Abstract
The tet(L) gene of Bacillus subtilis confers low-level tetracycline (Tc) resistance. Previous work examining the >20-fold-inducible expression of tet(L) by Tc demonstrated a 12-fold translational induction. Here we show that the other component of tet(L) induction is at the level of mRNA stabilization. Addition of a subinhibitory concentration of Tc results in a two- to threefold increase in tet(L) mRNA stability. Using a plasmid-borne derivative of tet(L) with a large in-frame deletion of the coding sequence, the mechanism of Tc-induced stability was explored by measuring the decay of tet(L) mRNAs carrying specific mutations in the leader region. The results of these experiments, as well as experiments with a B. subtilis strain that is resistant to Tc due to a mutation in the ribosomal S10 protein, suggest different mechanisms for the effects of Tc on translation and on mRNA stability. The key role of the 5" end in determining mRNA stability was confirmed in these experiments. Surprisingly, the stability of several other B. subtilis mRNAs was also induced by Tc, which indicates that addition of Tc may result in a general stabilization of mRNA.
The control of bacterial mRNA decay has been shown to be dependent, in part, on events at or near the 5" end. In Escherichia coli, the 5"-end dependence of mRNA decay is thought to be due to RNase E, the major decay-initiating endoribonuclease, whose cleavage activity is strongly influenced by the phosphorylation state and accessibility of the 5" end (8, 21).
In Bacillus subtilis, an RNase E sequence homologue is not identifiable in the genome. Nevertheless, indirect evidence for B. subtilis RNase E activity has been obtained (10), and we have shown, using the erythromycin (Em) resistance gene ermC mRNA as a model, that access to the 5" end is critical for determining mRNA half life; Em-induced stalling of a ribosome near the 5" end of ermC mRNA results in stabilization of the message (3, 11). The specific set of codons in the ermC leader region coding sequence is required for Em-induced ribosome stalling and resultant ermC mRNA stabilization (17, 22). Other B. subtilis mRNAs have not been found to be stabilized by addition of Em.
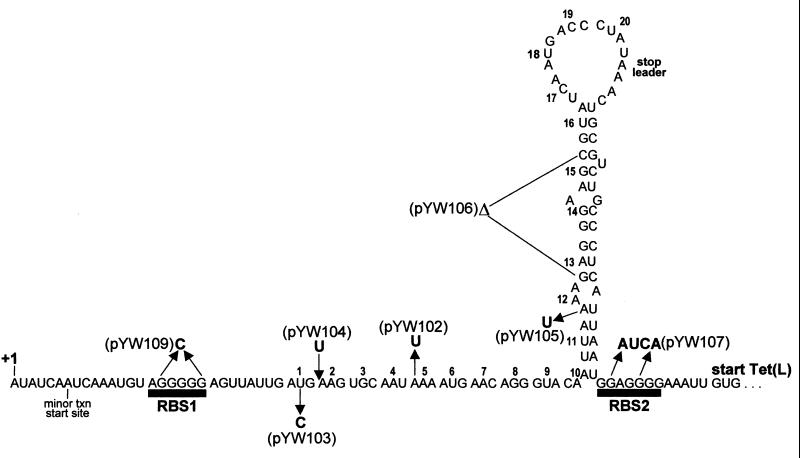
The B. subtilis chromosome contains a tetracycline (Tc) resistance gene (32), designated tet(L), encoding a multifunctional membrane protein that is a physiologically important monovalent cation/proton antiporter that catalyzes Na+(K+)/H+ antiport (18). Tet(L) protein is also capable of Tc/H+ exchange, and the presence of chromosomal tet(L) confers low-level resistance to Tc. Expression of tet(L) is inducible by Tc (7, 26), and inducibility is a function of the 124-nucleotide (nt) tet(L) leader region, which encodes a 20-amino-acid open reading frame (Fig. 1).
FIG. 1.
Nucleotide sequence of tet(L) leader region RNA. The sequence shown is from the major transcriptional start site (+1) to the start codon for the Tet(L) coding sequence. The minor transcriptional start site is indicated. Ribosome-binding sites RBS1 and RBS2 are shown, and the leader peptide codons are numbered 1 to 20. Mutations in the leader region are indicated alongside the name of the plasmid carrying each mutation. The Δ for pYW106 indicates a deletion of codons 13 to 15. The RNA structure shown is the putative structure that forms in the presence of Tc.
