Abstract
Mycobacteria are thought to have either one or two rRNA operons per genome. All mycobacteria investigated to date have an operon, designated rrnA, located downstream from the murA gene. We report that Mycobacteriun fortuitum has a second rrn operon, designated rrnB, which is located downstream from the tyrS gene; tyrS is very close to the 3" end of a gene (3-mag) coding for 3-methylpurine-DNA-glycosylase. The second rrn operon of Mycobacterium smegmatis was shown to have a similar organization, namely, 5" 3-mag-tyrS-rrnB 3". The rrnB operon of M. fortuitum was found to have a single dedicated promoter. During exponential growth in a rich medium, the rrnB and rrnA operons were the major and minor contributors, respectively, to pre-rRNA synthesis. Genomic DNA was isolated from eight other fast-growing mycobacterial species. Samples were investigated by Southern blot analysis using probes for murA, tyrS, and 16S rRNA sequences. The results revealed that both rrnA and rrnB operons were present in each species. The results form the basis for a proposed new scheme for the classification of mycobacteria. The approach, which is phylogenetic in concept, is based on particular properties of the rrn operons of a cell, namely, the number per genome and a feature of 16S rRNA gene sequences.
Mycobacterium tuberculosis and Mycobacterium leprae are major human pathogens which have the ability to grow slowly within host cells. The mechanisms used by these pathogens to control their growth rates is not well understood. Cell growth is dependent on protein biosynthesis, which, in turn, requires the synthesis of new ribosomes. The rate-determining step in ribosome synthesis is thought to be the rate of synthesis of rRNA (4, 8). Therefore, there is a need to identify factors important to transcription of rRNA (rrn) operons (12, 14) and the ways in which these transcripts are processed to form mature rRNA (16, 17).
Traditionally, mycobacteria are classified as either slow growers or fast growers according to the time taken (less than 7 days for fast growers, longer than 7 days for slow growers) for colonies to appear on a solid medium (15, 37). The slow-growing pathogens M. tuberculosis (6, 18, 36) and M. leprae (20, 21, 32) have a single rrn operon per genome, whereas most fast growers have two rrn operons per genome (2, 9). The apparent correlation between growth rate and the number of rrn operons per genome provides a possible explanation for the division of mycobacteria into fast- and slow-growing species.
However, slow-growing and fast-growing mycobacteria are all close relatives, as judged by the relatedness (95% or more) of their 16S rRNA sequences (26, 29, 35), so that comparative studies provide a way of identifying highly conserved sequence elements important to the synthesis of mature rRNA (13). Furthermore, it is valuable to identify the gene immediately upstream from the rrn operon under investigation (11, 13). First, this upstream gene serves to identify the operon. Second, upstream gene sequences may provide elements dedicated to the expression of the rrn operon, as we have found for mycobacterial rrnA operons (11, 13). The comparative approach may also provide insight into the phylogeny of mycobacterial rRNA operons.
Previously, we have found that in all the mycobacteria that have been studied, one rrn operon is located downstream from the murA gene, which codes for UDP-N-acetylglucosamine 1-carboxyvinyltransferase (EC 2.5.1.7), which is important to cell wall synthesis (13). The second operon of a representative fast grower, Mycobacterium smegmatis, was found to be located downstream from the tyrS gene, which codes for tyrosyl-tRNA synthetase (11, 27). Transcription of rrnB was found to be regulated by a single dedicated promoter, whereas transcription of rrnA was found to be regulated by three promoters (11).
Because little is known about the rrn operons of fast growers other than M. smegmatis, the rrn operons of Mycobacterium fortuitum and eight other mycobacterial species were investigated. M. fortuitum is classed as a pathogenic fast grower (15) which has two rrn operons per genome (9). One operon is a member of the rrnA family (13). We now report the organization and upstream sequence of the second rrn operon and its relation to the rrnB operon of M. smegmatis. The organizations of rrn operons of eight other fast-growing species were examined by restriction enzyme digestion of genomic DNA and Southern blotting using specific probes to identify each of the two operons present and their upstream genes. Our findings also suggest that characteristic features of mycobacterial rrn operons provide a way of classifying mycobacteria.
MATERIALS AND METHODS
Materials.
The following items were obtained from Sigma: acrylamide, salts and buffers, RNase, and DNase. Radiolabeled nucleoside and deoyribonucleoside triphosphates, restriction enzymes, and nylon filters were obtained from Amersham International.
Löwenstein-Jensen medium and Dubos medium enriched with Tween-albumin were obtained from Difco.
Bacterial strains.
The following mycobacterial type strains were used: M. fortuitum ATCC 6841T; M. smegmatis CIPT 1330010T, NCTC 8159; Mycobacterium agri CIPT 1320001T; Mycobacterium alvei CIPT 103464T; Mycobacterium diernhoferi CIPT 1170001T; Mycobacterium mucogenicum ATCC 49650T; Mycobacterium peregrinum ATCC 14467T; Mycobacterium porcinum CIPT 1460001T; Mycobacterium senegalense ATCC 10956T; and Mycobacterium wolinskyi ATCC 700010T (5). Mycobacteria were grown in Löwensterin-Jensen medium and further subcultured in Dubos medium enriched with Tween-albumin. Bacterial strains were stored at −70°C.
Cloning and identification of the rrnB operon upstream regions.
Mycobacterial DNA was isolated by the method described by Garcia et al. (10). Standard methods have been applied to prepare the M. fortuitum ATCC 6841T and M. smegmatis NCTC 8159 minilibraries in pUC18. Mycobacterial DNA was digested with BamHI or PstI for M. fortuitum and M. smegmatis, respectively, and fragments ranging from 2 to 4 kbp were cloned (30). An internal 16S rRNA PCR-amplified fragment was used as a probe for hybridization as described previously (9). Positive clones were detected by using standard methods (30) for colony hydridization.
The sequence of an insert was obtained by first mapping restriction endonuclease sites and then subcloning appropriate fragments, which were sequenced using both manual and automatic methods. A search of the data bank (1) led to the identification of the genes upstream from the rrnB operon, and the 16S rRNA gene sequences were identified with entries for each mycobacterium under investigation.
Southern blot analysis.
