Abstract
During complementary chromatic adaptation (CCA), cyanobacterial light harvesting structures called phycobilisomes are restructured in response to ambient light quality shifts. Transcription of genes encoding components of the phycobilisome is differentially regulated during this process: red light activates cpcB2A2, whereas green light coordinately activates the cpeCDE and cpeBA operons. Three signal transduction components that regulate CCA have been isolated to date: a sensor-photoreceptor (RcaE) and two response regulators (RcaF and RcaC). Mutations in the genes encoding these components affect the accumulation of both cpcB2A2 and cpeBA gene products. We have isolated and characterized a new pigmentation mutant called Turquoise 1. We demonstrate that this mutant phenotype is due to a dramatic decrease in cpeBA transcript abundance and results from a lesion in the cpeR gene. However, in this mutant cpeCDE RNA levels remain near those found in wild-type cells. Our results show that the coordinate regulation of cpeBA and cpeCDE by green light can be uncoupled by the loss of CpeR, and we furnish the first genetic evidence that different regulatory mechanisms control these two operons. Sequence analysis of CpeR reveals that it shares limited sequence similarity to members of the PP2C class of protein serine/threonine phosphatases. We also demonstrate that cpeBA and cpeCDE retain light quality responsiveness in a mutant lacking the RcaE photoreceptor. This provides compelling evidence for the partial control of CCA through an as-yet-uncharacterized second light quality sensing system.
Photosynthetic organisms closely regulate the composition of their light harvesting structures to maximize their photosynthetic efficiency. Much of this control is exerted through regulation of genes encoding light harvesting components. Cyanobacteria and eukaryotic red algae use macromolecular complexes called phycobilisomes (PBS) to capture light energy for photosynthesis (5, 6). PBS consist of light absorbing, chromophorylated phycobiliproteins, and nonchromophorylated, structural peptides termed linkers. PBS are composed of two domains; each contains both phycobiliproteins and linkers. The compact inner region, the core, anchors the PBS to the photosynthetic thylakoid membrane and transmits light energy to the photosynthetic reaction centers. The outer region is made up of structures called rods, which project from the core and harvest light energy.
PBS composition is regulated by numerous environmental factors, including light intensity and, in some species of cyanobacteria, light color. The PBS structural changes that occur in response to changes in the color of ambient light are termed complementary chromatic adaptation (CCA) (8, 9, 43). Most of our understanding of CCA and its regulation has come from the study of the filamentous cyanobacterium Fremyella diplosiphon (Calothrix sp. strain PCC 7601).
During CCA in F. diplosiphon, the composition of the PBS rods is dramatically altered, while the core remains essentially unchanged. CCA, a photoreversible process, is maximally responsive to red light (RL, ∼650 nm) and green light (GL, ∼540 nm) (19, 23, 46). In RL the rods are composed primarily of the phycobiliprotein phycocyanin (PC; maximum absorbance wavelength, ∼620 nm) and its associated linkers, whereas in GL rods contain primarily the phycobiliprotein phycoerythrin (PE; maximum absorbance wavelength, ∼566 nm) and its associated linkers. Thus, CCA allows the organism to maximally absorb the predominant ambient wavelength of light and maintain photosynthetic rates at relatively high levels under a range of light qualities (10). During CCA, PBS composition is controlled primarily via transcriptional regulation of three operons encoding PBS structural components (16, 20, 29, 31, 33, 34). cpeBA, encoding PE, and cpeCDE, encoding the PE linkers, are activated in GL and inactivated in RL. cpcB2A2H2I2D2 (abbreviated as cpcB2A2), which encodes both the RL-inducible PC (PCi) (B2A2) and its associated linkers (H2I2D2), is activated in RL and inactivated in GL.
Molecular genetic approaches were used to isolate three genes encoding CCA signal transduction pathway components: rcaE, rcaF, and rcaC (12, 25, 26). RcaE is a member of the sensor class, and RcaF and RcaC are members of the response regulator class of two-component regulatory systems. RcaE covalently binds a bilin chromophore and is a photoreceptor that controls, either by itself or as part of a complex, light quality sensing during CCA (K. Terauchi and D. M. Kehoe, unpublished data). rcaE null mutants are phenotypically greenish black and are called black (FdBk) mutants. In FdBk mutants, PE and PC levels are approximately equal (as measured by light absorption), regardless of the ambient light quality (25). rcaE, rcaF, and rcaC have been proposed to encode components that regulate both cpeBA and cpcB2A2, since lesions in genes encoding these proteins affect both PCi and PE accumulation (25, 26).
There also appear to be unique mechanisms regulating cpcB2A2 and cpeBA during CCA. Such evidence has been supplied by protein synthesis inhibitor experiments, fluence response studies, and analyses of the kinetics of RNA accumulation for these two operons (33, 34, 35). Support for this possibility has also been provided by isolation of F. diplosiphon green (FdG) mutants (7, 14, 24, 44). FdG mutants properly regulate PC levels under all light conditions but never accumulate measurable levels of PE. Although such mutants could result from mutations in the PE-specific portion of the CCA regulatory pathway, mutations in many other genes could also lead to this phenotype, since the abundance of PE is controlled by numerous environmental parameters and cellular processes, such as chromophore biosynthesis (24).
cpeBA and cpeCDE are not closely linked in the genome (20). However, similar RNA accumulation kinetics have been measured for these two operons in wild-type (WT) cells shifted between RL and GL (20). A central question in the study of CCA is how the expression patterns of cpeBA and cpeCDE are coordinated. Functional studies to identify light responsive elements in the cpeBA or cpeCDE promoter have not been reported to date. However, cpeBA promoter binding studies have led to the identification of several DNA-binding proteins: RcaA, RcaB, and PepB (40, 42). DNA footprint analyses demonstrated that RcaA and PepB both protect the region from positions −67 to −45 of the cpeBA promoter, suggesting that these are the same protein. However, PepB binding activity was equivalent in extracts from both GL- and RL-grown cells (40), whereas RcaA binding was only evident in protein extracts from cells grown in GL (42). The reason for this difference has not yet been resolved. The region from −67 to −45 of the cpeBA promoter contains a direct hexameric repeat (5"-TTGTTA-3") separated by four nucleotides. This direct repeat is not present in the promoter region of cpeCDE (42), although it has been identified in a modified form with five of the six nucleotides conserved, the repeats separated by 52 bp, and in the opposite orientation relative to their position in cpeBA (40). There is also a conserved 17-bp sequence present upstream of both operons (20). It is 83 bp 5" of the cpeBA transcription start site, and its reverse complement is 195 bp 5" of the cpeCDE transcription start site. The role of this sequence is unclear, although a repeated sequence found within this element has been noted to be quite common in the genome of this organism (32). Overall, the mechanism through which coordinated light regulation of cpeBA and cpeCDE is achieved remains unclear.
