Abstract
Reproductive cloning is uniformly rejected as a valid technology in humans because of the severely abnormal phenotypes seen in cloned animals. Gene expression aberrations observed in tissues of cloned animals have also raised concerns regarding the therapeutic application of “customized” embryonic stem (ES) cells derived by nuclear transplantation (NT) from a patient's somatic cells. Although previous experiments in mice have demonstrated that the developmental potential of ES cells derived from cloned blastocysts (NT-ES cells) is identical to that of ES cells derived from fertilized blastocysts, a systematic molecular characterization of NT-ES cell lines is lacking. To investigate whether transcriptional aberrations, similar to those observed in tissues of cloned mice, also occur in NT-ES cells, we have compared transcriptional profiles of 10 mouse NT- and fertilization-derived-ES cell lines. We report here that the ES cell lines derived from cloned and fertilized mouse blastocysts are indistinguishable based on their transcriptional profiles, consistent with their normal developmental potential. Our results indicate that, in contrast to embryonic and fetal development of clones, the process of NT-ES cell derivation rigorously selects for those immortal cells that have erased the “epigenetic memory” of the donor nucleus and, thus, become functionally equivalent. Our findings support the notion that ES cell lines derived from cloned or fertilized blastocysts have an identical therapeutic potential.
Keywords: expression profiling, nuclear transfer
Nuclear transfer allows for the derivation of genetically matched ES cell lines from somatic cells of diseased individuals that can be differentiated into a host of cell types for cell replacement therapy (1–4). The feasibility of this approach, sometimes referred to as somatic cell nuclear transfer has been demonstrated in animal models (3) and the clinical application of human nuclear transplantation (NT)-ES cells represents an attractive prospect for the treatment of various medical conditions (4).
In animals, however, aberrations in gene expression patterns, like the failure to activate genes essential for early embryonic development or to silence genes that are specific for the somatic donor cell type, affect the majority of preimplantation NT embryos, resulting in a high frequency of embryonic and fetal lethality and widespread gene dysregulation in clones at birth (4, 5–10). Furthermore, surviving clones frequently display severe phenotypic and transcriptional abnormalities (11–14). It has been shown that the differentiation state of the donor nucleus can influence the abnormal gene expression patterns in newborn clones, suggesting retention of the “epigenetic memory” of the somatic donor genome after NT throughout embryonic and fetal development (4, 5, 14–16).
The persistence of gene expression abnormalities in somatic tissues of cloned animals has raised the question whether ES cells derived by NT, in contrast to fertilization-derived ES cells, may pose risks regarding their therapeutic application (5, 17–19). The possibility that cloned ES cells or their derivatives may carry epigenetic alterations causing transcriptional changes in oncogenes or tumor-suppressor genes is of particular concern. However, it has been suggested previously that the process of ES cell derivation, which entails strong selection for in vitro proliferation, allows for the survival of only those cells that have lost the “epigenetic memory” of the respective donor nucleus, thus rendering ES cells derived from NT-blastocysts equivalent to those derived from fertilized ones (15). This notion is supported by evidence indicating that both NT- and fertilization-derived ES cell lines are functionally indistinguishable and can support development of entirely ES cell-derived mice after injection into tetraploid blastocysts (3, 20–23). Because data for the developmental potency of NT-ES cells are not sufficient to address the significant safety concerns regarding gene expression abnormalities, it is important to complement the biological evidence with a molecular characterization of ES cells derived from fertilized and NT blastocysts. Transcriptional profiles of fertilization-derived ES cells have been published in refs. 24–27. However, a systematic comparison of gene expression in NT-ES and fertilization-derived-ES (F-ES) cells is lacking.
