5 μM reserpine, and 106 mouse cells per ml were incubated with Hoechst and 100 μM verapamil to determine the SP gate. Chick cells were also analyzed with a phycoerythrin-conjugated mouse anti-chick CD45 (LT40) antibody (0.2 μg per 106 cells; Southern Biotechnology Associates), and mouse cells were analyzed with phycoerythrin-conjugated rat anti-mouse CD45 (30-F11). Cells were analyzed and sorted on a flow cytometer (Becton Dickinson).
Culture of Muscle SP Cells. Chick and quail SP cells were cultured alone or cocultured with primary quail or chick myoblasts, respectively. Avian primary myoblasts were obtained through four rounds of preplating muscle mononuclear cells on plastic dishes and culturing floating cells on collagen-coated dishes. Whether cultured alone or with primary myoblasts, SP cells were cultured on poly(l-lysine) HBr and collagen-coated Permanox slides initially in growth media (F10 medium containing 20% FBS, 1% Pen/Strep, 1% chick serum, and 2.0 ng/ml rhFGF, Basic) and upon confluency, switched to differentiation media (DMEM medium containing 2% horse serum, 1% Pen/Strep, and 1% l-glutamine).
All mouse SP cells were cocultured with C2C12 cells on poly(l-lysine) HBr- and collagen-coated Permanox slides in growth media (DMEM medium containing 20% FBS, 1% Pen/Strep, and 1% l-glutamine) and upon confluency were switched to differentiation media (same as above).
Immunohistochemistry. For identification of satellite cells, muscles were isolated and fixed in 4% paraformaldehyde (PFA), embedded in OCT compound, cryo-sectioned, and antibody labeled. Quail/chick chimera tissue was sequentially labeled with QCPN and laminin, Cy3 goat anti-mouse and Cy5 goat anti-rabbit, Alexa 488 directly conjugated Pax7 (Zenon labeling, Molecular Probes), PFA postfix, and DAPI (counterstain for nuclei). RIS-GFP chick tissue was sequentially labeled with Pax7 and GFP, Cy3 goat anti-mouse and Cy2 goat anti-rabbit, laminin, and Cy5 goat anti-rabbit. MCre or Pax3CreKI;Z/EG tissue was labeled sequentially with GFP, Cy2 goat anti-rabbit, PFA postfix, sodium citrate antigen retrieval, Pax7 and laminin, Cy3 goat anti-mouse, and Cy5 goat anti-rabbit. Monoclonal anti-Pax7 and QCPN were used at 2 μg/ml (Developmental Studies Hybridoma Bank), rabbit anti-laminin was used at 2 μg/ml (L9393, Sigma), rabbit anti-GFP was used at 10 μg/ml (A21311, Molecular Probes), and secondary antibodies were used at 3.5 μg/ml (Jackson ImmunoResearch).
To label sorted SP and MP cells, cells were fixed in 4% PFA, spotted on poly(l-lysine)-coated slides, and air dried. To label cultured cells, cells were fixed either in cold 100% methanol or 4% PFA. All cells were labeled with Pax7, QCPN, 1:1,000 dystrophin (CAP6–10; ref. 47), or 1:400 α-actinin (EA-53, Sigma) and detected with secondary antibodies.
Supplementary Material
Acknowledgments
We thank Alan Flint for assistance with the cell sorter and Emanuela Gussoni for technical assistance and review of the manuscript. L.M.K. is an Investigator of the Howard Hughes Medical Institute, J.A.E. and K.A.E. are supported by National Institutes of Health Grant R01 HL61475, and C.J.T. and G.K. are supported by National Institutes of Health grants.
Conflict of interest statement: No conflicts declared.
Abbreviations: SP, side population; psm, presomitic mesoderm; MP, main population; En, embryonic day n; Pn, postnatal day n; PFA, paraformaldehyde; RIS-GFP, replication-incompetent splice acceptor GFP.