In this report we analyze a member of a new class of F. diplosiphon pigmentation mutants, the Turquoise 1 (FdTq1) mutant. We demonstrate that in this genetic background the patterns of cpeBA and cpeCDE transcript accumulation have been altered and that these operons are no longer coordinately regulated. In addition, we have cloned, sequenced, and analyzed the gene responsible for the FdTq1 mutant phenotype. Our results suggest a possible mechanism through which cpeBA expression may, in part, be regulated.
MATERIALS AND METHODS
Growth and transformation.
Our F. diplosiphon WT is a shortened filament strain called Fd33 (originally SF33) (15). The FdBk mutant used here has been previously described (25). Strains were grown in BG-11 medium (1) with 10 mM HEPES (pH 8.0), either with or without 25 μg of kanamycin per ml (kan25) at 25 to 30°C. Liquid cultures were bubbled with air containing 3% CO2 while illuminated with 15 μmol of either red (Industrial F20T12/R; Lightbulbs Unlimited, Pomano Beach, Fla.) or green (General Electric F20T12-G) fluorescent light m−2 s−1. Growth was monitored and cell densities were normalized at an absorption wavelength of 750 nm (A750) by using a Beckman DU640B spectrophotometer. Plated cultures were grown in 40 μmol of cool white fluorescent light (General Electric F20T12-CW) m−2 s−1. Photon flux was measured with a Li-Cor LI-250 light meter connected to a LI109SA Quantum Sensor. Transformations were conducted by electroporation by using previously described methods (27).
FdTq1 mutant isolation and complementation.
FdTq1 was isolated as a spontaneous F. diplosiphon pigmentation mutant growing on an FdBk mutant plate. FdTq1 was complemented by transformation with an F. diplosiphon WT genomic plasmid library (27). Transformants were screened on plates containing kan25 for colonies with a restored FdBk phenotype. To establish whether the FdBk phenotype of these colonies was due to the introduced plasmid or a secondary mutation, cells were plated onto BG-11 with or without kan25 and examined for reversion. Plasmids were rescued from phenotypically complemented F. diplosiphon lines by extraction of total DNA, transformation into Escherichia coli DH5α, and selection for resistant colonies on Luria-Bertani plates (38) containing kan25.
DNA cloning, sequencing, and analysis.
Escherichia coli rescued plasmids were isolated by using a Qiaprep Miniprep kit (Qiagen, Valencia, Calif.). PCR amplifications were carried out using the Expand PCR Kit (Roche, Nutley, N.J.) with an MJ Research PTC-150 MiniCycler. Primers used in PCR amplifications to check for insertions in the regions containing cpeBA, cpeCDE, and cpeYZ (encoding the putative lyases for PE [24]) in the FdTq1 mutant were as follows: for cpeBA and cpeYZ (checked together), 5"-AGCCTGCTCC TTTCTCTAAT GG-3" (hybridizes to the noncoding strand 365 bp upstream of the start of translation of cpeB [20]) and 5"-CCGCTGAAAC TTCGTTGTCA TTT-3" (hybridizes to the coding strand 12 bp upstream of the end of cpeZ translation [31]); for cpeCDE, 5"-CCATTACCCA ATTTCCCATG CC-3" (hybridizes to the noncoding strand 290 bp upstream of the start of translation of cpeC [20]) and 5"-GCTTATCTGA ACCGTATTGC CAT-3" (hybridizes to the coding strand 33 bp downstream of the end of cpeE [21]).
Primers used for subcloning trqA, along with the DNA region upstream of open reading frame (ORF) 3, into pPL2.7 were as follows: BPCR1P, 5"-GCGCTGCAGT CAAGTCATAT AGATACAAAC TCA-3"; BPCR1K, 5"-GTGCTGCAGG CTTAATCCAA CCTACATCTG C-3"; BPCR2K, 5"-CTAGGTACCG TGTGGGTAAG CCTAGATAAA GGTAT-3"; and APCR1P, 5"-CACGGTACCT AGGGTCATAA CTAGATGAGG ATAAA-3". Primers BPCR1P and BPCR1K were used to PCR amplify the intergenic region upstream of ORF 3, and BPCR2K and APCR1P were used to amplify trqA. Primers ending with “P” had a PstI site at their 5" ends, and those ending with K had a KpnI site at their 5" ends. PCR products were combined and annealed at the KpnI site and then reamplified with BPCR1P and APCR1P. The resulting PCR product was cut with PstI and inserted into the PstI site of the shuttle plasmid pPL2.7 (13, 39). Sequencing of this subcloned region, and all other sequencing, was conducted by using an ABI Big Dye sequencing kit and an ABI Prism 3700 automated sequencer (Applied Biosystems, Foster City, Calif.). Database searches were carried out by using the PSI BLAST (2) and BLASTP (3) network services at the National Center for Biotechnology Information. Sequence alignments were performed by using both BLASTP (3) and CLUSTAL W (http://www2.ebi.ac.uk/clustalw).
The region of genomic DNA upstream of trqA was isolated by using a PCR-based approach. PCR amplifications were performed with an F. diplosiphon WT genomic plasmid library in pPL2.7 (27). Two sets of primers were used: primers that annealed to vector sequence near the genomic DNA insertion site (3" end of the primer toward the insertion) and primers that annealed to the coding strand or sequences upstream of trqA. The pPL2.7 primers also each contained sequence corresponding to a BamHI site at their 5" ends; their sequences were 5"-CGGGATCCCT GCCAGAATTC GCCCCTCT-3" and 5"-CGGGATCCCT TGCCATCCTA TGGAACTGC-3". The primers specific for the trqA region were 5"-GCGCTGCAGT TAGAGGCAGA TTCGGCAGGG-3", 5"-GCATAGGTTT CCAGGTTCAA CA-3", 5"-CCCAAGACTA TACTTTCGGC AA-3", 5"-CGTAAGCGGT GACTAACACA AA-3", 5"-ACATAAGTCA ATCACTGCTG GAT-3", and 5"-GGACATTTAG TGGGGTAGGC A-3". These primers were also used for sequencing of this region. Reactions were separated on 0.8% agarose TAE gels, and bands containing amplified fragments were excised, purified, and sequenced.
PCR amplifications of the rcaE or rcaF locus in WT and the FdBk mutant were carried out with primer EC100 (5"-ACGTGAGCTC GGATCCCAGA ACACCTCACC ATTCGGT-3"), which anneals ca. 500 bp upstream of the first methionine in rcaE and encodes a BamHI and SacI site at its 5" end, and primer RCAF-SP2 (5"-CCAATGGAGA ATTCTACTTA TTTG-3"), which anneals to the 3" end of the rcaF coding region. Southern blots were conducted according to standard methods (38) by using the PCR products generated with the EC100 and RCAF-SP2 primers.
RNA analysis.
