Abstract
engE, coding for endoglucanase E, one of the three major subunits of the Clostridium cellulovorans cellulosome, has been cloned and sequenced (Y. Tamaru and R. H. Doi, J. Bacteriol. 181:3270-3276, 1999). The N-terminal-half region of EngE possesses three repeated surface layer homology (SLH) domains, which are homologous to those of some bacterial S-layer proteins. Also, the C-terminal-half region consists of a catalytic domain of glycosyl hydrolase family 5 and a duplicated sequence (dockerin) for binding EngE to scaffolding protein CbpA. Our hypothesis is that the SLH domains serve in the role of anchoring to the cell surface. This model was investigated by using recombinant EngEs (rEngE) with and without SLH domains that were synthesized in Escherichia coli and cell wall preparations from C. cellulovorans. When rEngE and SLH polypeptides of EngE were incubated with cell wall fragments prepared by sodium dodecyl sulfate treatment, these proteins bound strongly to the cell wall. However, rEngEs without SLH domains lost their ability to bind to cell walls. When rEngE was incubated with mini-CbpA, consisting of two cohesin domains, and cell wall fragments, the mini-CbpA was able to bind to the cell wall with rEngE. However, the binding of mini-CbpA was dramatically inhibited by addition of a chelating reagent, such as EDTA, which prevents cohesin-dockerin interactions. These results suggest not only that the SLH domains of EngE can bind to the cell surface but also that EngE plays an anchoring role for cellulosomes through the interaction of its dockerin domain with a CbpA cohesin.
Clostridium cellulovorans is a mesophilic, anaerobic, spore-forming bacterium which stains as gram negative and which utilizes not only cellulose but also xylan, pectin, and several other carbon sources (5, 27). C. cellulovorans produces an extracellular cellulolytic multienzyme complex, which has been called the cellulosome (1, 5), that has a total molecular weight of about 1,000,000 and that is capable of hydrolyzing crystalline cellulose (5). The cellulosome of C. cellulovorans comprises three major subunits, scaffolding protein CbpA (26), exoglucanase ExgS (14), and endoglucanase EngE (29). In addition, the cellulosome contains several other enzyme subunits. We recently cloned and sequenced several cellulosomal subunits (5). One of these subunits, EngE, had a unique structure on the basis of the derived amino acid sequence (29), i.e., the N-terminal-half region of EngE possessed three repeated-sequence domains which were partially homologous to those of bacterial S-layer proteins. Also, the C-terminal-half region consisted of a catalytic domain of glycosyl hydrolase family 5 and a duplicated sequence (dockerin) for binding to scaffolding protein CbpA.
Specifically, the bacterial surface layer homology (SLH) domains appeared to have the important role of mediating binding between extracellular proteins and the cell surface. Several results showing cell wall association of SLH domains in vivo and in vitro were reported recently; e.g., a mutant having an SLH-defective protein of the S-layer (SlpA) in Thermus thermophilus lost its ability to bind to cell wall components such as peptidoglycan (18). Lemaire et al. reported that recombinant proteins containing SLH domains of C. thermocellum outer layer protein OlpB and S-layer protein SlpA could be bound to cell wall preparations (12, 13). Therefore, the interesting SLH domain structure of EngE suggests not only that EngE binds to scaffolding protein CbpA through its dockerin but also that it may bind to the cell surface through its SLH domains. To investigate this hypothesis, we carried out binding assays using recombinant EngEs (rEngEs) and mini-CbpA, consisting of a cellulose binding domain, one hydrophilic domain, and two cohesin domains of CbpA, synthesized in Escherichia coli, and cell wall preparations from C. cellulovorans.
In this paper, we provide evidence that the SLH domains of EngE are necessary to anchor EngE to the C. cellulovorans cell surface and are able to anchor mini-CbpA to the cell surface through its cohesin-dockerin interaction. This is the first report that indicates that a mesophilic cellulosomal enzyme may be able to anchor the cellulosome to the cell surface.
MATERIALS AND METHODS
Bacterial strains and media.
C. cellulovorans ATCC 35296 (27) was used as the source of chromosomal DNA and cell wall preparations. C. cellulovorans was grown anaerobically at 37°C in serum bottles containing a previously described medium (27) which included 5 g of cellobiose (Sigma) per liter. E. coli Novablue and BL21(DE3) (Novagen) were used as cloning hosts for production of recombinant proteins and were grown at 30 or 37°C in Luria-Bertani medium containing ampicillin (100 μg/ml; Sigma).