Mycobacterial DNA (2 μg) was digested with BamHI. The digests were separated by electrophoresis on horizontal gel slabs containing 0.75% (wt/vol) agarose and then transferred to nylon membrane filters (Amersham). Three different probes were used sequentially on the same blot to identify 16S rRNA, murA, and tyrS sequences. After the first and second hybridization reactions, radioactive probe sequences hydrogen bonded to immobilized DNA were removed by “melting” the complexes; a boiling solution of 0.5% (wt/vol) sodium dodecyl sulfate was poured onto the membrane, as recommended by the supplier (Amersham International). Autoradiographs confirmed that all the radioactivity was removed from the membrane. The probe for 16S rRNA sequences was previously described by Domenech et al. (9). Probes for murA and tyrS sequences were obtained by amplifying M. fortuitum DNA sequences using PCR. The oligonucleotides used for murA were mur (5" CCG-TTC-CCA-GGA-TTC-CCA-ACT-G 3") and c-mur (5" CGC-GGT-CGA-TAT-GAA-ACA-CAT-CG 3") (see GenBank accession number AJ295297 positions 893 to 1192 [300 bp]). The oligonucleotides used for tyrS were trs (5" GTG-AAC-ACC-GCA-GAT-GCC-GAT-GTG 3") and c-trs (5" CCC-GCA-ATG-TGG-CGC-TTG-CCA-CG 3"). The target for the tyrS probe corresponds to positions 1389 to 1881 (493 bp) of the M. fortuitum tyrS sequence (GenBank accession number AJ296160). The cycling profile consisted of 30 cycles of 1 min at 94°C, 1 min at 65°C, and 1 min at 72°C, followed by a final incubation at 72°C for 5 min. Probes were labeled and purified and hybridizations were carried out under stringent conditions as described previously (10).
Isolation of total RNA.
Mycobacteria were cultured in Lemco broth containing 0.1% Tween 80. Exponential-phase cells were collected and resuspended in guanidinium buffer. Total RNA was isolated by procedures described previously (11). Further DNase treatment was performed for the RNase protection assays in order to obtain DNA-free RNA.
Identification of transcription starting points.
Both primer extension and RNase protection methods were used, as described previously (23). The primer extension studies were carried out using the oligodeoxyribonucleotide JY15 (5" CAC ACT ATT GAG TTC TC 3"), whose target site lies within the highly conserved CL2 motif; this motif has been found within the leader regions of all transcripts of mycobacterial rrn operons studied so far (13). The primer JY15 was end labeled with [γ-32P]ATP by T4 polynucleotide kinase, and primer extension was performed using avian myeloblastosis virus reverse transcriptase (11).
The RNase protection assay method requires previous preparation of a radiolabeled RNA probe complementary to the RNA transcript by transcription of a minigene corresponding to the region under analysis (14). Minigenes were synthesized by PCR amplification of the M. fortuitum rrnB upstream region by using oligonucleotides rpa4F (5" ACC-GGG-CTC-TGA-CCT-GCG-GAA-ACG-AC 3") and JY15T (JY15 containing the T7 promoter sequence) and of the M. smegmatis rrnB upstream region by using oligonucleotides rpa4 (5" ACC-GCG-TCT-GAC-CAG-GGA-AAA-TAG 3") and rpa2 (5" T7 promoter region + GGC-GGA-CAA-AAA-ACA-ACA-AAC 3").
Radiolabeled transcription products were obtained by using the Riboprobe kit (Promega) as specified by the manufacturer; this yielded probes of 238 nucleotides for M. fortuitum and 273 nucleotides for M. smegmatis. The products were purified on a polyacrylamide gel electrophoresis (6% [wt/vol] polyacrylamide) sequencing gel. The appropriate bands were excised and eluted as described previously (12).
Hybridization was carried out in PES buffer (22) with radiolabeled samples (2.5 × 105 cpm) and 20 μg of DNA-free RNA; the reaction products were digested with different amounts of an RNase cocktail (Ambion). Stock RNase cocktail comprises both RNase T1 (20 U/μl) and RNase A (1 μg/μl), and an optimum concentration (up to a 10-fold dilution) was established for use in the RNA protection assays.
Products of both primer extension and RNase protection assays were analyzed by electrophoresis through 6% (wt/vol) polyacrylamide-8 M urea sequencing gels. Radioactive products were located by autoradiography either at 20°C or at −70°C using an intensifying screen. The gels were calibrated with either HaeIII-digested φX174 DNA molecular size markers (Promega, Madison, Wis.) or the products of appropriate DNA-sequencing reactions using primer JY15.
Sequencing of double-stranded DNA.
DNA sequences were determined by the dideoxy chain termination procedure with [α-35S]dATP (30).
RESULTS
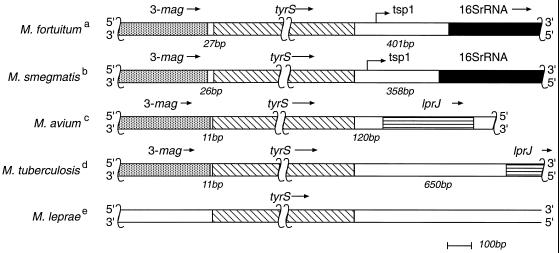
Previously, we reported that one rrn operon (rrnA) of M. fortuitum was located downstream from the murA gene (13). We now report that the second operon (rrnB) of this species is located downstream from a gene (tyrS) coding for a protein significantly similar to M. tuberculosis tyrosyl-tRNA-synthetase (protein Rv 1689) (6). The start codon for the tyrS gene was found to be 27 bp downstream from the stop codon of a gene (3-mag) coding for a protein significantly similar to M. tuberculosis 3-methylpurine-DNA-glycosylase (protein Rv 1688) (6). The short distance separating the coding regions of 3-mag and tyrS (Fig. 1) suggests that both genes are transcribed as a single unit. Further analysis of the tyrS gene of M. smegmatis revealed that, in this case too, tyrS was located close to a gene significantly similar to 3-mag; 27 bp was found to separate the coding regions of the two genes (Fig. 1). The 3-mag and tyrS genes are also found in close proximity in M. tuberculosis (6; http://genomic.sanger.ac.uk) and Mycobacterium avium (39) but not in M. leprae (7; http://genomic.sanger.ac.uk) (Fig. 1). However, M. leprae may be an exception because it is an atypical mycobacterium whose genome has suffered large deletions compared with M. tuberculosis and M. avium (7). Therefore, the location of tyrS immediately downstream from 3-mag is a common but not universal feature of the mycobacterial genome.
FIG. 1.