RNA was extracted from 50 ml of F. diplosiphon liquid cultures at mid-logarithmic-growth phase (A750, ∼0.6). Cells were quick cooled to 4°C by swirling in flasks submerged in liquid nitrogen and then centrifuged at 4,000 × g at 4°C for 10 min. Cell pellets were pipetted vigorously with 5 ml of Tri-Reagent (Molecular Research Center, Cincinnati, Ohio), and RNA isolation was performed according to the manufacturer's instructions. RNA was separated for Northern analysis by electrophoresis for 2.5 h at 100 V on 1% agarose-formaldehyde gels (38). RNA was transferred overnight from gels to Nytran Plus nylon membranes (Schleicher & Schuell, Keene, N.H.) using 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and then UV cross-linked as recommended by the manufacturer. Gel-purified PCR products were used as probes for cpeBA (from −70 to +600, relative to cpeB translation start [31]) and cpeCDE (from −310 to +928, relative to cpeC translation start [20]). Gel-purified restriction fragments were used as probes for cpcB2A2 (from −250 to +1020 relative to transcription start [17]) and rRNA (the ca. 5-kb PstI/EcoRI fragment containing the 23S ribosomal DNA from pAN4 [45]). Probes were labeled with [α-32P]dCTP and purified by passing over Centri-Sep spin columns (Princeton Separations, Adelphia, N.J.). RNA blots were hybridized at 65°C for 15 to 18 h in 10 ml of hybridization buffer (0.1 M NaH2PO4, pH 7.0; 0.5 M NaCl; 0.1 M Tris, pH 7.5; 2 mM EDTA; 0.1% sodium dodecyl sulfate [SDS]) and then washed two times for 10 min at 27°C with 50 ml of wash buffer 1 (10 mM NaH2PO4, pH 7; 20 mM EDTA; 0.1% SDS), once for 10 min at 50°C with wash buffer 1, and then once for 10 min at 50°C with wash buffer 2 (50 mM NaH2PO4, pH 7; 20 mM EDTA; 0.1% SDS). Quantification of probe hybridization was conducted using a Molecular Dynamics SP PhosphorImager. Blots were also exposed to Kodak X-Omat AR film at −80°C with an intensifying screen. Each blot was stripped and rehybridized using the rRNA probe (45). For each lane, the relative ribosomal value obtained from the PhosphorImager was used to normalize the test mRNA value, which was then expressed as a percentage of the WT GL value (for cpeCDE and cpeBA) or RL value (for cpcB2A2). Each result is the mean value of three to four independent experiments. Statistical analyses employed a two-tailed Student's t test.
RESULTS
The FdTq1 mutant fails to accumulate PE.
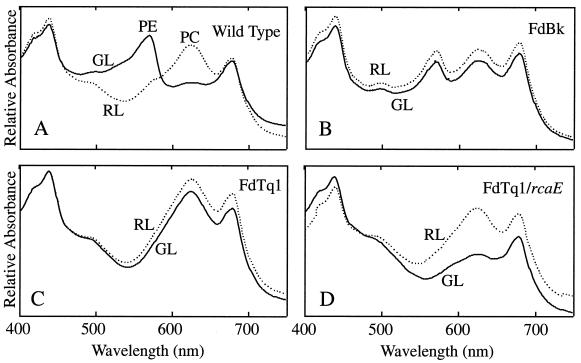
In WT cells, PC levels are high in RL and low in GL, while PE levels are low in RL and high in GL (Fig. 1A). In an FdBk mutant, which lacks functional RcaE (Terauchi and Kehoe, unpublished), both PE and PC accumulate to intermediate levels in both RL and GL (Fig. 1B). FdTq1 is a member of a new class of pigmentation mutants that have been generated in an FdBk mutant background and designated Turquoise (FdTq) mutants. Unlike the green-black color of their parent strain, FdTq mutants are blue-green. Whole-cell absorbance spectra taken of FdTq1 demonstrate that it accumulates high levels of PC and no detectable PE in both RL and GL (Fig. 1C). Under all light conditions it appears similar to WT cells grown in RL. The fact that FdTq1 was isolated from a FdBk mutant line suggested that its phenotype was the result of a mutation in a second genetic locus. To test this, the plasmid pDK4 (25), containing a WT copy of rcaE, was introduced into FdTq1. Transformed cells were grown in RL and GL and whole-cell absorbance measurements were made. In FdTq1 cells containing WT rcaE, light regulation of PC abundance was clearly restored, with levels in RL and GL that appear identical to those observed in WT cells (Fig. 1D). However, these transformed cells did not accumulate any detectable PE in either GL or RL. These results suggested that the FdTq1 phenotype was the result of a secondary mutation in a rcaE null mutant background that resulted in the loss of PE accumulation in all light conditions examined. Thus, the phenotype of FdTq1 transformed with pDK4 (Fig. 1D) is equivalent to that of FdG mutants, which appear to exist in an otherwise WT background (7, 14, 24, 44).
FIG. 1.
Absorption spectra of F. diplosiphon cells grown in liquid BG-11 in 15 μmol of RL (dashed line) and GL (solid line) m−2 s−1. The x axis is the wavelength from 400 to 750 nm, and the y axis is the relative absorbance. (A) WT cells (B) FdBk mutant cells (C) FdTq1 mutant cells (D) FdTq1 mutant cells transformed with a pDK4, which contains a WT copy of rcaE. Chlorophyll absorption peaks are at ca. 430 and 680 nm; PC and PE absorbance peaks are labeled.
All pigmentation mutations thus far complemented for this organism (over 100 total) have resulted from DNA insertion events (12, 24, 25, 26; D. M. Kehoe, unpublished data). Thus, the FdTq1 genes encoding the known PE apoproteins, linkers, lyases (cpeBA, cpeCDE, and cpeYZ), and flanking sequences were checked for mutations caused by DNA insertion events by using PCR amplification. Neither these operons nor their surrounding sequences had DNA insertions (data not shown).
Complementation of FdTq1.
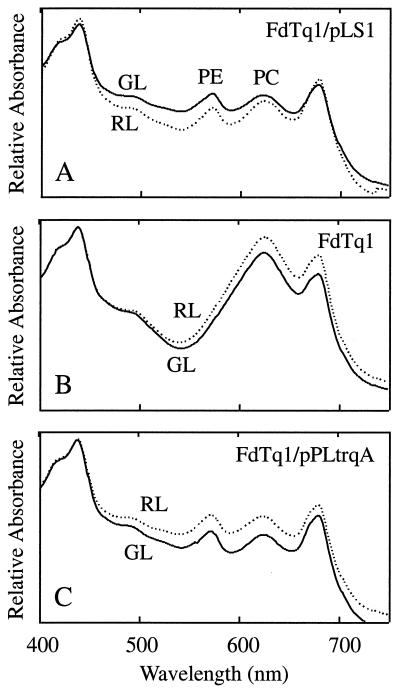
To determine the nature of the secondary mutation, FdTq1 cells were transformed with a WT F. diplosiphon genomic DNA plasmid library (27) and spread on plates containing kan25. Approximately 8,000 kan25-resistant transformed colonies were visually screened for a FdBk mutant phenotype. Ten partially or completely black colonies were identified, isolated, and restreaked onto BG-11 plates containing kan25. All of these segregated completely to the FdBk mutant phenotype and, when grown in liquid with kan25 selection, had whole-cell absorption spectral profiles (Fig. 2A) that were very similar to that of FdBk (Fig. 1B). All of the transformed lines rapidly reverted back to the FdTq1 phenotype upon removal from kan25 selection (Fig. 2B). A plasmid was rescued from one of the transformed lines into E. coli and designated pLS1. This plasmid contained a 1.6-kb fragment of F. diplosiphon genomic DNA. Reintroduction of purified pLS1 plasmid into FdTq1 conferred the FdBk pigment phenotype (data not shown).