Construction of pENGE, pSLHE, pDSLHE, and pEMCBP.
Four fragments containing 3,014, 1,470, 1,629, and 1,665 bp were amplified by PCR to create expression plasmids pENGE, pSLHE, pDSLHE, and pMCBP, respectively. The forward and reverse primers for pENGE, pSLHE, pDSLHE, and pEMCBP were designed to carry artificial restriction enzyme sites of NcoI and XhoI, respectively (Table 1). The amplified fragments were inserted between the NcoI and XhoI sites of pET22b (Novagen). The inserted fragments were sequenced to verify the lack of mutations caused by PCR. These expression plasmids were designed to allow fusion proteins at the N-terminal end with the PelA signal peptide and at the C-terminal end with the six-histidine tag from the vector.
TABLE 1.
Designs of primers used in this study
| Primer | Sequence (5"-3")a | Plasmid(s) | Direction | Positionb |
|---|---|---|---|---|
| EE-FA1 | CATGCCATGGCAGAAGCTAACTACACAACAAAAGGC | pENGE, pSLHE | Forward | 32-40 |
| EE-R1 | GGCTCGAGTATTGCTTTTTTTAAGAATGCAAG | pENGE, pDSLHE | Reverse | 1023-1030 |
| SLH-R3 | CCGCTCGAGTAAATCCATAGCAGAAAGACCTCT | pSLHE | Reverse | 508-515 |
| EE-F2 | CCATGCCATGGCACCAGAATTCCCTGCT | pDSLHE | Forward | 494-499 |
| MCBP-F | GTATACCATGGCAGCG | pEMCBP | Forward | 28-29 |
| MCBP-B | GGCTCGAGTATAGGATCTCCAAT | pEMCBP | Reverse | 572-578 |
Expression and purification of the recombinant proteins.
Four recombinant proteins were purified from each of the E. coli BL21 strains harboring expression plasmids. When the cultures reached an optical density at 600 nm of 0.5, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration 0.5 mM, and the cells were further cultivated at 30°C for 3 h. After cells were collected by centrifugation, they were resuspended in buffer 1 (50 mM phosphate, 300 mM NaCl, 10 mM imidazole, pH 8.0) and disrupted by sonication. Each of the cell extracts was applied to a Ni-nitrilotriacetic acid agarose affinity column (Qiagen). The recombinant proteins were eluted by buffer 1 with 250 mM imidazole, and the collected proteins were dialyzed against 10 mM phosphate buffer (pH 7.0). Each of dialyzed protein preparations was concentrated to 1.2 to 1.8 mg/ml by ultrafiltration (Ultra free biomax-30; Millipore).
Preparation of cell wall fragments of C. cellulovorans.
C. cellulovorans cells from 500 ml of a mid-exponential-phase culture were harvested by centrifugation at 12,100 × g for 10 min and were washed twice with 100 ml of 50 mM phosphate buffer (pH 7.0) and resuspended with 10 ml of the same buffer. An aliquot of the cell suspension was labeled as the whole-cell fraction (fraction F1). The cell suspension was disrupted by sonication, intact cells were removed by twice centrifuging at 1,940 × g for 5 min, and the suspensions were recentrifuged at 39,200 × g for 20 min. The resulting supernatant was labeled as the cell extract fraction (fraction F2), and the pellet, which was suspended in 5 ml of 50 mM phosphate buffer (pH 7.0), contained the crude cell wall fraction (fraction F3). Fraction F3 was treated with 1% sodium dodecyl sulfate (SDS) by heating at 100°C in a water bath for 20 min and was centrifuged at 16,000 × g for 20 min at room temperature. The supernatant consisted of the cell wall-associated proteins (fraction F4). The pellet was resuspended in 5 ml of 50 mM phosphate buffer (pH 7.0) after being washed three times with the phosphate buffer. This suspension consisted of cell wall fragments containing peptidoglycan and polysaccharide (fraction F5). To remove covalently bound cell wall polymers from the peptidoglycan layer, the method of Ries et al. (21) was used. Fraction F5 was treated with 48% hydrofluoric acid (HF) for 24 h at 4°C. After centrifugation at 16,000 × g for 20 min at 4°C, the pellet was washed three times with 50 mM phosphate buffer (pH 7.0).