The immediate neighbors of the tyrS gene of M. fortuitum; comparison with other mycobacteria. The same scale is used throughout. Superscript letters: a, this study, GenBank accession number AJ296160; b, this study, GenBank accession numbers AJ312210 and AJ312211; c, TIGR database (http://www.tigr.org); d, Sanger Centre (http://genomic.sanger.ac.uk); e, Sanger Centre (http://genomic.sanger.ac.uk). tyrS, putative tyrosyl tRNA synthetase gene (M. tuberculosis Rv 1689); 3-mag, putative 3-methylpurine-DNA-glycosylase gene (M. tuberculosis Rv 1688); lprJ, putative membrane lipoprotein gene (M. tuberculosis Rv 1690). The arrows indicate the direction of gene transcription. tsp1, transcription starting point for the rrnB operon.
Identification of the transcription starting point.
The precursor rRNA (pre-rRNA) fraction comprises transcripts originating from the transcription starting points of both the rrnA and rrnB operons and their processing products. The composition of this fraction is governed by the steady states established between the rates of transcription and the rates of processing. The relative abundances of pre-rRNA transcripts of M. fortuitum growing exponentially with a generation time of 4 h were investigated by the primer extension procedure. Products extending to the 5" ends of transcripts were obtained by using an end-labeled radioactive deoxyribonucleotide primer with a binding site located within the highly conserved CL2 sequence motif (see below). Transcripts of both rrnA and rrnB operons were identified because the CL2 motif is common to both. Attempts to identify primers specific for either rrnA or rrnB were not successful.
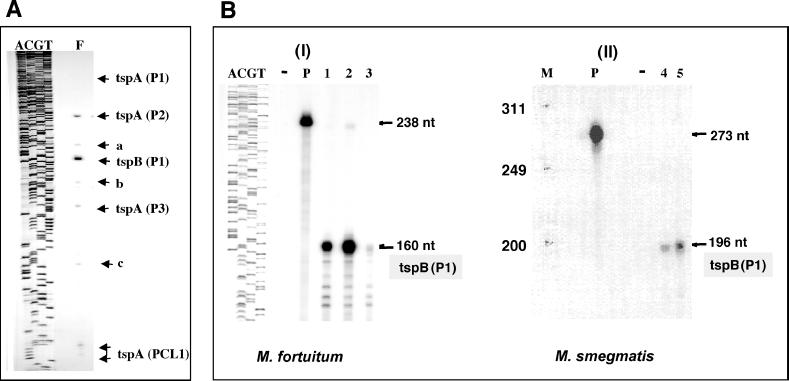
Eight products were identified (Fig. 2A). One product [tspA(P1)] was scarcely visible except on longer exposures of the photographic film (results not shown). The sizes of five products corresponded to the transcription starting points for putative promoters recognized by sigma 70-like initiation factors (13). Four products [tspA(P1), tspA(P2), tspA(P3), and tspA(PCL1)] were found to map to four putative promoters of the rrnA operon. A fifth product was found to map to a putative promoter of the rrnB operon. Similar products were identified previously (13).
FIG. 2.
Identification of transcription starting points. The RNA fractions of M. fortuitum growing exponentially with a generation time of 4 h and M. smegmatis growing with a doubling time of 2 h were investigated. The products of the primer extension and RNase protection assays were separated by electrophoresis through 6% (wt/vol) polyacrylamide-8 M urea gels. The radioactive products were revealed by autoradiography. (A) Products of the primer extension assay. ACGT denote DNA-sequencing reactions of rrnB sequences obtained with JY15 as the primer. The locations of the radioactive products are shown by the horizontal arrows. tspA(P1), etc., refer to the transcription starting points of potential promoters identified by their sequences. tspA denotes the rrnA operon, tspB denotes the rrnB operon, and PCL1 and (P1), etc., identify the promoter (see Fig. 3). The PCL1 promoter is identified by the highly conserved CL1 motif at its 3" end. a, b, and c are products which do not correspond to a classical promoter sequence and are thought to be products of pre-rRNA processing. (B) Products of RNase protection assays. ACGT is defined in panel A; P, probe; −, no-RNA control; 1, 2, 3, 4, and 5, different levels of RNases; M, HaeIII-digested φX174 DNA molecular size markers; nt, nucleotides; tspB(P1), product corresponding to the tsp for the P1 promoter of the rrnB operon. I, M. fortuitum; II, M. smegmatis.
The use of primers 32P labeled at their 5" ends ensures that the relative radioactivities of the products reflect their relative abundances because their radioactivities are directly proportional to the number of transcripts present irrespective of their lengths. The principal product, tspB(P1), was found to account for approximately 68% of the radioactivities of the products of all five promoters identified, revealing that the P1 promoter of rrnB gave rise to the majority of the pre-rRNA transcripts.
The remaining three products (a, b, and c), which do not map to known promoter sequences, are thought to be products of the pre-rRNA processing. However, the possibility that the products originated from unknown promoters cannot be excluded.
Confirmation that the product tspB(P1) originated from rrnB was obtained by the RNase protection procedure. Minigenes with promoters specific for T7 RNA polymerase were constructed using primers described in Materials and Methods. The target sites for these primers are identified in Fig. 3D. Radioactive transcripts, which were generated in vitro, were hybridized with the RNA fraction, the hybridization mixture was treated with RNase, and the products were separated by polyacrylamide gel electrophoresis (see Materials and Methods). Protected fragments of 160 and 196 nucleotides were expected for M. fortuitum and M. smegmatis, respectively, and principal products of the appropriate sizes were observed. The principal product for M. fortuitum was a radioactive fragment of 160 nucleotides (Fig. 2B, panel I). Parallel experiments were carried out with the RNA fraction of M. smegmatis, when the principal product was a radioactive fragment of 196 nucleotides (Fig. 2B, panel II). The locations of the transcription starting points of the rrnB operons are shown in Fig. 3.
FIG. 3.