FIG. 2.
Complemented FdTq1 mutant absorption spectra. (A) The FdTq1 mutant harboring the pLS1 plasmid was grown in liquid BG-11 containing kan25 in 15 μmol of RL (dashed line) and GL (solid line) m−2 s−1. (B) The FdTq1 mutant containing pLS1 after 4 days' growth in BG-11 without kan25 in 15 μmol of RL (dashed line) and GL (solid line) m−2 s−1. (C) The FdTq1 mutant was transformed with pPLtrqA (Fig. 3C) and grown in liquid BG-11 containing kan25 in 15 μmol of RL (dashed line) and GL (solid line) m−2 s−1. The chlorophyll absorption peaks are at ca. 430 and 680 nm; the PC and PE absorption peaks are labeled.
Sequencing and analysis of the pLS1 genomic DNA.
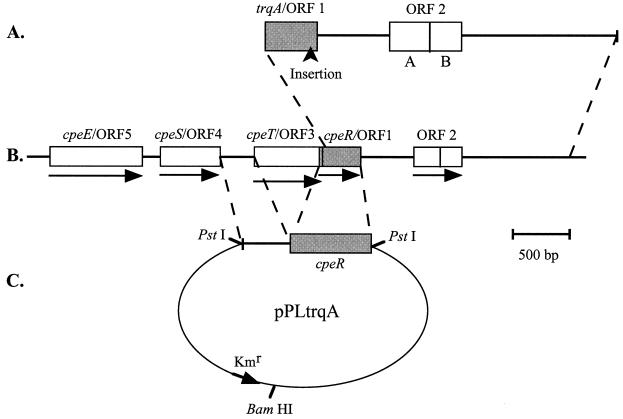
Sequencing of the 1.6-kb fragment of F. diplosiphon WT genomic DNA within pLS1 revealed ORFs that were initially designated ORF 1, ORF 2A, and ORF 2B (Fig. 3A). The ORF 1 sequence was 336 bp long and was located at one end of the genomic DNA fragment. ORF 2A was 206 bp long and was located 455 bp downstream of, and in the same orientation as, ORF 1. ORF 2B was 189 bp long, in the same orientation and reading frame as ORF 2A, and separated from ORF 2A by a single stop codon. The corresponding 1.6-kb region of genomic DNA was examined in the FdTq1 mutant and compared to the WT fragment. Only a single difference was identified: the FdTq1 sequence contained a 1.5-kb DNA insertion in ORF 1, 42 bp from its 3" end (between the 15th and 14th amino acids from the carboxyl terminus) (Fig. 3A). ORF 1 was given the designation trqA based on this result.
FIG. 3.
Genomic DNA fragments used for complementation of the FdTq1 mutant. (A) The F. diplosiphon genomic DNA fragment contained within pLS1 contained a 5" truncated form of trqA/ORF 1 and ORFs 2A and 2B. The location of a ca. 1.5-kb DNA insertion within trqA/ORF 1 is shown. (B) The region of the F. diplosiphon genome containing the pLS1 insert DNA (indicated by dashed lines). ORF numbers and GenBank designations (accession no. AF334109) are provided above each ORF, and the direction of transcription is indicated by an arrow for each. The cpeT/ORF 3 and cpeR/ORF 1 reading frames overlap. cpeE/OFR 5 is located at the 3" end of the cpeCDE operon. (C) Construction of a plasmid containing cpeR fused to the putative promoter sequence upstream of cpeT. This chimera (indicated by dashed lines) was cloned into the unique PstI site of pPL2.7 to make pPLtrqA, which was then transformed into the FdTq mutant.
The 5" end and upstream regions of trqA were not apparent in the genomic DNA insert of pLS1. To obtain sequence corresponding to this region, PCR amplification of clones within the F. diplosiphon WT genomic plasmid library was conducted with one primer that annealed to the noncoding strand near the 5" end of trqA and another annealed to one of two sites within the pPL2.7 shuttle plasmid near the genomic DNA insertion site. The PCR products generated were sequenced directly. This approach provided ca. 1.6 kb of sequence upstream of the 5" end of trqA found in pLS1. Assuming the first methionine is used during translation, 24 bp (encoding eight amino acids) of the 5" end of trqA was absent in the genomic DNA found in pLS1. Three additional ORFs (ORF 3, ORF 4, and ORF 5) were found upstream of trqA (Fig. 3B); the 3" end of ORF 3 overlapped the 5" end of trqA for 23 bp. ORF 4 and ORF 3 are separated by 308 bp, if we assume that the first methionine of ORF 3 is the translation start site.
To conclusively establish whether the insertion in trqA caused the FdTq1 mutant phenotype, trqA alone was introduced into FdTq1 mutant cells. We wanted to include the promoter of trqA, but because its coding region overlapped that of ORF 3 it was not possible to clearly define such a region immediately upstream of trqA. Both the size and expression level of trqA RNA in RL and GL were investigated by using Northern blots, but no transcript corresponding to trqA could be detected (data not shown). Thus, a 292-bp region between ORF 3 and ORF 4 was inserted upstream of trqA (Fig. 3B) since it represented the most likely promoter region for both ORF 3 and trqA. All of the ORF 3 coding sequence was removed except the last 120 bp. This chimera was then cloned into the PstI site of pPL2.7 (13) to make pPLtrqA (Fig. 3C) and transformed into FdTq1. The phenotype of FdTq1 transformed with pPLtrqA (Fig. 2C) was identical to that of the mutant complemented with pLS1 (Fig. 2A). This result, and the presence of a DNA insertion at the 3" end of trqA in FdTq1, leads us to conclude that the mutation in trqA is the secondary mutation in the FdBk mutant that caused the FdTq1 mutant phenotype.
BLASTP searches (3) were conducted for the five ORFs described above. No perfect sequence matches were found in GenBank for ORFs 1 to 4 when they were originally analyzed. However, analysis of ORF 5, which was sequenced last, demonstrated that it was cpeE (100% identity), which is located at the 3" end of the cpeCDE operon and encodes a PE linker peptide (21). Our subsequent BLASTP searches by using ORFs 1 to 4 showed that the sequences of ORF 4, ORF 3, and trqA (and their corresponding proteins) had been submitted to GenBank in the intervening period as cpeS (CpeS), cpeT (CpeT), and cpeR (CpeR) (accession no. AF334109 [unpublished], 100% identity for each) (Fig. 3B). Thus, trqA is cpeR. ORF 2A had also been submitted to GenBank, as an unknown protein (accession no. AF334109 [unpublished]). BLASTP searches using the combined ORF 2A and ORF 2B translated sequences demonstrated that this region was closely related to the crtE gene product geranylgeranyl diphosphate synthase in the thermophilic cyanobacterium Synechococcus elongatus (accession no. AB016093; expectation [E] value of 6 × 10−37) (36).