Using tet(L)-lacZ fusions containing various mutations in the tet(L) leader region, we developed a model for translational regulation that involves Tc-induced reinitiation of translation (27). Specifically, the translational signals for the Tet(L) coding sequence function poorly, so that basal-level translation is low. On the other hand, the translational signals for the tet(L) leader peptide function extremely efficiently, so that the leader peptide is translated constitutively at a high level. In the absence of Tc, ribosomes are continually translating the leader peptide, but this has either no effect or a negative effect on translation of the Tet(L) coding sequence. When a subinhibitory concentration (0.25 μg/ml) of Tc is added, Tc-bound ribosomes stall in the leader peptide coding sequence, allowing the leader region mRNA to assume a secondary structure (Fig. 1) that facilitates transfer of ribosomes from the leader peptide coding sequence to the Tet(L) coding sequence.
In the previous work, we measured a 12-fold translational induction, which represented only part of the total >20-fold induction by Tc. The results of tet(L)-lacZ transcriptional fusions suggested that induction of tet(L) gene expression included a component related to an increase in tet(L) mRNA.
In this report, we show that addition of a subinhibitory concentration of Tc to B. subtilis results in stabilization of tet(L) mRNA. The effect of Tc addition on the stability of various tet(L) RNAs containing leader region mutations was examined to probe the mechanism of Tc-induced stability. Addition of Tc also resulted in stabilization of several other cellular mRNAs.
MATERIALS AND METHODS
Bacterial strains.
The B. subtilis host was BD99, which is hisA1 thr-5 trpC2. The S10 mutant strain was isolated by Williams and Smith (31) after chemical mutagenesis of a thr-5 trpC2 parent strain and selection for Tc resistance. The preparation of B. subtilis growth media and competent B. subtilis cultures was done as described (12). Escherichia coli strain DH5α (16) was the host for plasmid constructions.
Plasmids.
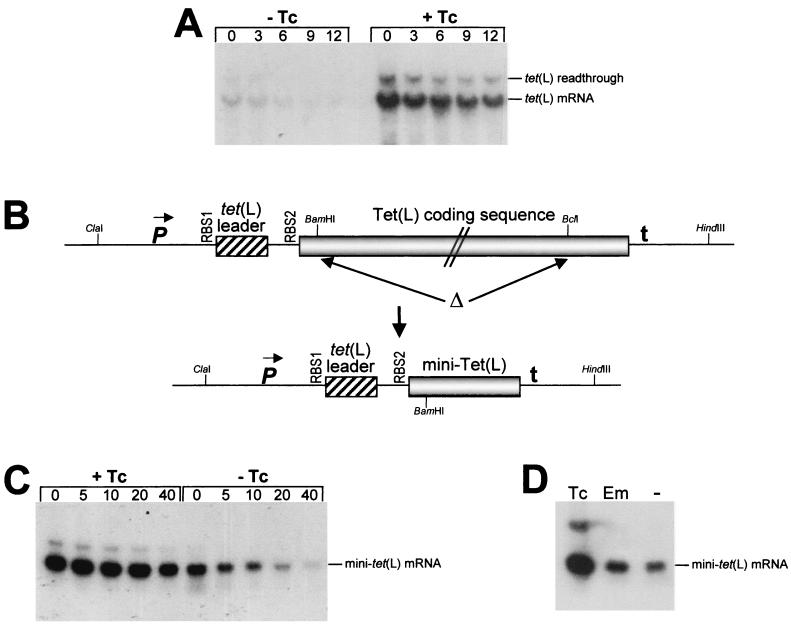
The B. subtilis tet(L) gene was cloned on a ClaI-HindIII fragment from pjbC1 (7) into the E. coli-B. subtilis shuttle vector pYH56 (6), replacing the ermC gene on pYH56. A BamHI site was introduced at the 12th codon of the Tet(L) coding sequence by site-directed mutagenesis (19), and a deletion of most of the Tet(L) coding sequence was accomplished by removing a 1.2-kb fragment, starting from this BamHI site to a BclI site located 105 bp from the end of the coding sequence (Fig. 2B). This gave plasmid pSS45. The hybrid BamHI/BclI site was converted to a BamHI site by ligating the following two PCR-generated fragments to HindIII- and ClaI-cleaved pSS45: from the upstream ClaI site to the BamHI site, and from the BamHI site to the downstream HindIII site. This gave plasmid pYW101, the wild-type mini-tet(L) plasmid.
FIG. 2.