In this study, we performed molecular and developmental tests to compare mouse ES cells derived from NT blastocysts with ES cells derived from fertilized blastocysts. Specifically, we examined developmental potency and gene expression profiles of five ES cell lines derived by nuclear transplantation from B and T cells or from fibroblasts and five fertilization-derived ES cell lines of matching genetic backgrounds. We report that ES cell lines cannot be classified as derived from either fertilized or NT blastocysts on the basis of their expression profiles. Our results indicate that gene expression differences attributed to genetic background are more prominent across the tested cell lines than those due to derivation of the respective ES cell line from a fertilized or an NT blastocyst. Our data support the notion that NT-ES cells have lost the “epigenetic memory” of their donor nucleus and have an identical developmental and therapeutic potential as ES cell lines derived from fertilized blastocysts.
Results
Developmental Potency of ES Cell Lines. To determine the developmental potency of NT- and fertilization-derived ES cell lines, we used tetraploid (4n) blastocyst complementation (28). This test is the most stringent for ES cell pluripotency, because virtually all cells in the resulting mouse are derived from the ES cells after their injection into a tetraploid host blastocyst, except for the persistence of a few scattered tetraploid cells (28–30). In contrast, after diploid blastocyst complementation, both the ES cells and cells from the host blastocyst contribute to the resulting chimera. We have shown previously that NT-ES cells derived from fibroblasts, lymphocytes, and olfactory neurons are pluripotent and can give rise to normal mice (3, 22, 23). We have now extended these studies and analyzed the developmental potential of two additional NT-ES cell lines derived from a T lymphocyte and a fibroblast donor nucleus, respectively. As summarized in Table 1, all NT-ES cell lines analyzed in this study generated live pups after tetraploid blastocyst complementation, exhibiting the same developmental potency as the fertilization-derived ES cell lines examined (20, 31).
Table 1. Summary of ES cell lines examined in this study and derived from fertilized eggs or by nuclear transfer.
| ES cell line | Nuclear donor | Genetic background | Developmental potency | Ref. |
|---|---|---|---|---|
| LN1 | B cell | C57BL/6 × DBA/2 F1 | 4n | 22 |
| LN2 | T cell | 129/SvJae × C57BL/6 F1 | 4n | 22 |
| LN3 | T cell | 129/SvJae × C57BL/6 F1 | 4n | |
| ESCC | Fibroblast | C57BL/6 × M. cast. F1 | 4n | |
| Rag2-/- | Fibroblast | 129/SvEv × C57BL/6 F1 | 4n | 3 |
| V6.5 | Fertilized egg | C57BL/6 × 129/SvJae F1 | 4n | 20 |
| J1 | Fertilized egg | 129/SvJae | 4n | 20 |
| F1.2-3 | Fertilized egg | 129/SvJae × M. cast. F1 | 4n | 20 |
| V26.2 | Fertilized egg | C57BL/6 | 4n | 20 |
| Elm3 | Fertilized egg | 129/terSv | 2n | 31 |
The cells were tested for developmental potency by tetraploid complementation (4n, ES cell injection into tetraploid blastocysts) or chimera formation (2n, ES cell injection into diploid blastocysts). Tetraploid complementation is the most stringent test for potency because the resulting animal is virtually entirely derived from the injected ES cells.
Transcriptional Profiles from NT- and F-ES Cell Lines Are Highly Similar. We posited that incomplete epigenetic reprogramming should manifest in transcriptional dysregulation of a subset of genes in most or all NT-ES cell lines. These alterations could cause either the up- or down-regulation of a group of transcripts common to all NT-ES cell lines or, alternatively, an elevated variability of gene expression levels across the NT-ES cell lines when compared to their fertilization-derived counterparts (F-ES cells). We used microarray technology to compare the expression levels of >31,000 transcripts in ES cell lines derived from fertilized and NT blastocysts.
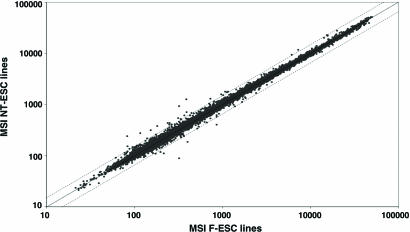
In an effort to assess whether, in analogy to cloned animals, systematic misexpression of genes could be observed in NT-ES cells, the mean probe signal levels of all NT-ES cell lines were compared to the corresponding mean signal values of all F-ES cell lines (Fig. 1). This analysis revealed a high degree of transcriptional similarity between NT- and F-ES cells (Pearson's coefficient of correlation: r = 0.9984). Most important, the examination failed to reveal any significantly deregulated transcripts in NT-ES cells, with Student's t test P values >0.1 for all 37 probes that displayed mean signal changes of >1.5-fold (see Table 2, which is published as supporting information on the PNAS web site, for P values). These results suggest that there is no subset of genes that is significantly up- or down-regulated in all NT-ES cell lines as compared to their fertilization-derived counterparts.