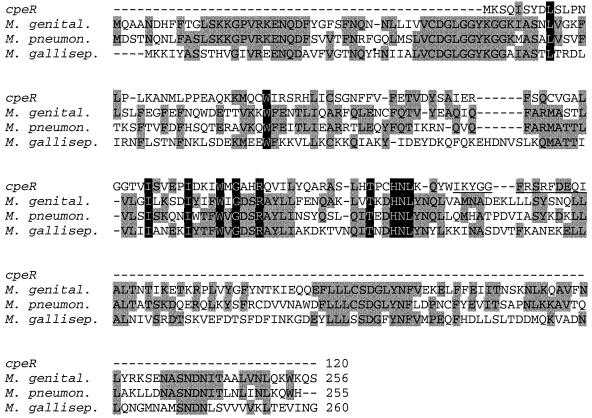
A BLASTP analysis of the translated protein sequence of CpeR provided the most significant alignment to a putative protein phosphatase from Mycoplasma genitalium (accession no. NP__072770 [22]). Additional searches were conducted by using CpeR and PSI-BLAST (2). After the second PSI-BLAST iteration, only three sequences other than CpeR itself produced alignments with an E value at or below 5 × 10−6. All of these were putative protein phosphatases from microorganisms; two of the three were categorized as PP2C class protein serine/threonine phosphatases. The alignment of these sequences with CpeR is shown in Fig. 4. CpeR, from amino acids 47 to 102, was 32% identical and 53% similar to the M. genitalium putative protein phosphatase after the second PSI-BLAST iteration.
FIG. 4.
Similarity of cpeR to PP2C class protein serine/threonine phosphatases from three bacterial Mycoplasma species: M. genital., M. genitalium putative protein phosphatase (accession no. NP_072770.1); M. pneumon., M. pneumoniae protein phosphatase 2C homolog (accession no. NP_109935.1); and M. gallisep., M. gallisepticum putative protein phosphatase (accession no. AAF36762.1). Alignment was performed with the CLUSTALW program with minor realignments made by using information from a PSI-BLAST analysis (second iteration). Residues conserved between two or three species are shown in gray; residues conserved in all four proteins are highlighted in black. Gaps (represented by dashes) have been inserted to maximize alignments. Total number of amino acids in each of the proteins is provided. Underlined amino acids in cpeR are those encoded downstream of the site of the DNA insertion in FdTq1.
The CpeR sequence, starting from the first methionine, was also analyzed by using various proteomics tools at the ExPASy Molecular Biology Server (http://www.expasy.ch/) and the San Diego Supercomputing Center's Biology WorkBench (http://workbench.sdsc.edu/). CpeR is composed of 120 amino acids and has an estimated molecular mass of 13.8 kDa with a theoretical pI of 9.2. InterPro Scan (http://www.ebi.ac.uk/interpro/scan.html) detected no significant similarities to CpeR in PROSITE, Pfam, or PRINTS domain databases. Both SAPS and GREASE (Kyte-Doolittle Hydropathy Profile) programs found that CpeR contained predominantly hydrophilic residues and no obvious potential membrane-spanning domains, clusters of positively or negatively charged residues, or large hydrophobic regions. The PELE protein structure prediction program identified three short regions (5 to 20 amino acids) of potential α-helices, and three possible β-strand stretches (5 to 10 amino acids) within CpeR. The majority of the protein was designated as random coil in this analysis. Finally, the HTH Program was unable to predict any helix-turn-helix motifs in CpeR.
CpeR is required for expression of cpeBA but not cpeCDE.
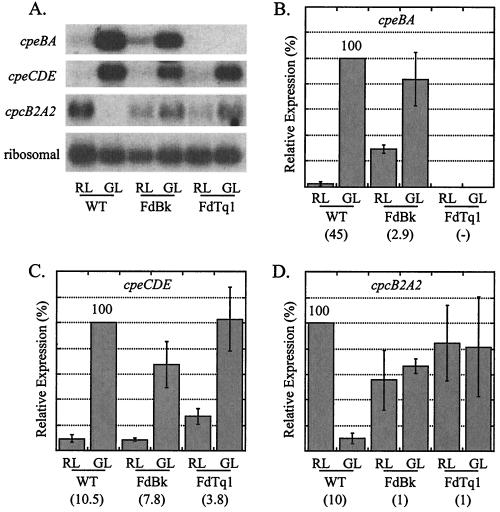
By using Northern blots, the effect of the cpeR mutation on transcript accumulation patterns in RL and GL was measured for cpeBA, cpeCDE, and cpcB2A2 (Fig. 5A). RNA was isolated from three lines (WT, an FdBk mutant, and the FdTq1 mutant) grown under each light condition. Three to four independent RNA isolations and hybridizations were conducted for each operon and all values were normalized by using ribosomal loading control values.
FIG. 5.
Northern blot analysis of cpeBA, cpeCDE, and cpcB2A2 expression in WT, the FdBk mutant, and the FdTq1 mutant. (A) Representative autoradiograms of the data presented in panels B to D showing hybridization to the probes listed on the left. RNA was isolated from the WT and the two mutant lines after growth in 15 μmol of RL and GL m−2 s−1. (B) Mean values from four independent experiments of cpeBA RNA, expressed as a percentage of the WT GL value (set to 100%). (C) Mean values from three independent experiments of cpeCDE RNA, expressed as a percentage of the WT GL value (set to 100%). (D) Mean values from four independent experiments of cpcB2A2 RNA, expressed as a percentage of the WT RL value (set to 100%). The numbers in parentheses are the fold induction between RL and GL for each operon in each cell line. Standard errors are shown; P values are provided in the text. Measurements were normalized by using ribosomal values prior to calculation of means.