Interaction of each recombinant protein with cell wall fragments of C. cellulovorans.
Binding experiments were carried out by incubating and cosedimenting each of the recombinant proteins with cell wall fragments (12). Each polypeptide (40 to 60 μg) was mixed with 100 μl of fraction F5 (75.1 μM diaminopimelate [DAPA]), and the reaction volume was brought to 300 μl with 50 mM phosphate buffer (pH 7.0). The reaction mixtures were incubated for 4 h at 30°C with shaking. For the binding test for mini-CbpA, after mini-CbpA and rEngE were preincubated for 1 h at 30°C, the cell wall fragments were added to the reaction mixture. The bound and free polypeptides were separated by centrifugation at 16,000 × g for 20 min at room temperature. The supernatant consisted of the soluble fraction. A wash fraction was obtained after washing the cell wall with 50 mM phosphate buffer (pH 7.0). The pellets, consisting of the insoluble cell wall fragments and attached proteins, were washed with the same buffer and then resuspended with 300 μl of the phosphate buffer. Each fraction was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
Analysis of the peptidoglycan of C. cellulovorans.
To determine the amino acids and the DAPA content in peptidoglycan, fraction F5 was analyzed. Fraction F5 was vacuum dried and hydrolyzed in 6 N HCl for 24 h at 110°C. Aliquots of the hydrolysate were analyzed by a Beckman 6300 amino acid analyzer.
SDS-PAGE and immunoblot analysis.
SDS-PAGE was performed on 12% polyacrylamide gels (10). Immunoblot analysis was performed by using anti-EngE antiserum. The procedure was done as described previously (29).
RESULTS AND DISCUSSION
Domain structure of EngE.
Analysis of the deduced amino acid sequence of EngE revealed a multidomain structure with a unique feature, i.e., a triplicated SLH domain at the N terminus, followed by a family 5 catalytic domain, a domain of unknown function, and a duplicated sequence (dockerin). The N-terminal half of EngE (residues 32 to 491) contained three highly conserved repeats of 163, 126, and 163 amino acids (29). Moreover, the latter half of the repeated region (residues 304 to 426) of EngE had high similarity to the N-terminal domain (residues 1 to 154) of S-layer protein RsaA from Caulobacter crescentus (21.1% identify and 69.5% similarity to the N-terminal region of RsaA). The N-terminal domain of RsaA was reported to be involved in cell surface anchoring (2, 29). Also, the N-terminal region of EngE (residues 63 to 272) possessed high similarity to the C-terminal region of RsaA (residues 784 to 907), which is involved in a secretion signal (2, 29). On the other hand, a comparison of the SLH domain of EngE with the SLH domains of several structural proteins from Clostridium thermocellum outer layer proteins OlpB, SlpA, and SdbA (7, 11, 13) and Bacillus anthracis S-layer proteins EA1 and Sap (6, 15) did not show significant homology. It was reported that SLH binding sites in different bacterial species are different and that these sites are not unique within the same species (4). We think that the homology between SLH domains may depend on the bacterial species and surface layer structures. Therefore, to determine whether or not the SLH domains of EngE have functions similar to those of RsaA, i.e., the functions of anchoring and targeting to the cell surface, we decided to test the binding ability of the SLH domains of EngE for C. cellulovorans cell wall preparation.
Localization of EngE in C. cellulovorans.
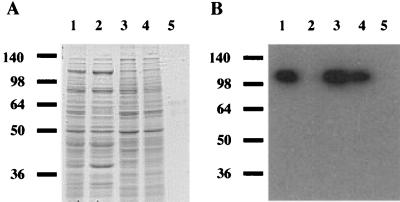
To identify the distribution of EngE in C. cellulovorans, we carried out immunoblot analyses using antiserum against EngE on prepared fractions. When fraction F3 was treated with SDS, many proteins associated with the cell wall were solubilized by the SDS (Fig. 1A). Such a treatment is able to remove the proteins that are noncovalently associated with the cell surface (16, 24). Thus, several proteins belonging to the C. cellulovorans cell envelope or present on the cell surface could be identified in fractions F3 and F4. On the other hand, no proteins were detected after Coomassie blue staining of the material extracted by treatment with SDS (Fig. 1A). Therefore, fraction F5 appears to consist of protein-free cell wall polymers containing peptidoglycan and polysaccharides. Furthermore, to know the proportion of components in the peptidoglycan, we analyzed the quantities of DAPA and amino acids in fraction F5. Alanine, glutamate, and DAPA were detected in hydrolysis products. The composition coincided with the peptidoglycan composition of clostridia (25). In immunoblot analysis using an EngE antibody, fractions F2 and F5 did not contain EngE; major bands, corresponding to 110 kDa, could be found in fractions F1, F3, and F4 (Fig. 1B). The signals were in good agreement with the molecular mass of EngE (29). These results indicated that EngE was associated with the C. cellulovorans cell envelope.