Fig. 1.
Analyses of expression profiles from cloned and fertilization-derived ES cell lines (NT-ESC and F-ESC, respectively). Mean signal intensities (MSI) of five NT-ESC lines were plotted against the corresponding MSI of five F-ESC lines. Dotted lines indicate 1.5-fold regulation.
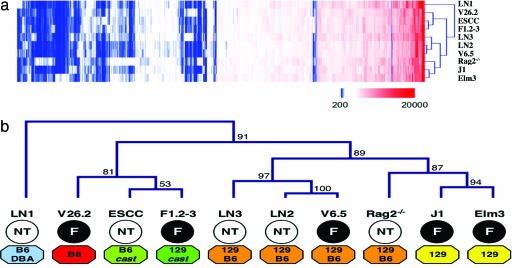
Transcriptional Differences Are Due to Genetic Background Rather than Type of Donor Blastocyst. Unsupervised hierarchical data set clustering was performed to assess differences and similarities between single ES cell line-expression profiles in an unbiased way (Fig. 2a). This grouping of transcriptional profiles according to their overall similarities is less sensitive to outliers than the average signal comparison approach and can identify transcriptionally similar subsets of cell lines. The resulting sample tree revealed no separation of NT- and F-ES cell lines (Fig. 2b). In fact, no direct clustering of any two NT-ES cell lines was observed, and of the F-ES cell lines, only J1 directly clustered with its subclone Elm3. In contrast, two NT-ES cell lines clustered with fertilization-derived ES cell lines, i.e., ESCC with F1.2–3 and LN2 with V6.5, respectively. These results suggest that most tested NT-ES cell lines displayed higher transcriptional similarity to an F-ES cell line than to other NT-ES cell lines. Genetic background annotation revealed separation of different genetic backgrounds between clusters, with the J1/Elm3 cluster including the only two inbred 129/Sv lines, the LN3/LN2/V6.5 cluster containing all three 129/SvJae × C57BL/6 lines and the ESCC/F1.2–3 cluster representing the only two ES cell lines with a Mus musculus domesticus × M. musculus castaneus background. Multiscale bootstrap resampling revealed that our data strongly support clusters containing cell lines of single genetic background like the LN3/LN2/V6.5 or the J1/Elm3 cluster (see Fig. 2b), The clustering results suggest that the most prominent differences in mRNA expression profiles between the ES cell lines analyzed can be attributed to genetic background rather than to the derivation of the respective ES cell line from an NT- or a fertilization-derived blastocyst.
Fig. 2.
Hierarchical clustering of individual ESC line expression profiles. (a) Heat map of clustering results (blue, no or very low expression; white, low expression; red, high expression). (b) Sample tree obtained from hierarchical clustering. ES cell line expression profiles cluster by genetic background (colored octagons) rather than by type of donor blastocyst (NT, cloned; F, fertilized; numbers next to nodes display multiscale bootstrap resampling probability based on 10,000 replications).