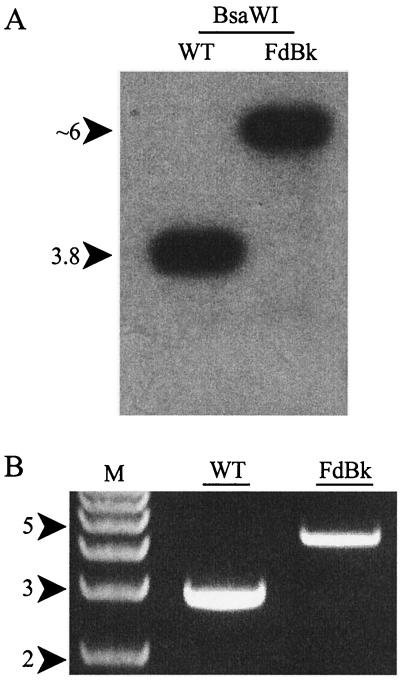
The expression of cpeBA in WT cells was 45 times higher in GL than in RL, but cpeBA RNA was not detected in RL or GL in the FdTq1 mutant (Fig. 5B). This finding is consistent with the pigmentation phenotypes of FdTq1 observed under these two light conditions (Fig. 1C). Surprisingly, for cpeBA in the FdBk mutant, some residual regulation by light quality remained: RNA levels in GL-grown cells were still 2.9-fold higher than in RL-grown cells (0.05 > P > 0.02). These data suggest that RcaE is exerting its control under both light conditions since, compared to the WT, cpeBA in the FdBk mutant appeared to be both less silenced in RL and less activated in GL. Since F. diplosiphon contains approximately six chromosomal copies per cell, PCR amplification and Southern blot analysis of the genomic region containing rcaE in the FdBk mutant were conducted to insure that no WT copies of the gene remained. Because a DNA insertion 69 bp downstream of the putative RcaE translation start site caused the rcaE mutation (K. Terauchi, A. R. Grossman, and D. M. Kehoe, unpublished data), this polymorphism is easily detected. Neither method detected any WT-size rcaE in this mutant (Fig. 6).
FIG. 6.
Check of the FdBk mutant for WT copies of rcaE. (A) Autoradiogram of a Southern blot of genomic WT or FdBk DNA cut with BsaW1 and hybridized with an rcaE probe. The DNA insertion in rcaE in the FdBk mutant results in decreased mobility that is distinguishable from the mobility of the same region in WT. (B) PCR amplification and agarose gel electrophoresis were used to examine the size of the rcaE region in the genome of the WT and the FdBk mutant.
The cpeCDE RNA level was 10.5 times higher in GL than in RL in WT cells (Fig. 5C). A portion of the WT level of RL and GL regulation of expression was also retained for this operon in the FdBk mutant. However, the non-RcaE-based light-sensing system(s) clearly contributed much more to the control of RL and GL responsiveness for cpeCDE than for cpeBA, since for this operon a 7.8-fold difference in transcript accumulation between the two light conditions remained in the FdBk mutant (Fig. 5C) (0.05 > P > 0.02). Measurement of cpeCDE transcript levels in FdTq1 highlighted a second major difference between the regulation mechanisms of these two operons. In this mutant, cpeCDE transcript was still 3.8-fold more abundant in GL than RL and accumulated to levels similar to those measured in FdBk cells (Fig. 5C). These results unambiguously demonstrate that, although CpeR is absolutely required for the expression of cpeBA in both RL and GL, it plays no obvious role in regulating cpeCDE expression in GL and a minor role, if any, in RL (P > 0.05 for significance of the difference between the means for cpeCDE levels in FdBk and FdTq1 mutant cells grown in RL).
For cpcB2A2 in WT cells, the RL expression level was ca. 10 times that measured for GL (Fig. 5D). FdBk mutant cells had no statistically significant difference in cpcB2A2 RNA levels when grown in RL and GL (P ≫ 0.5), and both levels were between 55 and 65% of the WT level in RL. This operon also continued to be expressed in the FdTq1 mutant but, as in the FdBk mutant, was not differentially regulated in RL and GL.
DISCUSSION
In this report we describe the isolation, characterization, and complementation of FdTq1, the first member of a newly described class of turquoise-colored F. diplosiphon mutants that were generated in an FdBk mutant background. Its color phenotype is novel, since it is equivalent to that of WT cells grown in RL (Fig. 1A) but is not altered by changes in light quality (Fig. 1C). By introducing plasmid-borne WT rcaE into this mutant, we have demonstrated that in a WT background, FdTq1 is phenotypically equivalent to the previously described FdG mutant class (7, 14, 24, 44), in which PC expression is normal but PE fails to accumulate in either RL or GL (Fig. 1D). Although many FdG mutants have been isolated, the only one that has been complemented to date was found to contain a lesion in the cpeYZ operon, which appears to encode subunits of a PE lyase (24).
We were able to complement FdTq1 with trqA (Fig. 2C), and we found a large DNA insertion in the 3" end of trqA in the FdTq1 mutant (Fig. 3A). These data demonstrate that the lesion in trqA is responsible for the shift from a FdBk mutant to the FdTq mutant phenotype. Although the amino-terminal eight amino acids of the putative protein encoded by trqA were not encoded by the F. diplosiphon genomic DNA in pLS1, this plasmid still complemented FdTq1. The most likely reasons for this are (i) that these residues are not required for protein function, (ii) that the truncated protein functions suboptimally but still complements FdTq1 in our assay, or (iii) that the translation start site is actually within the sequence contained in pLS1.
BLAST searches with trqA and the surrounding ORFs revealed that this region of the genome lies just downstream of the cpeCDE operon (Fig. 3B). Since the sequences of the ORFs in this region have been submitted to GenBank (accession no. AF334109), the designation cpeR supplants the name trqA. CpeR is most similar to a putative member of the PP2C class of protein phosphatases (22), which dephosphorylate serine and threonine residues and are found in both eukaryotes and prokaryotes (4, 18, 41). CpeR was most similar to the β5-β8 strand region of known protein phosphatases (18). However, the predicted size of CpeR is much smaller than previously described PP2C class proteins and it lacks most of the catalytic site and residues necessary for PP2C function (Fig. 4) (18). Thus, it is unlikely that, at least by itself, CpeR acts as a phosphatase and its function remains to be determined. It is also unclear whether CpeR is an element of the CCA or some other regulatory pathway. The three CCA signal transduction components isolated to date suggest that a two-component regulatory system makes up at least the initial part of this regulatory pathway (12, 25, 26). These pathways are typically regulated by histidine-aspartate phosphotransfer reactions. It is possible that CpeR acts in the downstream portion of this pathway, since two-component systems have been found to be integrated with mitogen-activated protein kinase pathways in both yeast and Arabidopsis (11, 28, 30, 37). The activities of two binding proteins that interact with the cpeBA promoter have been either shown (RcaA) or suggested (PepB) to be modified by their phosphorylation state (40, 42). Thus, the possibility that CpeR, in conjunction with one or more other factors, dephosphorylates a component(s) in the cpeBA-specific portion of the CCA pathway is an interesting one. Other than identifying relatively weak similarity to PP2C class proteins (Fig. 4), our analysis of the CpeR sequence failed to identify any outstanding features that might provide clues regarding its function. It appears to be primarily hydrophilic, possesses no obvious transmembrane stretches, and contains two to three potential α-helical sequences and the same number of β-strand-forming regions. It has no obvious helix-turn-helix motifs, which often serve as DNA-binding domains in transcription factors.