FIG. 1.
Localization of EngE on C. cellulovorans. (A) Coomassie blue-stained SDS gel; (B) immunoblot with anti-EngE antiserum on each cell wall fraction. Lanes 1, whole cells (fraction F1); lanes 2, cell extract (fraction F2); lanes 3, crude cell walls (fraction F3); lanes 4, cell wall proteins (fraction F4); lanes 5, SDS-extracted cell wall fragments (fraction F5). Molecular size markers (in kilodaltons) are shown at the left. Four and 10 μl of each fraction were used for immunoblot analysis and SDS-PAGE, respectively.
Binding of the SLH domains of EngE to the C. cellulovorans cell wall.
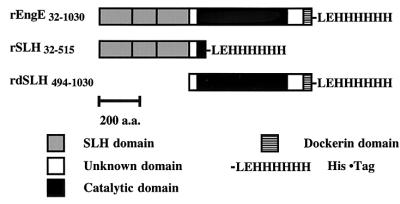
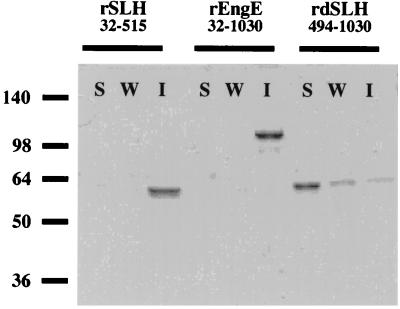
It was known that the SLH domains of several bacterial surface layer proteins had the ability to mediate attachment of proteins to cell walls (24). In addition, it was proposed that the attachment of SLH domains to cell walls in Bacillus stearothermophilus PV72/p2 and Thermoanaerobacterium thermosulfurigenes EM1 was mediated by secondary cell wall polysaccharides (3, 21). To appraise the interaction of the SLH domain of EngE with C. cellulovorans cell walls, we constructed rEngE with or without the SLH domains in E. coli. rEngE32-1030 and rSLH32-515, containing EngE SLH domains, and rdSLH494-1030, lacking EngE SLH domains, were constructed as fusion proteins with a six-histidine tag at the C terminus (Fig. 2). After purification by Ni2+ affinity chromatography, these recombinant proteins were tested for their ability to bind to the cell wall fraction, F5. As shown in Fig. 3, although rSLH32-515 and rEngE32-1030 with SLH domains were able to bind to the cell wall fragments, rdSLH494-1030 without its SLH domains lost the ability to bind to cell walls. These results strongly indicated that the SLH domains of EngE performed the role of anchoring to the C. cellulovorans cell surface. In addition, when the cell wall fragments were treated with HF, the binding of rSLH32-515 and rEngE32-1030 was abolished (data not shown). Similar effects of HF have been reported (3, 4, 12). HF is able to release secondary polymers, such as material containing GlcNAc, ManNAc (N-acetylmannosamine), MurNAc (N-acetylmuramic acid), glucose, galactose, and DAPA, by cleavage of phosphodiester linkages from peptidoglycan layers (3, 21). Thus, the results suggest that accessory polymers, acting as the adhesion component in fraction F5, are needed for attachment of SLH domains of EngE to the C. cellulovorans cell surface.
FIG.2.
Schematic representation of the domain structure of rEngE. The three rEngEs (rEngE32-1030, rSLH32-515, and rdSLH494-1030) were expressed by the pET system in E. coli BL21. Plasmid construction is described in Materials and Methods. These recombinant proteins were purified with a Ni affinity column. a.a., amino acids.
FIG. 3.
Binding of SLH domains of EngE to C. cellulovorans cell wall fragments. Each purified rEngE was incubated with fraction F5. Any insoluble material was precipitated and washed as described in Materials and Methods. Lanes S, soluble fraction; lanes W, wash fraction; lanes I, insoluble fraction. Molecular size markers (in kilodaltons) are shown at the left.