Northern blot analysis of full-length and mini-tet(L) mRNAs. (A) Decay of wild-type tet(L) mRNA in the absence and presence of Tc. Times (minutes) after rifampin addition are indicated above each lane. Total RNA was separated on a formaldehyde-agarose gel. Bands representing tet(L) mRNA and a tet(L) transcriptional readthrough product that is detected by the tet(L) probe (7) are indicated on the right. (B) Schematic diagram of construction of the mini-tet(L) gene. The top line represents the full-length tet(L) gene, with the promoter (P), leader region, Tet(L) coding sequence (not drawn to scale), and transcription terminator (t). The region deleted to give the mini-tet(L) gene (bottom line) is indicated by Δ. Relevant restriction sites are shown (see Materials and Methods). (C) Northern blot analysis of mini-tet(L) mRNA isolated in the presence and absence of Tc at times (in minutes) after rifampin addition. (D) Northern blot analysis of steady-state mini-tet(L) mRNA in the presence of Tc, Em, or no addition (−).
Mutations in the mini-tet(L) leader region carried on plasmids pYW102 to pYW107 (Fig. 1) were constructed by PCR amplification of the tet(L) leader regions of plasmids containing these mutations in the context of the full-length tet(L) gene (27), using an upstream primer that had a ClaI site and a downstream primer that had a BamHI site. Plasmid pYW109 was constructed by ligating the following two PCR-generated fragments to HindIII- and ClaI-cleaved pYW101: from the upstream ClaI site to the BsrGI site at the mutated ribosome-binding site 1 (RBS1) [replacement of the poly(G) tract from RBS1 with a C residue leaves a BsrGI recognition site (TGTACA)], and from the BsrGI site to the downstream HindIII site. All new constructs were verified by sequence analysis, performed by the Mount Sinai Department of Human Genetics DNA sequencing facility.
RNA analysis.
B. subtilis strains were grown in TTM medium (7), which consists of 100 mM Tris-HCl buffer (pH 7.0), 1 mM potassium phosphate, 0.01% MgSO4, 0.2% (NH4)2SO4, 50 μg each of l-threonine, l-histidine, and l-tryptophan per ml, 0.1% yeast extract, and 50 mM Tris malate as the carbon source. This growth medium was used to avoid perturbations in tet(L) gene expression that result from the presence of higher concentrations of Na+ and K+ (7). When cultures reached 130 Klett units, Tc (0.25 μg/ml) was added for 15 min before addition of rifampin to 150 μg/ml.
RNA isolation and Northern blot analysis, using either formaldehyde-agarose gels or 6% denaturing polyacrylamide gels, were performed as described (29). Riboprobes were synthesized by T7 RNA polymerase transcription in the presence of [α-32P]UTP, using isolated PCR fragments as templates. PCRs were performed using a downstream primer that contained, at its 5" end, the T7 RNA polymerase promoter sequence (TAATACGACTCACTATA), allowing transcription of the PCR product.
The tet(L) probe was complementary to tet(L) sequences starting upstream of RBS1 to 30 nucleotides downstream of the BamHI site. Riboprobes for the polC, sigA, and veg mRNAs were generated in a similar fashion, using PCR primers complementary to sites in the respective coding sequences. Complete details of riboprobe synthesis will be provided upon request.
Radioactivity in the bands on Northern blots was quantitated using a PhosphorImager instrument (Molecular Dynamics). Half-lives were determined by linear regression analysis on semilogarithmic plots of percent RNA remaining versus time. Reverse transcriptase products were generated using reagents from the Gibco-BRL Superscript preamplification system.
RESULTS
Tc-induced stabilization of tet(L) mRNA.
We had previously observed that addition of a subinhibitory concentration of Tc (0.25 μg/ml) results in an increase in the steady-state concentration of tet(L) mRNA (7). To determine whether this was due to an increase in transcription or an increase in mRNA stability, Northern blot analysis was performed on total RNA isolated from a wild-type B. subtilis strain at various times after rifampin addition in the presence or absence of Tc (Fig. 2A). The half-lives (average of three experiments) were 7.1 min in the absence of Tc and 20.0 min in the presence of Tc; i.e., addition of Tc resulted in a 2.8-fold increase in tet(L) mRNA stability. Thus, the increase in stability, together with the increase in translational level (approximately 12-fold), accounts for the >20-fold increase in Tet(L) expression in response to Tc induction (7, 27). We conclude that the previously observed increase in the amount of tet(L) mRNA in the presence of Tc is due to mRNA stabilization.