Genetic Background but Not Donor Blastocyst Type Can Be Predicted Based on Transcriptional Profiles. We investigated whether a supervised learning scheme could successfully be developed that would allow for the classification of the samples as “NT-ES cell” or “F-ES cell.” We chose to build class predictors by using both k-nearest neighbor and weighted voting algorithms, containing anywhere from 10 to 50 features. Class predictors were evaluated in a cross-validation process, where a predictor is built on data from all but one samples and then used to predict the class of the one sample that was left out. This process was then repeated for each sample. No predictor was found that could accurately classify samples as NT-ES cell or F-ES cell. In contrast, we were able to build accurate predictors for the classification of genetic backgrounds, demonstrating that samples could successfully be classified based on transcriptional profiles within our data set (see Table 3, which is published as supporting information on the PNAS web site, for three representative examples of predictors). We generated predictors that could classify samples as “129/Sv” versus “Other” (100% accuracy), “M. mus. × M. cast.” versus “M. mus. × M. mus.” (100% accuracy), and “129/Sv × C57BL/6” versus Other (100% accuracy, with Rag2–/– assigned “no call” because of low prediction confidence). It should be noted that because in the first two examples given above one class contained only two samples (J1 and Elm3 in class 129/Sv and ESCC and F1.2–3 in class M. mus. × M. cast.), the corresponding predictors might not perform reliably outside our data set. Our class prediction results suggest that genetic background rather than donor blastocyst type is the main parameter associated with gene expression differences among the ES cell lines examined.
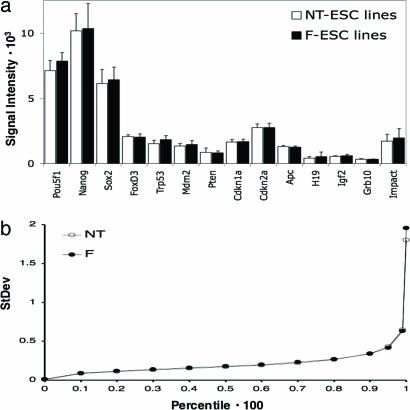
Similar Levels of Transcriptional Variability in NT-ES and F-ES Cells. To determine whether variability in gene expression was elevated in NT-versus F-ES cells, we assessed the expression levels of a number of genes, including stem cell-specific genes, donor cell-specific genes, oncogenes, tumor-suppressor genes and imprinted genes. Fig. 3a shows the expression levels of 14 representative genes (donor cell-specific genes were silent or showed similar levels of expression in NT- and F-ES cell lines, data not shown). Our analysis revealed no marked differences in mean expression levels or variability of gene expression across NT-versus F-ES cells for any of the genes we examined. The expression levels for H19 and Igf2, for example, showed slightly higher variability in the ES cell lines derived from fertilized blastocysts than in the NT-ES cell lines. We have previously reported that DNA methylation patterns in imprinted genes show high variability in ES cells in culture (32). The results of the present study imply that epigenetic instability affects both NT-ES and F-ES cells at the same level.
Fig. 3.
Analysis of variability in gene expression among cloned and fertilization-derived ES cell lines. (a) Comparison of gene expression levels in NT-ES (open bars) and F-ES (filled bars) cell lines. Columns display mean signal intensities; error bars display standard deviation. (b) Comparison of standard deviation levels across all probes in the data set. As a measure of gene expression variability, standard deviation levels were calculated for the log2 probe signal values for each group. Probes in each group (NT-ES and F-ES cell lines) were ordered by their standard deviation levels, and then standard deviation levels were compared at different percentiles. Data sets display closely matched standard deviation levels for different percentiles, indicating highly similar variability in gene expression between the two groups.
To examine whether a global increase in gene expression variability could be detected in the NT-ES cell lines compared to their fertilization derived counterparts, we sorted the standard deviations of all probe signal levels obtained in both groups and plotted the standard deviation values calculated for different percentiles of these sorted data sets (Fig. 3b). The resulting diagram indicates standard deviation levels between the data sets of NT- and F-ES cell lines to be highly similar. The values obtained for the 95th to the 100th percentile indicate a slightly higher variability in gene expression in the fertilization-derived cell lines. Our results suggest that the NT-ES cell lines examined here do not display an increased overall variability in gene expression levels. This finding is in contrast to measurements indicating that overall variability in gene expression among cell lines of the same genetic background is lower than among cell lines of different genetic background (Fig. 5, which is published as supporting information on the PNAS web site).