Large glycohydrolase CAC3469, which has high homology to EngE (17), has been identified recently from the sequence analysis of the Clostridium acetobutylicum genome. Unlike EngE, CAC3469 has an additional cell adhesion domain, which corresponds to the unknown domain (residues 840 to 970) of EngE (17, 29). Moreover, rdSLH494-1030, included in the unknown domain, was able to bind slightly to fraction 5 (Fig. 3). Thus, it appears that the unknown domain of EngE could be contributing a little to binding to the cell envelope.
The SLH domains of EngE were also similar to four hydrophilic domains (HLDs) of scaffolding protein CbpA of C. cellulovorans (5, 29). Scaffolding proteins in several cellulolytic clostridia, such as CipA of C. thermocellum (8), CipC of Clostridium cellulolyticum (20), and CipA of Clostridium josui (9), also possess unknown domains called X (1, 20), which are very similar to the HLDs (SLHs) of C. cellulovorans CbpA (5). So far, the function of these HLD domains remains obscure. We have proposed that the four HLDs in scaffolding protein CbpA may also have a role in anchoring to the cell surface, especially since there is similarity between the HLDs of CbpA and the SLH domains of EngE (5, 29).
Anchoring of mini-CbpA to the cell surface by SLH and dockerin domains of EngE.
A duplicated sequence (dockerin; residues 978 to 1030) resides in the C-terminal region of EngE (29). The dockerins, consisting of 22-amino-acid repeats, are well conserved in cellulosomal subunits from C. cellulovorans and other Clostridium species (1, 5). It is also known that the interaction between cohesins of the scaffolding protein CbpA and dockerin domains of enzymatic subunits is necessary to form cellulosomes in C. cellulovorans (5, 28, 30). Thus, we proposed that, through an interaction between the dockerin of EngE and the cohesin of CbpA, the SLH domains of EngE could help CbpA attach to the cell surface (29) and in this process could bind the whole cellulosome to the cell surface.
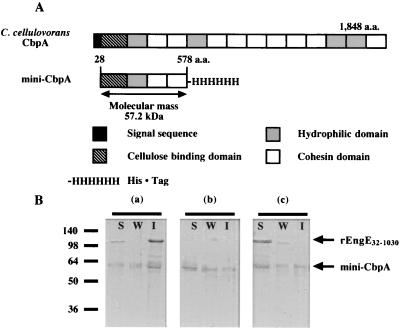
To test this hypothesis, we constructed a mini-CbpA that consisted of a cellulose binding domain, one HLD, and two cohesin domains (Fig. 4A). When rEngE32-1030 was incubated with mini-CbpA and fraction F5, the mini-CbpA was able to bind to the cell wall with rEngE32-1030 (Fig. 4B, a and b). The weak binding of mini-CbpA by itself to the cell wall (Fig. 4B, b and c) may be due to the single HLD present in mini-CbpA. However, the binding ability was dramatically inhibited by the addition of chelating reagent EDTA, which prevents the dockerin-cohesin interaction (Fig. 4B, c). It has been known that the dockerin domain of cellulosomal subunits has a putative calcium binding motif and that calcium ions are necessary for the binding between dockerin and cohesin domains of C. cellulovorans, C. cellulolyticum, and C. thermocellum (5, 19, 30). Therefore, these results indicated not only that the SLH domains of EngE can bind to the cell surface but also that EngE plays an anchoring role for cellulosomes through its interaction with the CbpA cohesins. The presence of EDTA influenced not only the cohesin-dockerin interaction but also indirectly the ability of rEngE32-1030 to bind to cell wall fragments (Fig. 4B, a and c).
FIG. 4.
(A) Schematic representation of C. cellulovorans CbpA and mini-CbpA. a.a., amino acids. (B) Binding of mini-CbpA to the cell surface by SLH domains of EngE. Purified rEngE32-1030 and mini-CbpA were incubated with fraction F5 and analyzed by SDS-PAGE. (a) Reaction of rEngE32-1030 and mini-CbpA; (b) reaction of mini-CbpA only; (c) reaction of rEngE32-1030 and mini-CbpA plus 20 mM EDTA. Any insoluble material was precipitated and washed as described in Materials and Methods. Lanes S, soluble fraction; lanes W, wash fraction; lanes I, insoluble fraction. Molecular size markers (in kilodaltons) are shown at the left.