Since the DNA insertion in cpeR in the FdTq1 mutant effectively ends CpeR only 14 amino acids from its carboxyl terminus, it is possible that FdTq1 cells contain a modified form of CpeR. If so, it appears to be functionally incapable of contributing to cpeBA expression. This insertion site may be in a critical region of the protein, since it is only four amino acids away from a block of three amino acids that are absolutely conserved in the three proteins with greatest similarity to CpeR (Fig. 4).
The disruption of cpeR had no significant effect on cpcB2A2 expression (Fig. 5D) but led to the failure to accumulate measurable levels of cpeBA RNA, which provides an explanation for the lack of accumulation of functional PE in these cells (Fig. 5B). cpeCDE transcript levels in FdTq1, however, were actually slightly higher than those measured in the FdBk mutant and WT backgrounds (Fig. 5C). Thus, CpeR is required for normal cpeBA expression but exerts little or no control over the accumulation of cpeCDE RNA. Previous analysis (20) has shown that the kinetics of cpeBA and cpeCDE RNA accumulation are similar during transitions between RL and GL, which suggested that their expression might be controlled through a common mechanism. The data presented here clearly demonstrate that either different mechanisms or different combinations of mechanisms control the expression of these two operons. We have not yet determined whether CpeR controls transcription of cpeBA or acts posttranscriptionally. If CpeR does affect cpeBA transcription, our findings would be consistent with the apparent lack of any unique, highly conserved sequence motifs shared between the promoters of these two operons (20, 40, 42) and, together, these data would strongly suggest that their regulation primarily operates through different pathways. Our findings do not preclude the possibility that common aspects of the regulation of these operons exist.
The FdBk mutant was originally identified by its intermediate levels of accumulation of PC and PE in both RL and GL (25). In this study, our examination of cpcB2A2, cpeBA, and cpeCDE RNA levels in the FdBk revealed unexpected and significant differences in the RL- and GL-sensing mechanisms for these operons. Under the conditions used here, cpcB2A2 RL- and GL-sensing occurs only through RcaE (Fig. 5D). The cpeBA operon is regulated primarily through RcaE (Fig. 5B), although the residual 7% RL and GL regulation represented a 2.9-fold level of GL induction. The RL-GL difference was statistically significant and observed in every experiment conducted. Thus, there is at least one additional system influencing the light quality responsiveness for this operon. Even more surprising was our finding that cpeCDE RNA levels were regulated by RL and GL at ca. 74% of the WT level (Fig. 5C). Thus, RcaE appears to have a relatively minor role in controlling the RL and GL responsiveness of the cpeCDE operon.
The residual threefold change in cpeBA transcript levels between RL and GL (Fig. 5B) might be expected to be visible in the FdBk mutant pigmentation phenotype. In fact, we occasionally observed slight variations in PE levels in this mutant (for example, see Fig. 1B), but typically these were no more than ca. 5% of the WT levels, a finding which correlates with the 7% RL-GL difference in cpeBA RNA levels in the FdBk mutant measured here (Fig. 5B). Since the linker proteins encoded by the cpeCDE operon are not chromophorylated, any RL versus GL differences in the abundance of these proteins in the FdBk mutant would not be visually detectable.
Because these findings constitute the first report of the cpeBA and cpeCDE operons being controlled by a light quality sensing system(s) other than that operating through RcaE, we carefully examined the FdBk mutant used in our studies for the presence of residual WT copies of rcaE. Both Southern blot (Fig. 6A) and PCR amplification approaches (Fig. 6B) identified only the form of rcaE containing the previously described DNA insertion in these cells (25). Western blot analysis of total protein from the FdBk mutant with anti-RcaE antibodies showed that no detectable RcaE is present in this mutant (Terauchi and Kehoe, unpublished). In addition, if any WT copies were present in the FdBk mutant, some difference in cpcB2A2 expression might be expected, since both its activation and inactivation are more sensitive to RL-GL shifts than is cpeBA (33).
Our analysis of the FdTq1 mutant provides clear genetic evidence that the cpeBA and cpeCDE transcript accumulation is controlled, at least in part, through different mechanisms. Although the mechanism through which CpeR regulates cpeBA expression remains to be elucidated, its sequence similarity to the PP2C class of protein phosphatases provides a starting point for the testing of its function. In addition, the data presented here demonstrate that there is dramatic variation in the degree to which the RcaE-controlled system regulates the RL and GL expression of the cpcB2A2, cpeBA, and cpeCDE operons. Collectively, these results clearly demonstrate that one or more important light-sensing mechanism(s), in addition to that controlled by RcaE, must regulate cpeBA and cpeCDE expression. The nature of the additional sensory system(s) that we have uncovered remains to be identified.
ADDENDUM IN PROOF
The altered patterns of cpeBA and cpeCDE transcript accumulation reported here for the FdTq1 mutant have also been found for a cpeR knockout mutant (J. Cobley, submitted for publication).
Acknowledgments
We thank Rick Alvey, Lina Li, Barbara Quinby, Arthur Grossman, Bettina Kehoe, Beronda Montgomery, and Emily Stowe-Evans for their thoughtful reviews and comments.
This work was supported by startup funds from Indiana University and National Science Foundation grant MCB-0084297.