Comparison of the intergenic domains of the rrnA and rrnB operons of M. fortuitum and M. smegmatis. (A and B) Comparison of the rrnA and rrnB operons of M. fortuitum (A) and M. smegmatis (B). The intergenic domains which extend from the 3" ends of the upstream genes to the 5" ends of the 16S rRNA genes are divided into promoter regions (PR) and core leader regions (core LR). The percent sequence similarities found between the promoter and core leader regions are shown beside double-headed arrows. Bent arrows labeled tsp1, etc., indicate transcription starting points for promoter P1, etc. CL1, conserved sequence motif found in rrnA operons; CL2, conserved sequence motif found in both rrnA and rrnB operons; stop 3", 3" end of the stop codon of the upstream gene indicated; 5" 16S rRNA, 5" end of the 16S rRNA gene. (C and D) Comparison of the sequences of the intergenic domains of the rrnA (C) and rrnB (D) operons of M. fortuitum and M. smegmatis. Dots represent identical nucleotides; dashes represent insertions-deletions. Termination codons are underlined. The CL1 motif, which is a feature of rrnA operons, and the CL2 motif are boxed (full lines). Broken lines enclose putative −10 and −35 boxes. Bold letters indicate the 5" end of the 16S rRNA coding region. rpa4F, rpa4, jy15t, and rpa2 indicate target sites for primers used in RNase ptotection experiments. Other symbols are defined in the legend to panels A and B.
The upstream gene-16S rRNA intergenic domains.
An intergenic domain is defined as extending from the 3" end of the upstream gene to the 5" end of the 16S rRNA gene. The intergenic domains of the four rrn operons of M. fortuitum and M. smegmatis are compared in Fig. 3. Each domain is conveniently divided into promoter and core leader regions. The promoter regions (213 to 275 bp) span one or more promoters and extend from the 3" ends of the stop codons of the upstream genes to the 5" ends of the conserved CL2 motifs. The core leader regions (164 to 176 bp) are common to all pre-rRNA transcripts originating from a particular promoter region, and they extend from the 5" ends of the CL2 motifs to the 5" ends of the 16S rRNA coding regions.
The lengths of the promoter regions of the rrnA operons reflect the number of promoters present (Fig. 3A to C). It was suggested (13) that the variability in the sizes of the promoter regions of rrnA operons has arisen through sequence duplications and that M. fortuitum has undergone one more event than M. smegmatis. The alignment (Fig. 3C) suggests that one M. fortuitum promoter, P3, may have arisen by duplication of the PCL1 promoter as judged by the location of its −35 box in relation to the M. smegmatis sequence.
The P1 promoter sequences are located within the murA gene, near the 3" ends. The promoter region of the rrnA operon of M. fortuitum comprises 275 bp and includes three promoters, P2, P3, and PCL1, corresponding to tspP2, tspP3, and tspPCL1, respectively. The distance between tspP1 and tspP2 was found to be 87 bp, compared with 76 bp for the distances between tspP2 and tspP3 and between tspP3 and tspPCL1. The promoter region of the rrnA operon of M. smegmatis (Fig. 3B) was found to comprise 211 bp and to include two promoters, P2 and PCL1, corresponding to tspP2 and tspPCL1, respectively (11). The distances between tspP1 and tspP2 and between tspP2 and tspPCL1 were found to be 99 and 76 bp, respectively.
The initiation of transcription is thought to involve the RNA polymerase holoenzyme binding to a 65- to 70-bp section of DNA extending from 45 to 50 bp upstream to 20 bp downstream from the tsp (19). The observed separation between tsps of the two rrnA operons of 76 to 99 bp is close to the minimum separation required (65 to 70 bp) for adjacent promoters to be capable of binding an RNA polymerase holoenzyme complex independently of one another. Thus, almost the entire promoter regions of the rrnA operons of M. smegmatis and M. fortuitum have the capacity to interact with RNA polymerase. In contrast, the promoter regions of the two rrnB operons comprise approximately 213 bp, but each has a single promoter (Fig. 3A, B, and D). The function of the ca. 140 bp not involved in binding to RNA polymerase remains to be identified.
Comparisons (including insertions-deletions) of the gene sequences of the intergenic domains of the four rrn operons were carried out to identify motifs important to the control of transcription and to the formation and processing of pre-rRNA. The results are summarized in Fig. 3. A frame of reference (Table 1) was first established by calculating the sequence identities (inferred protein sequences and gene sequences) between the upstream gene shown in Fig. 1 and between 16S rRNA genes sequences. When M. fortuitum sequences were taken as the standards (100%), the minimum number of sequence identities was found to be 68.4% for 3-MAG and 73.3% for TYRS in the inferred amino acid sequences and 58.7% for 3-mag, 56.9% for tyrS, and 95.4% for 16S rRNA in the gene sequences.
TABLE 1.
Comparison of gene sequences of M. fortuitum and other mycobacteriaa
| Species | % Similarity (% amino acid identity) to:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-mag
|
tyrS
|
16S rRNA
|
||||||||||||||
| M. fortuitum | M. smegmatis | M. avium | M. tuberculosis | M. leprae | M. fortuitum | M. smegmatis | M. avium | M. tuberculosis | M. leprae | M. fortuitum | M. smegmatis | M. avium | M. tuberculosis | M. leprae | ||
| M. fortuitum | 100 | 100 | 100 | |||||||||||||
| M. smegmatis | 58.7 (77) | 100 | 80.5 (86.8) | 100 | 98 | 100 | ||||||||||
| M. avium | 71.4 (68.4) | 55.6 (63.7) | 100 | 76.7 (76.8) | 63.2 (75.4) | 100 | 95.5 | 95.5 | 100 | |||||||
| M. tuberculosis | 63.1 (69) | 53.3 (79.8) | 75.8 (75) | 100 | 75.3 (79.4) | 64.6 (74.9) | 81.3 (86.7) | 100 | 96.1 | 95.9 | 98.5 | 100 | ||||
| M. leprae | NDb | ND | ND | ND | 100 | 56.9 (73.3) | 48.5 (71.3) | 60.19 (79.5) | 76 (82.2) | 100 | 95.4 | 95.5 | 98.5 | 98.3 | 100 | |
See Fig. 1.
ND, not determined.
Pairwise comparisons of the promoter regions of the four rrn operons led to the following values for sequence identities: rrnA and rrnB of M. fortuitum, 21% (Fig. 3A); rrnA and rrnB of M. smegmatis, 30% (Fig. 3B); rrnA of M. fortuitum and rrnA of M. smegmatis, 48% (Fig. 3C), which includes the conserved PCL1 motif and promoter elements); rrnB of M. fortuitum and rrnB of M. smegmatis, 67% (Fig. 3D). Thus, the promoter region of the rrnB operon of M. fortuitum is closer in sequence to the rrnB operon of M. smegmatis (67% identities) than to its own rrnA operon (21% identities). The identities found between promoter regions of the rrnB operons (67%) lie between 59 and 80% of those found between 3-mag and tyrS gene sequences, respectively, of M. fortuitum and M. smegmatis.