Surface-associated proteins can be solubilized with agents that break hydrogen bounds (e.g., guanidine hydrochloride and urea) and with metal-chelating agents (e.g., EDTA and EGTA) (16, 24). When fraction F5 was treated with EDTA and subjected to the binding test, the cell wall binding ability of rEngE32-1030 was drastically decreased (data not shown). In Caulobacter crescentus, calcium ions were involved with the S-layer assembly process, which was disrupted by metal ion chelators, such as EGTA (31). Thus, the interaction between SLH domains of EngE and cell wall components may also require metal ions that are attached to the cell surface layer.
In C. thermocellum, several proteins mediating attachment to the cell surface were found from sequence analyses (7). The manner of anchoring cellulosomes to the cell surface has been known to involve several proteins, such as OlpB, ORF2p, and SdbA, which also possess repeated SLH domains and type II cohesin domains (7, 11). Although the type I cohesin domain in OlpA was unable to bind the C-terminal type II dockerin domain of CipA, the type II cohesin domains of OlpB, ORF2p, and SdbA were necessary to bind to the type II dockerin of CipA (11, 22, 23). The mechanism for anchoring mini-CbpA to the cell surface by interactions between cohesin and dockerin domains and the SLH domains of EngE is similar to that for CipA and OlpB in C. thermocellum. On the other hand, so far, the mechanism for anchoring the cellulosome to the cell surface in mesophilic clostridia such as C. cellulovorans, C. cellulolyticum, and C. josui has been unclear. Interestingly, mini-CbpAs were able to bind to cell wall fragments not only through the cohesin-dockerin interaction but also slightly by themselves (Fig. 4B, b). Therefore, C. cellulovorans may possess several systems for anchoring the cellulosomes to the cell surface: (i) through the SLHs of EngE; (ii) through the interaction by the HLDs (SLHs) in CbpA, and (iii) through anchoring proteins such as C. thermocellum OlpB and SdbA. Further studies will be required for us to clearly understand the mechanisms for anchoring cellulosomes to the C. cellulovorans cell surface.
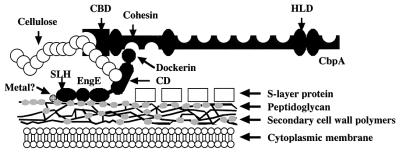
In Fig. 5, we present an updated anchoring model for cell surface attachment of scaffolding protein CbpA through the EngE molecule. Note that EngE binds to CbpA through the cohesin-dockerin interaction and to the cell surface via its SLHs.
FIG. 5.
Model showing attachment of EngE to the CbpA of C. cellulovorans and the cell surface. The three repeated SLH domains of the N terminus of EngE integrate into the cell wall layer containing peptidoglycan, while the C terminus of EngE is bound to CbpA through its dockerin domain. The question mark indicates that some interactions between SLH domains of EngE and cell wall containing peptidoglycan may be bridged by divalent ions but the data are not conclusive. CBD, cellulose binding domain; CD, catalytic domain.
Acknowledgments
This research was supported in part by grant DE-DDF03-92ER20069 from the U.S. Department of Energy.