REFERENCES
- 1.Allen, M. M. 1968. Simple conditions for growth of unicellular blue-green algae on plates. J. Phycol. 4:1-4. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bakal, C. J., and J. E. Davies. 2000. No longer an exclusive club: eukaryotic signaling domains in bacteria. Trends Cell Biol. 10:32-38. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, A., and L. Bogorad. 1971. Properties of subunits and aggregates of blue-green algal biliproteins. Biochemistry 10:3625-3634. [DOI] [PubMed] [Google Scholar]
- 6.Bennett, A., and L. Bogorad. 1973. Complementary chromatic adaptation in a filamentous blue-green alga. J. Cell Biol. 58:419-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruns, B. U., W. R. Briggs, and A. R. Grossman. 1989. Molecular characterization of phycobilisome regulatory mutants in Fremyella diplosiphon. J. Bacteriol. 171:901-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant, D. (ed.). 1994. The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 9.Bryant, D. A. 1981. The photoregulated expression of multiple phycocyanin species: general mechanism for control of phycocyanin synthesis in chromatically adapting cyanobacteria. Eur. J. Biochem. 119:425-429. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, D. 1996. Complementary chromatic adaptation alters photosynthetic strategies in the cyanobacterium Calothrix. Microbiology 142:1255-1263. [DOI] [PubMed] [Google Scholar]
- 11.Chang, C., S. F. Kwok, A. B. Bleecker, and E. M. Meyerowitz. 1993. Arabidopsis ethylene-response gene ERT1: similarity of product to two component regulators. Science 262:539-544. [DOI] [PubMed] [Google Scholar]
- 12.Chiang, G. G., M. R. Schaefer, and A. R. Grossman. 1992. Complementation of a red-light indifferent cyanobacterial mutant. Proc. Natl. Acad. Sci. USA 89:9415-9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang, G. G., M. R. Schaefer, and A. R. Grossman. 1992. Transformation of the filamentous cyanobacterium Fremyella diplosiphon by conjugation or electroporation. Plant Physiol. Biochem. 30:315-325. [Google Scholar]
- 14.Cobley, J. G., and R. D. Miranda. 1983. Mutations affecting chromatic adaptation in the cyanobacterium Fremyella diplosiphon. J. Bacteriol. 153:1486-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobley, J. G., E. Zerweck, et al. 1993. Construction of shuttle plasmids which can be efficiently mobilized from Escherichia coli into the chromatically adapting cyanobacterium Fremyella diplosiphon. Plasmid 30:90-105. [DOI] [PubMed] [Google Scholar]
- 16.Conley, P. B., P. G. Lemaux, and A. Grossman. 1985. Cyanobacterial light-harvesting complex subunits encoded in two red light-induced transcripts. Science 230:550-553. [DOI] [PubMed] [Google Scholar]
- 17.Conley, P. B., P. G. Lemaux, and A. Grossman. 1988. Molecular characterization and evolution of sequences encoding light-harvesting components in the chromatically adapting cyanobacterium Fremyella diplosiphon. J. Mol. Biol. 199:447-465. [DOI] [PubMed] [Google Scholar]
- 18.Das, A. K., N. R. Helps, P. T. W. Cohen, and D. Barford. 1996. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 Å resolution. EMBO J. 15:6798-6809. [PMC free article] [PubMed] [Google Scholar]
- 19.Diakoff, S., and S. Scheibe. 1973. Action spectra for chromatic adaptation in Tolypothrix tenuis. Plant Physiol. 51:382-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Federspiel, N. A., and A. R. Grossman. 1990. Characterization of the light-regulated operon encoding the phycoerythrin-associated linker proteins from the cyanobacterium Fremyella diplosiphon. J. Bacteriol. 172:4072-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Federspiel, N. A., and L. Scott. 1992. Characterization of a light-regulated gene encoding a new phycoerythrin-associated linker protein from the cyanobacterium Fremyella diplosiphon. J. Bacteriol. 174:5994-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser, C. M., et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 23.Haury, J. F., and L. Bogorad. 1977. Action spectra for phycobiliprotein synthesis in a chromatically adapting cyanophyte Fremyella diplosiphon. Plant Physiol. 60:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahn, K., D. Mazel, J. Houmard, N. Tandeau de Marsac, and M. R. Schaefer. 1997. A role for cpeYZ in cyanobacterial phycoerythrin biosynthesis. J. Bacteriol. 179:998-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehoe, D. M., and A. R. Grossman. 1996. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273:1409-1412. [DOI] [PubMed] [Google Scholar]
- 26.Kehoe, D. M., and A. R. Grossman. 1997. New classes of mutants in complementary chromatic adaptation provide evidence for a novel four-step phosphorelay system. J. Bacteriol. 179:3914-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kehoe, D. M., and A. R. Grossman. 1998. Use of molecular genetics to investigate complementary chromatic adaptation: advances in transformation and complementation. Methods Enzymol. 297:279-290. [Google Scholar]
- 28.Kieber, J. J., M. Rothenberg, G. Roman, K. A. Feldman, and J. R. Ecker. 1993. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72:427-441. [DOI] [PubMed] [Google Scholar]
- 29.Lomax, T. L., P. B. Conley, J. Schilling, and A. R. Grossman. 1987. Isolation and characterization of light-regulated phycobilisome linker polypeptide genes and their transcription as a polycistronic mRNA. J. Bacteriol. 169:2675-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda, T., S. M. Wurgler-Murphy, and H. Saito. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242-245. [DOI] [PubMed] [Google Scholar]
- 31.Mazel, D., G. Guglielmi, J. Houmard, W. Sidler, D. A. Bryant, and N. Tandeau de Marsac. 1986. Green light induces transcription of the phycoerythrin operon in the cyanobacterium Calothrix 7601. Nucleic Acids Res. 14:8279-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazel, D., J. Houmard, A. M. Castets, and N. Tandeau de Marsac. 1990. Highly repetitive DNA sequences in cyanobacterial genomes. J. Bacteriol. 172:2755-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oelmüller, R., A. R. Grossman, and W. R. Briggs. 1988. Photoreversibility of the effect of red and green light pulses on the accumulation in darkness of mRNAs coding for phycocyanin and phycoerythrin in Fremyella diplosiphon. Plant Physiol. 88:1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oelmüller, R., P. B. Conley, N. Federspiel, W. R. Briggs, and A. R. Grossman. 1988. Changes in accumulation and synthesis of transcripts encoding phycobilisome components during acclimation of Fremyella diplosiphon to different light qualities. Plant Physiol. 88:1077-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oelmüller, R., A. R. Grossman, and W. R. Briggs. 1989. Role of protein synthesis in regulation of phycobiliprotein mRNA abundance by light quality in Fremyella diplosiphon. Plant Physiol. 90:1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohto, C., C. Ishida, H. Nakane, M. Muramatsu, T. Nishino, and S. Obata. 1999. A thermophilic cyanobacterium Synechococcus elongatus has three different class I prenyltransferase genes. Plant Mol. Biol. 40:307-321. [DOI] [PubMed] [Google Scholar]
- 37.Posas, R., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG-1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two component” osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Schaefer, M. R., G. G. Chiang, J. G. Cobley, and A. R. Grossman. 1993. Plasmids from two morphologically distinct cyanobacterial strains share a novel replication origin. J. Bacteriol. 175:5701-5705. [DOI] [PMC free article] [PubMed]
- 40.Schmidt-Goff, C. M., and N. A. Federspiel. 1993. In vivo and in vitro footprinting of a light-regulated promoter in the cyanobacterium Fremyella diplosiphon. J. Bacteriol. 175:1806-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi, L., M. Potts, and P. J. Kennelly. 1998. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 22:229-253. [DOI] [PubMed] [Google Scholar]
- 42.Sobczyk, A., G. Schyns, N. Tandeau de Marsac, and J. Houmard. 1993. Transduction of the light signal during complementary chromatic adaptation in the cyanobacterium Calothrix sp. PCC 7601: DNA-binding proteins and modulation by phosphorylation. EMBO J. 12:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tandeau de Marsac, N. 1977. Occurrence and nature of chromatic adaptation in cyanobacteria. J. Bacteriol. 130:82-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tandeau de Marsac, N. 1983. Phycobilisomes and complementary adaptation in cyanobacteria. Bull. Inst. Pasteur 81:201-254. [Google Scholar]
- 45.Tomioka, N., K. Shinozaki, and M. Sugiura. 1981. Molecular cloning and characterization of ribosomal RNA genes from a blue-green alga, Anacystis nidulans. Mol. Gen. Genet. 184:359-363. [DOI] [PubMed] [Google Scholar]
- 46.Vogelmann, T. C., and J. Scheibe. 1978. Action spectrum for chromatic adaptation in the blue-green alga Fremyella diplosiphon. Planta 143:233-239. [DOI] [PubMed] [Google Scholar]