We wished to test whether various leader region mutations affect Tc-induced mRNA stabilization. However, deletion of the tet(L) gene (30) and possibly other changes in tet(L) expression (7) lead to secondary mutations, which appear to be necessary for the fitness of the strain under standard laboratory conditions (30). Therefore, an examination of the effects of mutations in the tet(L) leader region is best performed in a strain containing the wild-type tet(L) gene. In order to assess the effects of leader region mutations on mRNA stability, a version of the tet(L) gene was constructed that had a large deletion in the Tet(L) coding sequence. This construct was carried on plasmid pYW101 and contained the tet(L) promoter region and complete leader region, followed by the first 12 codons of the Tet(L) coding sequence fused to the last 35 codons (Fig. 2B). Northern blot analysis of decay of mini-tet(L) mRNA showed a 2.5-fold induction of mRNA stability by Tc (Fig. 2C, Table 1), similar to that observed with the wild-type tet(L) mRNA. Thus, mini-tet(L) mRNA serves as a substitute for the full-length tet(L) mRNA in examining effects of leader region mutations on Tc-induced stability.
TABLE 1.
Quantitation of Northern blot analyses of mini-tet(L) mRNA decaya
| Plasmid | Description of leader region | Half-life (min)
|
Fold increase in half-life | |
|---|---|---|---|---|
| Without Tc | With Tc | |||
| pYW101 | Wild type | 17.2 ± 2.3 | 41.8 ± 2.9 | 2.5 |
| pYW107 | RBS2 mutation | 18.0 ± 7.8 | 34.0 ± 5.7 | 1.9b |
| pYW106 | Deletion of codons 13-15 | 15.5 ± 0.7 | 35.0 ± 7.1 | 2.3 |
| pYW104 | Stop at codon 2 | 20.5 ± 2.1 | 62.7 ± 6.3 | 3.0 |
| pYW102 | Stop at codon 5 | 27.3 ± 3.9 | 51.0 ± 12.7 | 1.9b |
| pYW105 | Stop at codon 12 | 22.5 ± 0.5 | 50.5 ± 9.1 | 2.2 |
| pYW103 | Mutated initiation codon | 7.4 ± 0.4 | 26.8 ± 9.0 | 3.6 |
| pYW109 | RBS1 mutation | 3.2 ± 0.2 | 3.6 ± 0.1 | 1.2b |
For all Northern blot analyses, data are from experiments in which the linear regression gave an R2 value of >0.90. Data are the averages of three experiments, except where indicated.
Average of two experiments.
We have shown previously that there are two transcriptional start sites for the tet(L) gene, situated 6 nucleotides apart (see Fig. 1). The level of transcription from the upstream start site is approximately fivefold greater than from the downstream start site (2). It was possible that the addition of Tc was influencing the site of transcription initiation, e.g., favoring the downstream start site, which would give a different 5" end that was less sensitive to decay. Therefore, reverse transcriptase analysis was performed on mini-tet(L) mRNA isolated in the presence and absence of Tc. No difference in the relative amounts of transcription from the two start sites was observed (data not shown).
We have observed a stabilizing effect of Em on ermC mRNA, which is caused by ribosome stalling in the ermC leader peptide coding sequence. To test for the specificity of Tc-induced stability, we compared the effects of Tc and Em addition. Northern blot analysis of steady-state mini-tet(L) mRNA isolated in the presence of either Tc or Em or no addition demonstrated that there was no effect of Em addition (Fig. 2D). This was confirmed by Northern blot analysis of mini-tet(L) isolated after rifampin addition in the presence and absence of Em (data not shown). In either case, the half-life of mini-tet(L) mRNA was between 16 and 20 min. Thus, stabilization of mini-tet(L) mRNA is specific for Tc.
tet(L) leader region mutations.
The nucleotide sequence of the tet(L) leader region, beginning with the major, upstream transcriptional start site, is shown in Fig. 1. The putative RNA secondary structure is the one predicted by our model for translational reinitiation. That is, in the absence of Tc, translation from RBS1 is at a very high level but translation from RBS2 is at a low basal level. When Tc is added, a Tc-bound ribosome stalls while translating a particular sequence of codons in the leader peptide coding sequence, resulting in assumption of the secondary structure shown and reinitiation of translation at a high level from RBS2 (27).
Since addition of Tc results in a large increase in Tet(L) translation, it was possible that the observed induced stability was due to increased ribosome density on the tet(L) message. To test this, a mini-tet(L) mRNA was constructed that carried a mutation in RBS2. The nucleotide changes shown for pYW107 in Fig. 1 have been found to almost completely abolish Tet(L) expression (27). However, Northern blot analysis of mini-tet(L) mRNA carrying the RBS2 mutation also showed Tc-induced stability, which was slightly less than that of the wild type (1.9-fold; Table 1). Therefore, the effect of Tc on mRNA stability is not due to an increase in Tet(L) translation.