Discussion
In this study, we have compared the gene expression patterns of ES cells derived from either fertilized or nuclear transfer blastocysts. Our results revealed no marked differences in gene expression profiles between F-ES and NT-ES cell lines but showed differences between ES cell lines of different genetic background. We observed no elevated levels of transcriptional variability in the tested NT-ES cell lines as compared to F-ES cells. Our results provide molecular evidence substantiating the biological observations that F-ES and NT-ES cells have an identical developmental potential as most stringently tested by their ability to generate ES cell mice by tetraploid complementation (3, 20–23).
Previous data have shown that embryos developed from somatic donor cells after nuclear transfer exhibit marked differences in gene expression when compared to embryos obtained from fertilized eggs (7, 10, 14). This evidence has led to the suggestion that the blastocyst retains an “epigenetic memory” of its donor nucleus and that this retention is the cause for the abnormal subsequent fetal development of the NT blastocyst (15). In contrast, after explantation in vitro, such epigenetic differences are erased during the process of ES cell derivation, rendering both NT- and F-ES cell lines functionally indistinguishable (15).
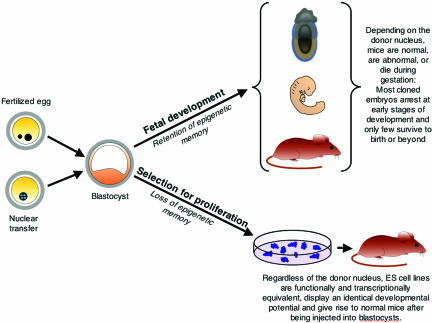
Our findings are consistent with the notion that ES cell derivation is a highly selective process for the rare cells that are able to proliferate under tissue culture conditions. In fact, it has been reported that most inner cell mass (ICM) cells of blastocysts extinguish the expression of Oct4, a key pluripotency gene, and cease dividing after the blastocysts have been explanted in culture (33). Only a small fraction of the explanted cells maintain Oct4 expression and proliferate to give rise to continuously growing immortal cell lines that are designated as “embryonic stem cells.” The important point is that, regardless of whether a given ES cell line was derived from a fertilized or from an NT blastocyst, the ICM cells had to undergo the same stringent selection for in vitro survival and proliferation. We suggest that the result of this process is the loss of the “epigenetic memory” of the donor nucleus, regardless of its origin, in the surviving ES cells, which are, therefore, functionally and transcriptionally equivalent (Fig. 4).
Fig. 4.
Retention of epigenetic memory in blastocysts and loss of epigenetic memory in NT-ES cell lines. The outcome of postimplantation development depends strictly on the origin of the donor nucleus. Fertilized blastocysts develop into normal mice with high efficiency, whereas blastocysts derived after nuclear transplantation from donor cells, such as ES cells, fibroblasts, immune cells, or neurons, develop into abnormal mice with an efficiency that depends on the differentiation status of the respective donor nucleus (15, 16). For example, cloned blastocysts derived from ES cell donor nuclei develop to birth with high efficiency, whereas those derived from fibroblasts, immune cells or neurons develop to birth with low or very low efficiency. In contrast to postimplantation development, the process of deriving embryonic stem cells entails rigorous selection for in vitro proliferation and results in the loss of the epigenetic memory of the donor nucleus.
The derivation and culture of wild-type ES cells has been shown to result in the imbalanced expression of imprinted genes (32). It thus remains to be formally demonstrated that tissues derived from cultured ES cells are equivalent to their native counterparts. However, extensive analyses of chimeric animals over the past 2 decades did not reveal any obvious defects or tumor-forming potentials of ES cell-derived somatic cells.
Based on the abnormal phenotypes and gene expression aberrations seen in cloned animals it has been argued that ES cells derived by somatic cell nuclear transfer from a patient may be unsafe for therapeutic use (5, 17, 19). The results described in this paper are relevant for this debate because they complement previous observations of functional equivalence of NT-ES and F-ES cells (3, 20–23). Thus, if ES cells derived from fertilized blastocysts are useful for therapeutic application, so are “customized” ES cells derived by nuclear transfer.