References
- 1.Bischoff, R. & Franzini-Armstrong, C. (2004) in Myology, eds. Engel, A. G. & Franzinin-Armstrong, C. (McGraw–Hill, New York), Vol. 1, pp. 66–86. [Google Scholar]
- 2.Mauro, A. (1961) J. Biophys. Biochem. Cytol. 9, 493–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charge, S. B. & Rudnicki, M. A. (2004) Physiol. Rev. 84, 209–238. [DOI] [PubMed] [Google Scholar]
- 4.Asakura, A., Komaki, M. & Rudnicki, M. A. (2001) Differentiation 68, 245–253. [DOI] [PubMed] [Google Scholar]
- 5.Wada, M. R., Inagawa-Ogashiwa, M., Shimizu, S., Yasumoto, S. & Hashimoto, N. (2002) Development (Cambridge, U.K.) 129, 2987–2995. [DOI] [PubMed] [Google Scholar]
- 6.Oustanina, S., Hause, G. & Braun, T. (2004) EMBO J. 23, 3430–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seale, P., Sabourin, L. A., Girgis-Gabardo, A., Mansouri, A., Gruss, P. & Rudnicki, M. A. (2000) Cell 102, 777–786. [DOI] [PubMed] [Google Scholar]
- 8.Goodell, M. A., Brose, K., Paradis, G., Conner, A. S. & Mulligan, R. C. (1996) J. Exp. Med. 183, 1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asakura, A., Seale, P., Girgis-Gabardo, A. & Rudnicki, M. A. (2002) J. Cell Biol. 159, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gussoni, E., Soneoka, Y., Strickland, C. D., Buzney, E. A., Khan, M. K., Flint, A. F., Kunkel, L. M. & Mulligan, R. C. (1999) Nature 401, 390–394. [DOI] [PubMed] [Google Scholar]
- 11.Bachrach, E., Li, S., Perez, A. L., Schienda, J., Liadaki, K., Volinski, J., Flint, A., Chamberlain, J. & Kunkel, L. M. (2004) Proc. Natl. Acad. Sci. USA 101, 3581–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevallier, A., Kieny, M. & Mauger, A. (1977) J. Embryol. Exp. Morphol. 41, 245–258. [PubMed] [Google Scholar]
- 13.Christ, B., Jacob, H. J. & Jacob, M. (1977) Anat. Embryol. (Berlin) 150, 171–186. [DOI] [PubMed] [Google Scholar]
- 14.Kardon, G., Campbell, J. K. & Tabin, C. J. (2002) Dev. Cell 3, 533–545. [DOI] [PubMed] [Google Scholar]
- 15.Rees, E., Young, R. D. & Evans, D. J. (2003) Dev. Biol. 253, 264–278. [DOI] [PubMed] [Google Scholar]
- 16.Armand, O., Boutineau, A. M., Mauger, A., Pautou, M. P. & Kieny, M. (1983) Arch. Anat. Microsc. Morphol. Exp. 72, 163–181. [PubMed] [Google Scholar]
- 17.Gros, J., Manceau, M., Thome, V. & Marcelle, C. (2005) Nature 435, 954–958. [DOI] [PubMed] [Google Scholar]
- 18.Bober, E., Franz, T., Arnold, H. H., Gruss, P. & Tremblay, P. (1994) Development (Cambridge, U.K.) 120, 603–612. [DOI] [PubMed] [Google Scholar]
- 19.Goulding, M., Lumsden, A. & Paquette, A. J. (1994) Development (Cambridge, U.K.) 120, 957–971. [DOI] [PubMed] [Google Scholar]
- 20.Relaix, F., Rocancourt, D., Mansouri, A. & Buckingham, M. (2004) Genes Dev. 18, 1088–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engleka, K. A., Gitler, A. D., Zhang, M., Zhou, D. D., High, F. A. & Epstein, J. A. (2005) Dev. Biol. 280, 396–406. [DOI] [PubMed] [Google Scholar]
- 22.Kassar-Duchossoy, L., Giacone, E., Gayraud-Morel, B., Jory, A., Gomes, D. & Tajbakhsh, S. (2005) Genes Dev. 19, 1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Relaix, F., Rocancourt, D., Mansouri, A. & Buckingham, M. (2005) Nature 435, 948–953. [DOI] [PubMed] [Google Scholar]
- 24.De Angelis, L., Berghella, L., Coletta, M., Lattanzi, L., Zanchi, M., Cusella-De Angelis, M. G., Ponzetto, C. & Cossu, G. (1999) J. Cell Biol. 147, 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaBarge, M. A. & Blau, H. M. (2002) Cell 111, 589–601. [DOI] [PubMed] [Google Scholar]