We tested whether formation of the secondary structure shown in Fig. 1 was required for the induction of tet(L) mRNA stability, since it has been shown that 5"-proximal RNA secondary structure can influence the accessibility of a message to initiation of mRNA decay (4, 13). A deletion derivative of the leader region that lacked codons 13 to 15, which would disallow formation of the predicted secondary structure, was constructed. Northern blot analysis of this mutant, pYW106, showed that induction of stability by Tc was essentially wild type (Table 1). Therefore, the assumption of secondary structure is not relevant to the effect of Tc on mini-tet(L) stability.
We hypothesized that stalling of a ribosome in the leader peptide coding sequence could result in stabilization of mRNA, in much the same way as stalling of an Em-bound ribosome stabilizes ermC mRNA. To test this, mini-tet(L) mRNAs with nonsense codons introduced at the 2nd, 5th, and 12th positions of the leader peptide coding sequence were constructed (pYW104, pYW102, and pYW105, respectively), and their decay was analyzed. The results (Table 1) showed that addition of Tc could still induce stability in the mini-tet(L) mRNAs with premature leader stop codons.
The lack of any effect of mutating the second codon to a nonsense codon or deleting codons 13 to 15 is very different from what we observed previously when examining translational induction (27). These two mutations resulted in a complete loss of translational induction. Therefore, the mechanisms of Tc-induced mRNA stabilization and Tc-induced translation are distinct.
Since translation of the leader peptide coding sequence was shown not to be required for induced stability, we tested whether initiation of translation was required. The initiation codon for the leader peptide was changed from AUG to the nonfunctional ACG to give plasmid pYW103. Although the stability of mini-tet(L) mRNA encoded by pYW103 was significantly less than that of the wild type (7 versus 17 min), the addition of Tc resulted in a relatively greater increase in mRNA stability (3.6-fold; Table 1). Presumably, the shorter half-life of the uninduced mini-tet(L) mRNA magnifies the effect of Tc. Finally, the poly(G) tract of the RBS1 sequence was replaced with a C nucleotide (pYW109). Mini-tet(L) mRNA encoded by pYW109 was far less stable than the wild type (3 versus 17 min). Despite this short uninduced half-life, which would easily allow detection of an effect of Tc, no significant stabilizing effect was observed (Table 1). We conclude that ribosome binding at RBS1 but not initiation of leader peptide translation is required for Tc-induced tet(L) mRNA stability.
Induced stabilization of other mRNAs.
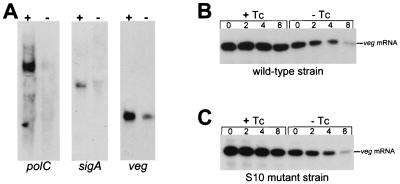
We asked whether Tc-induced mRNA stability was specific for tet(L) mRNA. Three chromosomally encoded messages were chosen for analysis: polC mRNA, a ca. 4.4-kb mRNA encoding the DNA polymerase III α-subunit; sigA mRNA, a ca. 1.2-kb mRNA encoding the RNA polymerase ςsgr;A factor; and veg mRNA, a ca. 0.3-kb mRNA encoding a protein of unknown function. The veg gene has been used as a model for gene expression in B. subtilis (20, 23). These three were chosen because they are transcribed constitutively and represent a wide range of mRNA sizes.
Northern blot analysis of RNA isolated from B. subtilis in the presence or absence of Tc, using riboprobes specific for these genes, showed a significant increase in the steady-state amount of all three mRNAs (Fig. 3A). The decay of veg mRNA after rifampin addition was analyzed, and a four- to sixfold increase in veg mRNA stability was observed in the presence of Tc (Fig. 3B). Thus, the stabilizing effect of Tc is not limited to tet(L) mRNA but appears to affect diverse mRNAs.
FIG. 3.
Effect of Tc on stability of other mRNAs. (A) Northern blot analysis of the indicated mRNAs isolated from a wild-type strain grown in the presence (+) or absence (−) of Tc. (B and C) Northern blot analysis of veg mRNA at times (in minutes) after rifampin addition, isolated from a wild-type strain or an S10 mutant strain grown in the presence and absence of Tc. Total RNA was separated on a 6% denaturing polyacrylamide gel.