Materials and Methods
ES Cell Derivation and Culture. Derivation of ES cell lines after nuclear transplantation was carried out as described in ref. 24 by using primary tail-tip fibroblasts or lymphocytes as donor cells. ES cells were cultured on γ-irradiated primary feeder fibroblasts in DMEM [15% FBS/1,000 U/ml of leukemia inhibitory factor (LIF)]. Two independent cultures of each cell line were assayed. After feeder cell depletion by preplating, ES cells were pelleted, snap frozen in liquid nitrogen and stored at –80°C until RNA extraction.
Manufacture of Microarrays. The Array-Ready Oligo Set version 3.0 (Operon Technologies, Alameda, CA), containing 31,769 oligonucleotide probes, was printed onto CodeLink glass slides (Amersham Pharmacia). Printing and postprint processing of glass slides was carried out according to the manufacturer's recommendations.
Microarray Target Preparation and Hybridization. RNA was extracted from frozen cell pellets by using RNeasy Mini columns (Qiagen) and stored at –80°C. Five micrograms of total RNA from each cell population were reverse transcribed with Superscript II reverse transcriptase (Invitrogen) by using a T7-poly-d(T) primer (TCTAGTCGACGGCCAGTGAATTGTAATACGACTCAC TATAGGGCGT21N). Second-strand synthesis was performed by using DNA Polymerase I and Ribonuclease H (Invitrogen). Double-stranded cDNA was transcribed into unlabeled cRNA in vitro by using the Microarray RNA Target Synthesis Kit (T7) (Roche). Two micrograms of the resulting cRNA were chemically labeled with either Cy5 or Cy3 by using the Micromax ASAP RNA labeling kit (PerkinElmer) with the following modification to the manufacturer's recommendations: 1 μl of Cy-dye labeling solution was used per 2 μg of cRNA. One of the two independent samples generated from each individual cell line was labeled with Cy5, the other one with Cy3. Reactions were incubated at 85°C for 15 min and labeled cRNA was purified over Microcon YM-50 columns (Millipore). Purified labeled cRNAs were hybridized to glass arrays by using a commercially available 2× hybridization buffer system (Agilent Technologies) for 14 h at 60°C. The two cRNA samples from each cell line were hybridized with samples from two other cell lines to minimize hybridization artifacts across cell lines (e.g., V6.5-Cy3 vs. LN1-Cy5, V6.5-Cy5 vs. LN2-Cy3, and LN2-Cy5 vs. J1-Cy3). Arrays were hybridized, washed, and dried according to the 60-mer oligo microarray processing protocol (Agilent Technologies).
Data Processing and Statistical Analysis. Arrays were scanned with a GenePix400B Scanner (Axon Instruments) and signal intensities were extracted by genepix pro 6 software (Axon Instruments). Data normalization within and between arrays was performed with the limma software package of the Bioconductor project (34). We chose quantile normalization after confirming that all data sets showed highly similar signal distribution (for normalization details, see Data Set 1, which is published as supporting information on the PNAS web site). Features flagged as noninformative by the signal extraction software were removed from the data sets, resulting in 31,648 probes considered informative across all cell lines. Signal values of two independent samples were averaged for each cell line. Raw data and normalized expression data sets have been submitted to the ArrayExpress database and are available under the accession no. M-Exp-501. Statistical power was calculated based on common variance as outlined in ref. 35. We determined that in this experiment our power to detect a gene expression difference of >1.5-fold between the two groups NT-ES cells and F-ES cells at a two-sided significance level of 0.1 was 96.2%. For hierarchical clustering, data sets were clipped at signal values of 200 and 65,000 and filtered for probes that displayed a ratio of minimum/maximum signal values ≥2 across all cell lines. Average linkage clustering by Euclidean distance was performed by using the tm4 software (36). Single-linkage and complete-linkage clustering yielded similar results. To assess the statistical significance of the clustering results, we performed multiscale bootstrap resampling with 10.000 replications by using the r module pvclust (37). Predictor construction and evaluation was carried out as described by using genecluster2 software (38). K-nearest neighbor and weighted voting algorithms were used for predictor construction.