The core leader regions of the rrnA and rrnB operons of the same species (Fig. 3A and B) were found to be almost identical (98% identities), whereas sequence identities of 80% were found when operons of different species were compared (Fig. 3C and D). This high level of sequence conservation is thought to reflect the functional roles of the core leader regions in the transcription of the operon (13) and in the formation of secondary structures and in pre-rRNA processing (16, 17).
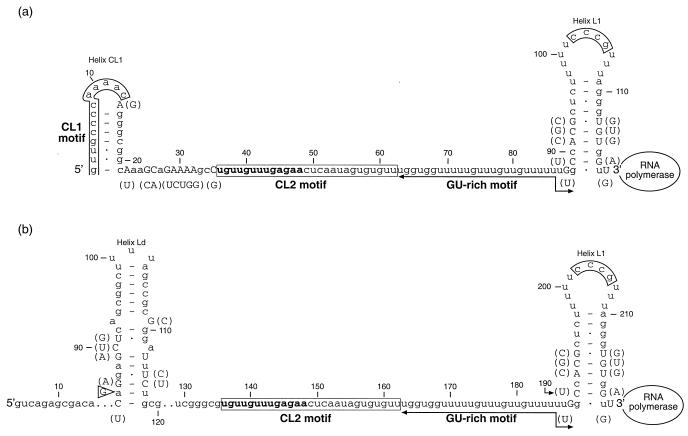
Features of pre-rRNA in the early stages of transcription of both rrnA and rrnB operons are illustrated in Fig. 4. The diagram represents partial pre-rRNA transcripts extending from tspCL1 (rrnA) or tspP1 (rrnB) to helix L1 of the core leader regions. The CL1 motif, which is part of the form of the PCL1 promoter implicated in the formation of an initiation complex with RNA polymerase, is a characteristic feature of rrnA operons. However, this motif is unlikely to be essential for either the regulation of transcription or pre-RNA processing because it is absent from transcripts of rrnB.
FIG. 4.
Comparison of features of the secondary structures of partial transcripts of M. fortuitum and M. smegmatis rrnA (A) and rrnB (B) operons. The sections presented extend from the transcription starting points of the PCL1 promoters (rrnA) and P1 promoters (rrnB) to the 3" ends of helices L1 of the core leader regions. The locations of RNA polymerase denote the extents of the transcription processes. The nucleotide positions are measured from the nearest transcription starting points (tspCL1 for rrnA and tspP1 for rrnB). The M. fortuitum sequences are presented. M. smegmatis sequences are shown as variations of M. fortuitum sequences. a, c, g, u, nucleotides present in both species; A, C, G, U, nucleotides present in M. fortuitum. (A), (C), (G), (U), nucleotides present in M. smegmatis. (A), etc., are placed either beside or below A, etc. <, deletion of the adjacent nucleotide from the M. fortuitum sequence; >, etc., insertion of the nucleotide shown relative to the M. fortuitum sequence. The CL1, CL2, and 5"-CCCG-3" motifs are boxed. The bold letters at the 5" end of the CL2 motif indicate the putative BoxA element.
Helix Ld (Fig. 4b) appears to be a feature of the two rrnB operons. The evidence for this stem-loop structure is based on three considerations. First, no alternative of equivalent stability was identified. Second, the stem-loop structures proposed for M. fortuitum and M. smegmatis have features in common. Both of the proposed stem-loop structures are stabilized by 16 A-U, G-C, and G-U base pairs. Ten base pairs are common to both species and four of the remaining six base pairs result from two compensating changes and four neutral changes arising from transitions (three C ↔ U and one A ↔ G). Third, indirect evidence for a helix involving positions 97 to 118 (Fig. 4b) was obtained from primer extension studies. A primer extension product specific for the M. smegmatis rrnB operon was sought by synthesizing a primer complementary to positions 97 to 118, but no product was obtained. It is inferred that the primer was not able to form a DNA-RNA complex because the primer could not compete successfully for the target sequence, which more readily formed a helix through interactions between residues 82 to 99 and residues 103 to 118.
The features of the core leader regions illustrated in Fig. 4 (namely, the CL2 motif, the UG-rich element extending from the 3" end of CL2 to the 5" end of helix L1, and helix L1) are all found in the 11 representative Mycobacterium species which have been studied (13); the core leader regions of Mycobacterium abscessus and Mycobacterium chelonae, two rapidly growing species each with a single rrn operon per genome, contain these features and no others (13). Hence, it is inferred that the sections of the core leader regions depicted in Fig. 4 span features essential for function. These features may play a role in preventing premature termination of transcription (antitermination), in forming pre-rRNA secondary structure, and in processing precursor-16S rRNA to yield mature 16S rRNA.
At least part, and possibly all, of the CL2 motif may be involved in the prevention of premature termination of transcription (antitermination). Antitermination in E. coli is thought to involve a sequence of 12 to 25 nucleotides of pre-rRNA, which resembles an antiterminator motif first identified in phage λ (3). Part of this sequence, termed BoxA, was reported as forming a complex with RNA polymerase through interaction with N-utilizing substance (NUSB) and ribosomal protein S10 (25) in the phage λ model. Sequences similar to the E. coli BoxA motif have been identified in a large number of rrn operons and so appear to have been conserved during bacterial evolution (3). The CL2 motif (16) is implicated in rrn antitermination because of its BoxA sequence at its 5" end (Fig. 4).
No specific function has been identified for the UG-rich element. However, the sequence comprising both the CL2 motif and the UG-rich element has the capacity to interact with sequences downstream from the 3" end of 16S rRNA to form a long-stem structure stabilized by more than 40 A-U, G-C, and G-U base pairs (16, 17). Helix L1 was identified by its location within the core leader region, by the presence of a 5"-(C/U)(C/U)(C/U)(G/A)-3" quartet in the loop region, and by compensating base changes in the stem (13). The function of helix L1 has not been established, and a role in transcriptional regulation cannot be excluded. Thus, the potential of the whole of the core leader region shown in Fig. 4 for regulating transcription remains to be investigated.
Locations of rrn operons in other fast-growing mycobacterial species.