The effect of Tc addition on veg mRNA was also analyzed in a strain of B. subtilis that has a mutation in the gene encoding ribosomal protein S10. This strain was originally isolated in a screen for Tc-resistant B. subtilis strains, following ethylmethane sulfonate mutagenesis (31). Ribosomes isolated from the S10 mutant strain were shown to be resistant to the effects of Tc on translation in vitro, and it was assumed that Tc does not bind well to ribosomes that contain the S10 mutant protein. If the effect of Tc on mRNA stability was through binding of Tc to ribosomes, we expected that this effect should not be observed in the S10 mutant strain. However, the results of Northern blot analysis of veg mRNA in the S10 mutant strain demonstrated a similar Tc-induced stabilization as in the wild type (Fig. 3C). Mini-tet(L) mRNA was also stabilized in the S10 mutant strain as in the wild type (data not shown). Thus, although the S10 mutation makes the ribosome resistant to the effect of Tc on translation (31), it does not influence the effect of Tc on mRNA stabilization. This suggests that Tc does, in fact, bind to the S10 mutant ribosomes, although it does not affect function.
DISCUSSION
The results reported here demonstrate an additional effect of Tc on gene expression in a gram-positive organism. It is well documented that binding of Tc to a ribosome inhibits translation by blocking the binding of aminoacyl-tRNA to the ribosomal A site (14, 15). We show here that another effect of Tc is to increase mRNA stability. Although we have not proven directly that the stabilizing effect of Tc is related to its binding to ribosomes, we infer this from the fact that inactivation of the RBS1 sequence eliminated the stabilizing effect of Tc.
Previous work on Em-induced stability of ermC mRNA has shown that initiation of mRNA decay in B. subtilis occurs primarily from the 5" end (3, 11). If the only effect of Tc were to inhibit elongation, one could suggest that the stabilizing effect of Tc is due to slowing of a ribosome by inhibiting the first elongation step after ternary complex formation, which would position the ribosome near the 5" end and block access of a decay-initiating RNase. However, data from leader region mutations of the mini-tet(L) mRNA demonstrated that the effect of Tc on mRNA stability was a function of the ribosome-binding site (RBS) alone; insertion of a stop codon immediately after the leader peptide initiation codon, and even inactivation of the initiation codon itself, did not eliminate the stabilizing effect of Tc, while deletion of RBS1 completely abolished this effect (Table 1).
We speculate that binding of Tc to a ribosome causes the ribosome to engage in a more stable interaction with the RBS, thus providing greater protection against initiation of decay from the 5" end. In this regard, it is interesting that an analysis of UV cross-linking of 16S rRNA nucleotides revealed three cross-links that are affected by addition of Tc (25). Two of these are at the top of helix 44, which lies close to the 3′-proximal helix 45 that is next to the anti-RBS sequence (24).
The results with Tc-induced stability of mini-tet(L) mRNA in the S10 mutant strain (Fig. 3C) are consistent with the stabilizing effect of Tc being independent of the effect of Tc on translation. In vitro studies of translation with ribosomes containing the mutant S10 protein demonstrated resistance to the inhibitory effect of Tc (31). Nevertheless, addition of Tc to the S10 mutant strain gave a similar mRNA stabilization as in the wild-type (Fig. 3C and 3D). This result is consistent with our hypothesis that Tc-induced stability is due to a Tc-induced alteration of the ribosome-RBS interaction, rather than through an effect on translation.
It was shown recently that binding of Tc to the Thermus thermophilus 30S ribosomal subunit did not involve proteins but, rather, specific residues of 16S rRNA (5). One of the proteins that lies nearest to the primary Tc binding site is the S10 protein, specifically the loop composed of residues 50 to 60 (5). We used PCR to amplify the rpsJ gene, which encodes the S10 protein, from B. subtilis wild-type and S10 mutant strains in order to determine the actual mutation site. A comparison of the sequences showed that there was a mutation in codon 46 of the rpsJ coding sequence gene, resulting in a Lys to Glu change. Thus, it is possible that this mutation confers resistance to the inhibitory effects on translation caused by Tc binding to nearby rRNA nucleotide residues, but does not alter the effect of Tc on mRNA stability. (It should be noted, however, that residues 45 to 56 of the B. subtilis S10 protein constitute a region of the protein that has the least similarity to the T. thermophilus S10 protein sequence, suggesting caution in making inferences about binding of Tc to B. subtilis ribosomes from the data on T. thermophilus ribosomes.)
Of particular interest was the fact that Tc-induced stability was observed even when translation of the body of the message was eliminated (RBS2 mutation on pYW107; Table 1). This reinforces the conclusion from our studies on induced ermC mRNA stabilization, which indicated a primary role of the 5" end in determining mRNA half-life, independent of translation of the coding sequence (3, 11). Furthermore, the insertion of a stop codon as 5"-proximal as the second codon of the leader peptide coding sequence had little effect on mini-tet(L) mRNA stability, with or without Tc induction. Only when the leader peptide initiation codon was mutated to a nonfunctional ACG start codon was there a significant effect on uninduced mini-tet(L) mRNA half-life (decrease from about 17 min to about 7 min).