REFERENCES
- 1.Bayer, E. A., L. J. W. Shimon, Y. Shoham, and R. Lamed. 1998. Cellulosome--structure and ultrastructure. J. Struct. Biol. 124:221-234. [DOI] [PubMed] [Google Scholar]
- 2.Bingle, W., J. F. Nomellini, and J. Smit. 1997. Linker mutagenesis of the Caulobacter crescentus S-layer protein: toward a definition of an N-terminal anchoring region and a C-terminal secretion signal and the potential for heterologous protein secretion. J. Bacteriol. 179:601-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brechtel, E., and H. Bahl. 1999. In Thermoanaerobacterium thermosulfurigenes EM1 S-layer homology domains do not attach to peptidoglycan. J. Bacteriol. 181:5017-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chauvaux, S., M. Matuschek, and P. Béguin. 1999. Distinct affinity of binding sites for S-layer homologous domains in Clostridium thermocellum and Bacillus anthracis cell envelopes. J. Bacteriol. 181:2455-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi, R. H., and Y. Tamaru. 2001. The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity. Chem. Rec. 1:24-32. [DOI] [PubMed] [Google Scholar]
- 6.Etienne-Toumelin, I., J. C. Sirard, E. Duflot, M. Mock, and A. Fouet. 1995. Characterization of the Bacillus anthracis S-layer: cloning and sequencing of the structural gene. J. Bacteriol. 177:614-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujino, T., P. Béguin, and J.-P. Aubert. 1993. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J. Bacteriol. 175:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerngross, U. T., M. P. Romaniec, T. Kobayashi, N. S. Huskisson, and A. L. Demain. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 9.Kakiuchi, M., A. Isui, K. Suzuki, T. Fujino, E. Fujino, T. Kimura, S. Karita, K. Sakka, and K.Ohmiya. 1998. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 180:4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Leibovitz, E., and P. Béguin. 1996. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemaire, M., H. Ohayon, P. Gounon, T. Fujino, and P. Béguin. 1995. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177:2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemaire. M., I. Miras, P. Gounon, and P. Béguin. 1998. Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology 144:211-217. [DOI] [PubMed] [Google Scholar]
- 14.Liu, C.-C., and R. H. Doi. 1998. Properties of exgS, a gene for a major subunit of the Clostridium cellulovorans cellulosomes. Gene 211:39-47. [DOI] [PubMed] [Google Scholar]
- 15.Mesnage, S., E. Tosi-Couture, M. Mock, P. Gounon, and A. Fouet. 1997. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 23:1147-1155. [DOI] [PubMed] [Google Scholar]
- 16.Messner, P., and U. B. Sleytr. 1988. Separation and purification of S-layers from gram-positive and gram-negative bacteria, p. 97-104. In I. C. Hancock and I. R. Poxton (ed.), Bacterial cell surface techniques. John Wiley & Sons, New York, N.Y.
- 17.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olabarria, G., J. L. Carrascosa, M. A. de Pedro, and J. Berenguer. 1996. A conserved motif in S-layer proteins is involved in peptidoglycan binding in Thermus thermophilus. J. Bacteriol. 178:4765-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pages, S., A. Belaich, J. P. Belaich, E. Morag, R. Lamed, Y. Shoham, and E. A. Bayer. 1997. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29:517-527. [PubMed] [Google Scholar]
- 20.Pages, S., A. Belaich, H. P. Fierobe, C. Tardif, C. Gaudin, and J. P. Belaich. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ries, W., C. Hotzy, L. Schocher, U. B. Sleytr, and M. Sára. 1997. Evidence that the N-terminal part of the S-layer protein from Bacillus stearothermophilus PV72/p2 recognizes a secondary cell wall polymer. J. Bacteriol. 179:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salamitou, S., M. Lemaire, T. Fujino, H. Ohayon, P. Gounon, P. Béguin, and J. P. Aubert. 1994. Subcellular localization of Clostridium thermocellum ORF3p, a protein carrying a receptor for the docking sequence borne by the catalytic components of the cellulosome. J. Bacteriol. 176:2828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salamitou, S., O. Raynaud, M. Lemaire, M. Coughlan, P. Béguin, and J. P. Aubert. 1994. Recognition specificity of the duplicated segments present in Clostridium thermocellum endoglucanase CelD and in cellulosome-integrating protein CipA. J. Bacteriol. 176:2822-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sára, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoseyov, O., and R. H. Doi. 1990. Essential 170 kDa subunit for degradation of crystalline cellulose of Clostridium cellulovorans cellulase. Proc. Natl. Acad. Sci. USA 87:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sleat, R., R. A. Mah, and R. Robinson. 1984. Isolation and characterization of an anaerobic, cellulolytic bacterium. Clostridium cellulovorans sp. nov. Appl. Environ. Microbiol. 48:88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takagi, M., S. Hashida, M. A. Goldstain, and R. H. Doi. 1993. The hydrophobic repeated domain of the Clostridium cellulovorans cellulose-binding protein (CbpA) has specific interactions with endoglucanases. J. Bacteriol. 175:7119-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamaru, Y., and R. H. Doi. 1999. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit EngE. J. Bacteriol. 181:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamaru, Y., S. Karita, A. Ibrahim, H. Chan, and R. H. Doi. 2000. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 182:5906-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker, G. S., N. Karunaratne, N. Ravenscroft, and J. Smit. 1994. Characterization of mutants of Caulobacter crescentus defective in surface attachment of the paracrystalline surface layer. J. Bacteriol. 176:6312-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]