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health R01-HD045022 and R37-CA84198. K.H. was supported by a Genzyme Postdoctoral Fellowship.
Conflict of interest statement: No conflicts declared.
Abbreviations: F-ES, fertilization-derived-ES, NT, nuclear transplantation.
Data deposition: The microarray data have been deposited in the ArrayExpress database, www.ebi.ac.uk/arrayexpress (accession no. M-Exp-501).
References
- 1.Munsie, M. J., Michalska, A. E., O'Brien, C. M., Trounson, A. O., Pera, M. F. & Mountford, P. S. (2000) Curr. Biol. 10, 989–992. [DOI] [PubMed] [Google Scholar]
- 2.Wakayama, T., Tabar, V., Rodriguez, I., Perry, A. C., Studer, L. & Mombaerts, P. (2001) Science 292, 740–743. [DOI] [PubMed] [Google Scholar]
- 3.Rideout, W. M., 3rd, Hochedlinger, K., Kyba, M., Daley, G. Q. & Jaenisch, R. (2002) Cell 109, 17–27. [DOI] [PubMed] [Google Scholar]
- 4.Hochedlinger, K. & Jaenisch, R. (2003) N. Engl. J. Med. 349, 275–286. [DOI] [PubMed] [Google Scholar]
- 5.Kohda, T., Inoue, K., Ogonuki, N., Miki, H., Naruse, M., Kaneko-Ishino, T., Ogura, A. & Ishino, F. (2005) Biol. Reprod. 73, 1302–1311. [DOI] [PubMed] [Google Scholar]
- 6.Humpherys, D., Eggan, K., Akutsu, H., Friedman, A., Hochedlinger, K., Yanagimachi, R., Lander, E. S., Golub, T. R. & Jaenisch, R. (2002) Proc. Natl. Acad. Sci. USA 99, 12889–12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bortvin, A., Eggan, K., Skaletsky, H., Akutsu, H., Berry, D. L., Yanagimachi, R., Page, D. C. & Jaenisch, R. (2003) Development (Cambridge, U.K.) 130, 1673–1680. [DOI] [PubMed] [Google Scholar]
- 8.Boiani, M., Eckardt, S., Scholer, H. R. & McLaughlin, K. J. (2002) Genes Dev. 16, 1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, S., Chung, Y. G., Williams, J. W., Riley, J., Moley, K. & Latham, K. E. (2003) Biol. Reprod. 69, 48–56. [DOI] [PubMed] [Google Scholar]
- 10.Smith, S. L., Everts, R. E., Tian, X. C., Du, F., Sung, L. Y., Rodriguez-Zas, S. L., Jeong, B. S., Renard, J. P., Lewin, H. A. & Yang, X. (2005) Proc. Natl. Acad. Sci. USA 102, 17582–17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhind, S. M., Taylor, J. E., De Sousa, P. A., King, T. J., McGarry, M. & Wilmut, I. (2003) Nat. Rev. Genet. 4, 855–864. [DOI] [PubMed] [Google Scholar]
- 12.Ogura, A., Inoue, K., Ogonuki, N., Lee, J., Kohda, T. & Ishino, F. (2002) Cloning Stem Cells 4, 397–405. [DOI] [PubMed] [Google Scholar]
- 13.Yanagimachi, R. (2002) Mol. Cell Endocrinol. 187, 241–248. [DOI] [PubMed] [Google Scholar]
- 14.Ng, R. K. & Gurdon, J. B. (2005) Proc. Natl. Acad. Sci. USA 102, 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaenisch, R. (2004) N. Engl. J. Med. 351, 2787–2791. [DOI] [PubMed] [Google Scholar]
- 16.Jaenisch, R., Hochedlinger, K., Blelloch, R., Yamada, Y., Baldwin, K. & Eggan, K. (2004) Cold Spring Harbor Symp. Quant. Biol. 69, 19–27. [DOI] [PubMed] [Google Scholar]
- 17.Fulka, J., Jr., Miyashita, N., Nagai, T. & Ogura, A. (2004) Nat. Biotechnol. 22, 25–26. [DOI] [PubMed] [Google Scholar]
- 18.Armstrong, L., Lako, M., Dean, W. & Stojkovic, M. (November 10, 2005) Stem Cells, 10.1634/stemcells.2005-0350. [DOI] [PubMed]