- 26.Ferrari, G., Cusella-De Angelis, G., Coletta, M., Paolucci, E., Stornaiuolo, A., Cossu, G. & Mavilio, F. (1998) Science 279, 1528–1530. [DOI] [PubMed] [Google Scholar]
- 27.Bittner, R. E., Schofer, C., Weipoltshammer, K., Ivanova, S., Streubel, B., Hauser, E., Freilinger, M., Hoger, H., Elbe-Burger, A. & Wachtler, F. (1999) Anat. Embryol. (Berlin) 199, 391–396. [DOI] [PubMed] [Google Scholar]
- 28.McKinney-Freeman, S. L., Majka, S. M., Jackson, K. A., Norwood, K., Hirschi, K. K. & Goodell, M. A. (2003) Exp. Hematol. (Charlottesville, VA) 31, 806–814. [DOI] [PubMed] [Google Scholar]
- 29.Montanaro, F., Liadaki, K., Schienda, J., Flint, A., Gussoni, E. & Kunkel, L. M. (2004) Exp. Cell Res. 298, 144–154. [DOI] [PubMed] [Google Scholar]
- 30.Rivier, F., Alkan, O., Flint, A. F., Muskiewicz, K., Allen, P. D., Leboulch, P. & Gussoni, E. (2004) J. Cell Sci. 117, 1979–1988. [DOI] [PubMed] [Google Scholar]
- 31.Cepko, C. L., Fields-Berry, S., Ryder, E., Austin, C. & Golden, J. (1998) Curr. Top. Dev. Biol. 36, 51–74. [DOI] [PubMed] [Google Scholar]
- 32.Goulding, M. D., Chalepakis, G., Deutsch, U., Erselius, J. R. & Gruss, P. (1991) EMBO J. 10, 1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown, C. B., Engleka, K. A., Wenning, J., Min Lu, M. & Epstein, J. A. (2005) Genesis 41, 202–209. [DOI] [PubMed] [Google Scholar]
- 34.Buckingham, M., Bajard, L., Chang, T., Daubas, P., Hadchouel, J., Meilhac, S., Montarras, D., Rocancourt, D. & Relaix, F. (2003) J. Anat. 202, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldman, J. L. & Stockdale, F. E. (1992) Dev. Biol. 153, 217–226. [DOI] [PubMed] [Google Scholar]
- 36.Hartley, R. S., Bandman, E. & Yablonka-Reuveni, Z. (1992) Dev. Biol. 153, 206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armand, O. & Kieny, M. (1984) Arch. Anat. Microsc. Morphol. Exp. 73, 75–90. [PubMed] [Google Scholar]
- 38.Kelly, A. M. & Zacks, S. I. (1969) J. Cell Biol. 42, 135–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halevy, O., Piestun, Y., Allouh, M. Z., Rosser, B. W., Rinkevich, Y., Reshef, R., Rozenboim, I., Wleklinski-Lee, M. & Yablonka-Reuveni, Z. (2004) Dev. Dyn. 231, 489–502. [DOI] [PubMed] [Google Scholar]
- 40.Huntington, N. D. & Tarlinton, D. M. (2004) Immunol. Lett. 94, 167–174. [DOI] [PubMed] [Google Scholar]
- 41.Sherwood, R. I., Christensen, J. L., Conboy, I. M., Conboy, M. J., Rando, T. A., Weissman, I. L. & Wagers, A. J. (2004) Cell 119, 543–554. [DOI] [PubMed] [Google Scholar]
- 42.Conboy, I. M., Conboy, M. J., Wagers, A. J., Girma, E. R., Weissman, I. L. & Rando, T. A. (2005) Nature 433, 760–764. [DOI] [PubMed] [Google Scholar]
- 43.Brazelton, T. R., Nystrom, M. & Blau, H. M. (2003) Dev. Biol. 262, 64–74. [DOI] [PubMed] [Google Scholar]
- 44.Liu, J. P., Laufer, E. & Jessell, T. M. (2001) Neuron 32, 997–1012. [DOI] [PubMed] [Google Scholar]
- 45.Chen, C. M., Smith, D. M., Peters, M. A., Samson, M. E., Zitz, J., Tabin, C. J. & Cepko, C. L. (1999) Dev. Biol. 214, 370–384. [DOI] [PubMed] [Google Scholar]
- 46.Novak, A., Guo, C., Yang, W., Nagy, A. & Lobe, C. G. (2000) Genesis 28, 147–155. [PubMed] [Google Scholar]
- 47.Byers, T. J., Lidov, H. G. & Kunkel, L. M. (1993) Nat. Genet. 4, 77–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.