It seems clear that the process of ribosome binding and initiation of translation, even without subsequent elongation, helps to confer wild-type stability on mRNA in B. subtilis. On the other hand, abolition of Tc induction was seen only when RBS1 was deleted, and this resulted in a further reduction of uninduced half-life to about 3 min. Thus, it appears that both RBS binding and initiation of translation contribute to basal mRNA stability, and binding of Tc enhances the effect of ribosome binding.
In earlier reports on the relationship of translation and mRNA decay in E. coli, various antibiotics were used to either freeze ribosomes on mRNA or inhibit initiation and allow ribosomes to run off. Tc, which stabilizes polysomes, was found to stabilize mRNAs (9, 28). However, in these studies the concentrations of antibiotics used could rapidly inhibit all protein synthesis (e.g., Tc at 100 μg/ml). As such, there is likely to be little relationship between those findings and the stabilization of B. subtilis mRNA induced by the subinhibitory concentration of Tc used in our experiments.
Since Tc-regulated transcription is currently used for controlled gene expression in many eukaryotic systems (1), we thought it important to demonstrate that the effect of Tc on mRNA stability that we observed in B. subtilis did not also occur in higher organisms. In fact, Northern blot analysis of β-actin mRNA from Saccharomyces cerevisiae cultures treated with Tc showed no difference in mRNA stability from untreated controls (unpublished results).
Presumably, binding of Tc to the bacterial ribosome is needed specifically for the stabilizing effect. We have not investigated whether this effect occurs in other gram-positive organisms. Nevertheless, it is interesting to speculate whether use of subinhibitory levels of Tc in clinical or agricultural settings might result in mRNA stabilization, thereby affecting cellular function.
Acknowledgments
This work was supported by Public Health Service grants GM-48804 and GM-52837 from the National Institutes of Health.
We acknowledge the contributions of S. J. Stasinopoulos, who constructed pSS45 and performed initial Northern blot analyses of tet(L) mRNA decay in the presence of Tc.
REFERENCES
- 1.Baron, U., and H. Bujard. 2000. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 327:401-421. [DOI] [PubMed] [Google Scholar]
- 2.Bechhofer, D. H., and S. J. Stasinopoulos. 1998. tetA(L) mutants of a tetracycline-sensitive strain of Bacillus subtilis with the polynucleotide phosphorylase gene deleted. J. Bacteriol. 180:3470-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechhofer, D. H., and K. H. Zen. 1989. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J. Bacteriol. 171:5803-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvet, P., and J. G. Belasco. 1992. Control of RNase E-mediated RNA degradation by 5"-terminal base pairing in E. coli. Nature 360:488-491. [DOI] [PubMed] [Google Scholar]
- 5.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, J., A. A. Guffanti, and T. A. Krulwich. 1997. A two-gene ABC-type transport system that extrudes Na+ is induced by ethanol or protonophore. Mol. Microbiol. 23:1107-1120. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, J., A. A. Guffanti, W. Wang, T. A. Krulwich, and D. H. Bechhofer. 1996. Chromosomal tetA(L) gene of Bacillus subtilis: Regulation of expression and physiology of a tetA(L) deletion strain. J. Bacteriol. 178:2853-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acids Res. Mol. Biol. 62:55-108. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, P. S., K. R. Lynch, M. L. Walsh, J. M. Hill, and H. L. Ennis. 1977. Use of antibiotics to determine ribosome-messenger RNA interactions necessary for in vivo stability of specific messengers. J. Mol. Biol. 114:569-573. [DOI] [PubMed] [Google Scholar]
- 10.Condon, C., H. Putzer, D. Luo, and M. Grunberg-Manago. 1997. Processing of the Bacillus subtilis thrS leader mRNA is RNase E-dependent in Escherichia coli. J. Mol. Biol. 268:235-242. [DOI] [PubMed] [Google Scholar]
- 11.DiMari, J. F., and D. H. Bechhofer. 1993. Initiation of mRNA decay in Bacillus subtilis. Mol. Microbiol. 7:705-717. [DOI] [PubMed] [Google Scholar]