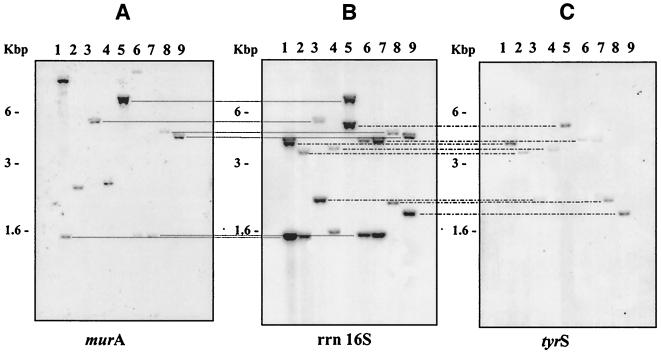
Both M. fortuitum and M. smegmatis were shown to have one rrn operon (rrnA) located downstream from murA and the second operon (rrnB) located downstream from tyrS. The possibility that the rrn operons were similarly located in other mycobacteria known to have two rrn operons per genome was investigated by Southern blotting. The enzyme BamHI cleaves Mycobacterium 16S rRNA genes at a site approximately 840 bp downstream from their 5" ends. Thus, a BamHI fragment which has this cleavage site at the 3" end will also contain sequences upstream from the 5" end of the 16S rRNA gene. The probability that a fragment will span both upstream gene and 16S rRNA sequences will depend on the location of the upstream BamHI site, as may be inferred from Fig. 1. Restriction enzyme digests of genomic DNA isolated from M. fortuitum and eight other mycobacterial species were separated by gel electrophoresis, and the fragments were transferred to membranes and hybridized with appropriate radiolabeled probes specific for 16S rRNA sequences, murA sequences, or tyrS sequences. The hybrids formed between immobilized DNA and the probes were identified by autoradiography (Fig. 5).
FIG. 5.
Identification of mycobacterial 16S rRNA gene sequences and their linkage with murA and tyrS genes. Mycobacterial DNA was digested with BamHI, and the fragments were separated by electrophoresis through 0.75% (wt/vol) agarose and transferred to a nylon membrane. 32P-labeled probes were used to identify murA (A), 16S rRNA gene (B), and tyrS (C) sequences. Fragment sizes are shown on the left. The mycobacterial DNA samples were isolated from M. agri (lane 1), M. alvei (lane 2), M. diernhoferi (lane 3), M. fortuitum (lane 4), M. mucogenicum (lane 5), M. peregrinum (lane 6), M. porcinum (lane 7), M. senegalense (lane 8), and M. wolinskyi (lane 9). The dotted lines link radioactive bands of identical sizes.
When BamHI fragments were probed with 32P-labeled 16S rRNA sequences, two radioactive bands were revealed in each digest (Fig. 5B), demonstrating that two rrn operons were present in the genomes of each of the species studied.
Three different hybridization patterns were observed when murA sequences were used as the probe (Fig. 5A). A single fragment, corresponding to one of the two fragments revealed by the probe for 16S rRNA sequences, was identified in five species: M. diernhoferi (lane 3), M. mucogenicum (lane 5), M. porcinum (lane 7), M. senegalense (lane 8) and M. wolinskyi (lane 9). Two fragments, one of which corresponds to one of the two fragments revealed by 16S rRNA sequences, were identified in two species: M. agri (lane 1) and M. peregrinum (lane 6). One fragment which does not correspond to either of the two fragments shown to contain 16S rRNA sequences was identified in the remaining two species: M. alvei (lane 2) and M. fortuitum (lane 4). M. fortuitum is known to have one rrn operon downstream from murA (13). The three hybridization patterns are explained by the positions of the 5"-end BamHI site in relation to the target sequence for the murA probe, namely, either upstream of the target sequence (first pattern, lanes 3, 5, 7, 8, and 9), within the target sequence (second pattern, lanes 1 and 6), or downstream from the target sequence (third pattern, lanes 2 and 4), as is known to be found in M. fortuitum.
The tyrS probe (Fig. 5C) identified one of the pair of fragments identified in Fig. 5B. Thus, in each of the species investigated, one particular fragment spanned both tyrS and 16S rRNA sequences. In each case, the 5"-end BamHI site lay upstream from the tyrS target sequence.
Ten mycobacterial species possessing two rrn operons per genome (M. fortuitum, M. smegmatis, and eight others) (Fig. 5) have been investigated. All 10 species were found to have one rrn operon (rrnB) located downstream from tyrS. An rrn operon (rrnA) is present in all mycobacteria examined to date (see, for example, reference 13), and M. alvei is unlikely to be an exception, even though direct evidence for this conclusion is lacking (Fig. 5A). We infer that the 10 species studied are representative of fast growers possessing two rrn operons per genome; each member of this class has one rrnA operon and one rrnB operon.
Two species, M. peregrinum and M. senegalense, are members of the M. fortuitum clade (15). Hence, it is likely that the other member of the clade, Mycobacterium farcinogenes, has rrnA and rrnB operons closely related to those of M. fortuitum. Another species investigated, M. diernhoferi, forms a two-membered clade with Mycobacterium neoaurum. The last-named species is known to have one operon that is a member of the rrnA family (13), and it is inferred that the second operon is a member of the rrnB family. The other species studied are regarded as single-member clades (15).
The information obtained from Fig. 5 for the organization of the rrn operons of these representative fast-growing species and their close relatives supports the notion that they are all members of a particular mycobacterial subgroup.
DISCUSSION
M. fortuitum has two rrn operons per genome. Previously, one operon (rrnA) was shown to be located downstream from murA (13). We have now shown that the second operon (rrnB) is located downstream from tyrS. The organization of the two rrn operons of M. fortuitum matches the organization of the two rrn operons of M. smegmatis (11). The intergenic domains of the rrnB operons of M. fortuitum and M. smegmatis were found to have more than 73% sequence homologies, and both rrnB operons were found to be regulated by a single dedicated promoter (Fig. 3D). In contrast, the intergenic domains of the rrnA of M. fortuitum and M. smegmatis operons have few sequence homologies (Fig. 3C). The region between the 3" end of the murA coding region and the 5" end of the conserved CL2 motif is termed the hypervariable multiple-promoter region (13). Southern blot analysis of eight other mycobacterial species, each with two rrn operons per genome, revealed that one operon (rrnA) was located downstream from murA and the other (rrnB) was located downstream from tyrS.
To date, all the mycobacterial species investigated have one rrn operon (rrnA) located downstream from murA; for species with two rrn operons per genome, the second operon is located downstream from tyrS. We infer that the duplication-deletion of an rrn operon is a rare event in the evolution of the genus. The rarity of the event suggests to us that it should be given prominence in describing the phylogeny of mycobacteria.