- 19.Doerflinger, R. (2004) Origins 34, 367–373. [PubMed] [Google Scholar]
- 20.Eggan, K., Akutsu, H., Loring, J., Jackson-Grusby, L., Klemm, M., Rideout, W. M., 3rd, Yanagimachi, R. & Jaenisch, R. (2001) Proc. Natl. Acad. Sci. USA 98, 6209–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, J., Ishii, T., Feinstein, P. & Mombaerts, P. (2004) Nature 428, 393–399. [DOI] [PubMed] [Google Scholar]
- 22.Hochedlinger, K. & Jaenisch, R. (2002) Nature 415, 1035–1038. [DOI] [PubMed] [Google Scholar]
- 23.Eggan, K., Baldwin, K., Tackett, M., Osborne, J., Gogos, J., Chess, A., Axel, R. & Jaenisch, R. (2004) Nature 428, 44–49. [DOI] [PubMed] [Google Scholar]
- 24.Ivanova, N. B., Dimos, J. T., Schaniel, C., Hackney, J. A., Moore, K. A. & Lemischka, I. R. (2002) Science 298, 601–604. [DOI] [PubMed] [Google Scholar]
- 25.Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. & Melton, D. A. (2002) Science 298, 597–600. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka, T. S., Kunath, T., Kimber, W. L., Jaradat, S. A., Stagg, C. A., Usuda, M., Yokota, T., Niwa, H., Rossant, J. & Ko, M. S. (2002) Genome Res. 12, 1921–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharov, A. A., Piao, Y., Matoba, R., Dudekula, D. B., Qian, Y., VanBuren, V., Falco, G., Martin, P. R., Stagg, C. A., Bassey, U. C., et al. (2003) PLoS Biol. 1, E74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy, A., Gocza, E., Diaz, E. M., Prideaux, V. R., Ivanyi, E., Markkula, M. & Rossant, J. (1990) Development (Cambridge, U.K.) 110, 815–821. [DOI] [PubMed] [Google Scholar]
- 29.Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J. C. (1993) Proc. Natl. Acad. Sci. USA 90, 8424–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Z. Q., Kiefer, F., Urbanek, P. & Wagner, E. F. (1997) Mech. Dev. 62, 137–145. [DOI] [PubMed] [Google Scholar]
- 31.Liu, X., Wu, H., Loring, J., Hormuzdi, S., Disteche, C. M., Bornstein, P. & Jaenisch, R. (1997) Dev. Dyn. 209, 85–91. [DOI] [PubMed] [Google Scholar]
- 32.Humpherys, D., Eggan, K., Akutsu, H., Hochedlinger, K., Rideout, W. M., 3rd, Biniszkiewicz, D., Yanagimachi, R. & Jaenisch, R. (2001) Science 293, 95–97. [DOI] [PubMed] [Google Scholar]
- 33.Buehr, M., Nichols, J., Stenhouse, F., Mountford, P., Greenhalgh, C. J., Kantachuvesiri, S., Brooker, G., Mullins, J. & Smith, A. G. (2003) Biol. Reprod. 68, 222–229. [DOI] [PubMed] [Google Scholar]
- 34.Smyth, G. K. & Speed, T. (2003) Methods 31, 265–273. [DOI] [PubMed] [Google Scholar]
- 35.Wei, C., Li, J. & Bumgarner, R. E. (2004) BMC Genomics 5, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., Braisted, J., Klapa, M., Currier, T., Thiagarajan, M., et al. (2003) BioTechniques 34, 374–378. [DOI] [PubMed] [Google Scholar]
- 37.Shimodaira, H. (2002) Syst. Biol. 51, 492–508. [DOI] [PubMed] [Google Scholar]
- 38.Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., Coller, H., Loh, M. L., Downing, J. R., Caligiuri, M. A., et al. (1999) Science 286, 531–537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.