- 12.Dubnau, D., and R. Davidoff-Abelson. 1971. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 56:209-221. [DOI] [PubMed] [Google Scholar]
- 13.Emory, S. A., P. Bouvet, and J. G. Belasco. 1992. A 5"-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 6:135-148. [DOI] [PubMed] [Google Scholar]
- 14.Geigenmüller, U., and K. H. Nierhaus. 1986. Tetracycline can inhibit tRNA binding to the ribosomal P site as well as to the A site. Eur. J. Biochem. 161:723-726. [DOI] [PubMed] [Google Scholar]
- 15.Gottesman, M. E. 1967. Reaction of ribosome-bound peptidyl transfer ribonucleic acid with aminoacyl transfer ribonucleic acid or puromycin. J. Biol. Chem. 242:5564-5571. [PubMed] [Google Scholar]
- 16.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hue, K. K., and D. H. Bechhofer. 1991. Effect of ermC leader region mutations on induced mRNA stability. J. Bacteriol. 173:3732-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krulwich, T. A., J. Jin, A. A. Guffanti, and D. H. Bechhofer. 2001. Functions of tetracycline efflux proteins that do not involve tetracycline. J. Mol. Microbiol. Biotechnol. 3:237-246. [PubMed] [Google Scholar]
- 19.Kunkel, T. A., K. Bebenek, and J. McClary. 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204:125-139. [DOI] [PubMed] [Google Scholar]
- 20.LeGrice, S. F., C. C. Shih, F. Whipple, and A. L. Sonenshein. 1986. Separation and analysis of the RNA polymerase binding sites of a complex Bacillus subtilis promoter. Mol. Gen. Genet. 204:229-236. [DOI] [PubMed] [Google Scholar]
- 21.Mackie, G. A. 1998. Ribonuclease E is a 5"-end-dependent endonuclease. Nature 395:720-723. [DOI] [PubMed] [Google Scholar]
- 22.Mayford, M., and B. Weisblum. 1989. ermC leader peptide: amino acid sequences critical for induction by translational attenuation. J. Mol. Biol. 206:69-79. [DOI] [PubMed] [Google Scholar]
- 23.Moran, C. P., Jr., N. Lang, S. F. LeGrice, G. Lee, M. Stephens, A. L. Sonenshein, J. Pero, and R. Losick. 1982. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol. Gen. Genet. 186:339-346. [DOI] [PubMed] [Google Scholar]
- 24.Mueller, F., and R. Brimacombe. 1997. A new model for the three-dimensional folding of Escherichia coli 16 S ribosomal RNA. I. Fitting the RNA to a 3D electron microscopic map at 20 Å. J. Mol. Biol. 271:524-544. [DOI] [PubMed] [Google Scholar]
- 25.Noah, J. W., M. A. Dolan, P. Babin, and P. Wollenzien. 1999. Effects of tetracycline and spectinomycin on the tertiary structure of ribosomal RNA in the Escherichia coli 30 S ribosomal subunit. J. Biol. Chem. 274:16576-16581. [DOI] [PubMed] [Google Scholar]
- 26.Sakaguchi, R., and K. Shishido. 1988. Molecular cloning of a tetracycline-resistance determinant from Bacillus subtilis chromosomal DNA and its expression in Escherichia coli and B. subtilis. Biochim. Biophys. Acta 949:49-57. [DOI] [PubMed] [Google Scholar]
- 27.Stasinopoulos, S. J., G. A. Farr, and D. H. Bechhofer. 1998. Bacillus subtilis tetA(L) gene expression: evidence for regulation by translational reinitiation. Mol. Microbiol. 30:923-932. [DOI] [PubMed] [Google Scholar]
- 28.Walker, A. C., M. L. Walsh, D. Pennica, P. S. Cohen, and H. L. Ennis. 1976. Transcription-translation and translation-messenger RNA decay coupling: Separate mechanisms for different messengers. Proc. Natl. Acad. Sci. USA 73:1126-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, W., and D. H. Bechhofer. 1997. Bacillus subtilis RNase III gene: cloning, function of the gene in Escherichia coli, and construction of Bacillus subtilis strains with altered rnc loci. J. Bacteriol. 179:7379-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, W., A. A. Guffanti, Y. Wei, M. Ito, and T. A. Krulwich. 2000. Two types of Bacillus subtilis tetA(L) deletion strains reveal the physiological importance of TetA(L) in K+ acquisition as well as in Na+, alkali, and tetracycline resistance. J. Bacteriol. 182:2088-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams, G., and I. Smith. 1979. Chromosomal mutations causing resistance to tetracycline in Bacillus subtilis. Mol. Gen. Genet. 177:23-29. [DOI] [PubMed] [Google Scholar]
- 32.Wilson, C. R., and A. E. Morgan. 1985. Chromosomal-DNA amplification in Bacillus subtilis. J. Bacteriol. 163:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]