A particular feature of 16S rRNA sequences was also taken into account, namely, variation in the lengths of helix 18 (for a definition, see the scheme of secondary structure proposed by Kempsell et al. (18). As shown in Fig. 6, the gene sequences coding for the shorter and longer versions of Helix 18 differ by about 14 bp. The longer version of helix 18 was first considered to be a signature for slowly growing mycobacteria by Stahl and Urbance (35), and their proposal has been widely accepted (15, 26, 33).
FIG. 6.
Nucleotide sequences of helix 18: comparison of mycobacteria representing different classes. Features of the secondary structure of helix 18 (positions 435 to 488 of the M. tuberculosis sequence) are boxed. Dot, identical nucleotide; dash, deletion-insertion (the deletions giving rise to the shorter helix are enclosed by dotted lines); An overline denotes possible participation in nonstandard base pairing, e.g., between A and G; =A, nonpaired residue; Superscript letters: a, accession number X58890; b, accession number X52918; c, accession number X53999; d, accession number L08169; e, accession number X53896; f, accession number X52925; g, accession number X82235, identical to the M. chelonae (X82236) sequence; h, accession number X52933, identical to the M. alvei (AF023664), M. mucogenicum (X80771), M. farcinogenes (Y11581), M. peregrinum (AF130308), and M. senegalense (Y11582) sequences and others; i, accession number Y08453, identical to the M. diernhoferi (X5593) and M. wolinskyi (Y12871) sequences and others.
A search of 852,000 sequences (1) with the M. tuberculosis sequence (Fig. 6) identified several 16S rRNA sequences with scores of P ≤ 4 × 10−4, and all were mycobacterial species having the longer version of helix 18. When sequences of the shorter version of helix 18 were used to search the same gene bank, a different outcome was obtained. Identical (or nearly identical) sequences were identified in the 16S rRNA genes of members of other genera such as Arthrobacter, Dietzia, Geodermatophilus, Nocardioides, Rhodoccus, and Tsukamurella. Therefore, the shorter version of helix 18 is a motif that is not genus specific, perhaps because 32 of the 48 positions are highly conserved.
On the basis of our current knowledge, the above-mentioned criteria generate four classes of mycobacteria: I-l, species with one rrn operon (rrnA) per genome and a long helix 18 (for example, M. leprae [20, 21] and M. tuberculosis [6]); I-s, species with one rrn operon (rrnA) per genome and a short helix 18 (for example, M. abscessus and M. chelonae [9, 13]); II-l, species with two rrn operons per genome and a long helix 18 (for example, M. celatum [28] and M. terrae [24]); and II-s, species with two rrn operons (rrnA and rrnB) per genome and a short helix 18 (for example, the species reported in this study [Fig. 5], namely, M. agri, M. fortuitum, M. peregrinum, M. senegalense, M. diernhoferi, M. alvei, M. wolinskyi, M. mucogenicum, M. porcinum, and M. smegmatis). Additional classes may be needed if mycobacterial rrn operons are identified which have locations other than downstream from murA or tyrS.
A highlight of the proposed scheme is that the number of rrn operons per genome does not correlate with growth rate. For example, M. chelonae (class I-s), which is now known to have a single rrn operon per genome (9), is classified as a fast grower (15, 37), whereas M. terrae (class II-l), which has two rrn operons per genome (24), is classed as a slow grower. Furthermore, M. smegmatis (class II-s) mutants with a single functional rrn operon per genome (either rrnA or rrnB) can grow as fast as the wild type, which has two (rrnA and rrnB) functional operons (31).
The correlation between slow growth and a long helix 18 was first proposed by Stahl and Urbance (35). However, several species, namely, Mycobacterium simiae, Mycobacterium genavense, Mycobacterium intermedium, and Mycobacterium triviale, are classed as slow growers (see, for example, reference 15), but each has a short helix 18 (34). It is unlikely that the length of helix 18 has an effect on ribosome function, because this helix forms part of variable region 3 (V3) of the 16S rRNA (18), where substantial variations in length and sequence appear to be tolerated without adverse effects on ribosome function being reported.
In conclusion, the proposed scheme, which is based on the numbers and organizations of rrn operons within genomes, does not conflict with the phylogenetic tree based on 16S rRNA sequence homologies proposed by Goodfellow and Magee (15). Our proposal is based on new information and so provides greater precision in classification than was possible previously. The scheme is capable of further development as new information becomes available and so will lead to further insights into the phylogenetic relationships between mycobacteria.
Acknowledgments
We are grateful to V. Vincent, who kindly provided some of the mycobacterial type strains. We thank Simon A. Cox for help in the preparation of the manuscript.
This work is supported as part of the European Commission Science Research and Development Programme, contract ERBIC 18CT 9720253. M.J.G. received support from the Spanish Ministry of Health (FIS 00/0473E). J.A.G.-y-M. received support from COFAA, EDI, and CEGEPI-980660, IPN, Mexico, and CONACYT (grant 27576-M).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lippman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bercovier, H., O. Kafri, and S. Sela. 1986. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem. Biophys. Res. Commun. 136:1136-1141. [DOI] [PubMed] [Google Scholar]
- 3.Berg, K. L., C. Squires, and C. L. Squires. 1989. Ribosomal RNA operon antitermination: function of leader and spacer region BoxB-BoxA sequences and their conservation in diverse microorganisms. J. Mol. Biol. 209:345-348. [DOI] [PubMed] [Google Scholar]
- 4.Bremer, H., and P. P. Dennis. 1987. Modulation of chemical composition and other parameters of the cell growth rate, p. 1527-1542. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington D.C.
- 5.Brown, B. A., B. Springer, V. A. Steingrube, R. W. Wilson, G. E. Pfyffer, M. J. Garcia, et al. 1999. Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov., two new rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 49:1439-1511. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, et al. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 8.Colston, M. J., and R. A. Cox. 1999. Mycobacterial growth and dormancy, p. 198-219. In C. Ratledge and J. Dale (ed.), Mycobacteria: molecular biology and virulence. Blackwell Scientific Publications Ltd., Oxford, United Kingdom.
- 9.Domenech, P., M. C. Menendez, and M. J. Garcia. 1994. Restriction fragment length polymorphism of 16S rRNA genes in the differentiation of fast-growing mycobacterial species. FEMS Microbiol. Lett. 116:19-21. [DOI] [PubMed] [Google Scholar]
- 10.Garcia, M. J., C. Guilhot, R. Lathigra, M. C. Menendez, P. Domenech, C. Moreno, B. Gicquel, and C. Martin. 1994. Insertion sequence IS1137, a new IS3 family element from Mycobacterium smegmatis. Microbiology 140:2821-2828. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-y-Merchand, J. A., M. J. Colston, and R. A. Cox. 1996. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology 142:667-674. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-y-Merchand, J. A., M. J. Colston, and R. A. Cox. 1998. The role of multiple promoters in transcription of rDNA: the effects of growth conditions on precursor rRNA synthesis in mycobacteria. J. Bacteriol. 180:5756-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-y-Merchand, J. A., M. J. Garcia, S. Gonzalez-Rico, M. J. Colston, and R. A. Cox. 1997. Strategies used by pathogenic and nonpathogenic mycobacteria to synthesize rRNA. J. Bacteriol. 179:6949-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-y-Merchand, J. A., M. J. Colston, and R. A. Cox. 1999. Effects of growth conditions on expression of mycobacterial murA and tyrS genes and contributions of their transcripts to precursor rRNA synthesis. J. Bacteriol. 181:4617-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodfellow, M., and J. G. Magee. 1998. Taxonomy of mycobacteria, p. 1-75. In P. R. J. Gangadharam and P. A. Jenkins (ed.), Mycobacteria, vol. 1. Basic aspects. Chapman & Hall, London, United Kingdom. [Google Scholar]
- 16.Ji, Y.-E., M. J. Colston, and R. A. Cox. 1994. Nucleotide sequence and secondary structures of precursor 16S rRNA of slow-growing mycobacteria. Microbiology 140:123-132. [DOI] [PubMed] [Google Scholar]
- 17.Ji, Y.-E., M. J. Colston, and R. A. Cox. 1994. The ribosomal RNA (rrn) operons of fast-growing mycobacteria: primary and secondary structures and their relation to rrn operons of pathogenic slow-growers. Microbiology 140:2829-2840. [DOI] [PubMed] [Google Scholar]
- 18.Kempsell, K. E., Y.-E. Ji, I. C. E. Estrada, M. J. Colston, and R. A. Cox. 1992. The nucleotide sequence of the promoter, 16S rRNA and spacer region of the ribosomal RNA operon of Mycobacterium tuberculosis and comparison with Mycobacterium leprae precursor rRNA. J. Gen. Microbiol. 138:1717-1727. [DOI] [PubMed] [Google Scholar]
- 19.Krummel, B., and M. J. Chamberlin. 1989. RNA chain initiation by Escherichia coli RNA polymerase. Structural transitions of the enzyme in early ternary complexes. Biochemistry 28:7829-7842. [DOI] [PubMed] [Google Scholar]
- 20.Liesack, W., C. Pitulle, S. Sela, and E. Stackebrandt. 1990. Nucleotide sequence of the 16S rRNA from Mycobacterium leprae. Nucleic Acids Res. 18:5558.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liesack, W., S. Sela, H. Bercovier, C. Pitulle, and E. Stackebrandt. 1991. Complete nucleotide sequence of the Mycobacterium leprae 23S and 5S rRNA genes plus flanking regions and their potential in designing diagnostic oligonucleotide probes. FEBS Lett. 281:114-118. [DOI] [PubMed] [Google Scholar]
- 22.Mironov, V. N., M. Van Montagu, and D. Inzé. 1995. High throughput RNase protection assay. Nucleic Acids Res. 23:3359-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Movahedzadeh, F., J. A. Gonzalez-y-Merchand, and R. A. Cox. 2001. Transcription start-site mapping, p. 105-124. In T. Parish and N. G. Stoker (ed.), Mycobacterium tuberculosis protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 24.Ninet, B., M. Monod, S. Emler, J. Pawlowski, C. Metral, P. Bohner, R. Auckenthaler, and B. Hirschel. 1996. Two different 16S rRNA genes in a mycobacterial strain. J. Clin. Microbiol. 34:2531-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nodwell, J. R., and J. Greenblatt. 1993. Recognition of BoxA antitermination factors NUSB and ribosomal protein S10. Cell 72:261-268. [DOI] [PubMed] [Google Scholar]
- 26.Pitulle, C., M. Dorsch, J. Kazda, and E. Stackebrandt. 1992. Phylogeny of rapidly growing members of the genus Mycobacterium. Int. J. Syst. Bacteriol. 42:337-343. [DOI] [PubMed] [Google Scholar]
- 27.Predich, M., L. Doukhan, G. Nair, and I. Smith. 1995. Characterization of RNA polymerase and two sigma-factor genes from Mycobacterium smegmatis. Mol. Microbiol. 15:355-366. [DOI] [PubMed] [Google Scholar]
- 28.Reischl, U., K. Feldmann, L. Naumann, B. J. M. Gaugler, B. Ninet, B. Hirschel and S. Emler. 1998. 16S rRNA sequence diversity in Mycobacterium celatum strains caused by the presence of two different copies of 16S rRNA gene. J. Clin. Microbiol. 36:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogall, T., J. Wolters, T. Flohr, and E. C. Böttger. 1990. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int. J. Syst. Bacteriol. 40:323-330. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sander, P., T. Prammananan, and E. C. Böttger. 1996. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 22:841-848. [DOI] [PubMed] [Google Scholar]
- 32.Sela, S., and J. E. Clark-Curtis. 1991. Cloning and characterization of the Mycobacterium leprae putative ribosomal RNA promoter in Escherichia coli. Gene 98:123-127. [DOI] [PubMed] [Google Scholar]
- 33.Springer, B., E. C. Böttger, P. Kirschner, and R. J. Wallace. 1995. Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int. J. Syst. Bacteriol. 45:262-267. [DOI] [PubMed] [Google Scholar]
- 34.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Böttger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl, D. A., and J. W. Urbance. 1990. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J. Bacteriol. 172:116-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki, Y., A. Nagata, Y. Ono, and I. Yamada. 1988. Complete nucleotide sequence of the 16S rRNA gene of Mycobacterium bovis BCG. J. Bacteriol. 170:2886-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wayne, G. L., and G. P. Kubica. 1986. The mycobacteria, p. 1435-1457. In P. H. A. Sneath, N. S. Mair, and M. E. Sharpe (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md. [Google Scholar]