Abstract
An in vitro transposition system, developed to facilitate gene disruption in Deinococcus radiodurans R1, has been used to inactivate the gene designated dr1819 in uvrA-1+ and uvrA-1 backgrounds. dr1819 encodes a protein with homology to a UV DNA damage endonuclease expressed by Schizosaccharomyces pombe. Interruption of dr1819 greatly sensitizes the uvrA-1 strain but not the uvrA-1+ strain to UV light, indicating that the dr1819 gene product is a component in a DNA repair pathway that can compensate for the loss of nucleotide excision repair in this species. Clones of dr1819 will restore UV resistance to UVS78, a uvrA-1 uvsE strain, indicating that dr1819 and uvsE are the same locus.
The Deinococcus radiodurans R1 genome encodes homologues of all components of the nucleotide excision repair pathway of Escherichia coli, including two UvrA-like proteins, UvrA-1 and UvrA-2 (34). The uvrA-1 gene product has 52% identity with the UvrA proteins of E. coli and Micrococcus luteus and appears to be the protein that functions in nucleotide excision repair (1, 26). Inactivation of uvrA-1 sensitizes an otherwise wild-type strain of D. radiodurans to mitomycin C.
There are two well-characterized null mutations of the uvrA-1 gene of D. radiodurans R1. Strain 262 carries a 1,300-bp insertion sequence (IS) element, designated IS2621, that inserted 986 bp from the 3" end of the uvrA-1 gene (26). Strain 302 has a 144-bp deletion that removes the first 34 bp of the uvrA-1 coding sequence and 110 bp of upstream sequence (26). Each mutation completely eliminates D. radiodurans' ability to grow in the presence of 60 ng of mitomycin C per ml (21, 32). The E. coli uvrA gene will complement the D. radiodurans uvrA-1 alleles, restoring mitomycin C resistance (1) to strains 262 and 302.
Unlike the uvrA strains of E. coli, the uvrA-1 strains of D. radiodurans exhibit nearly wild-type levels of resistance to UV light. A second locus, uvs, must be inactivated before uvrA-1 strains become UV sensitive (9, 10, 22). The uvs gene product, also called endonuclease β, appears to be part of an alternative excision repair pathway that completely compensates for the loss of nucleotide excision repair. Three mutations have been described that inactivate endonuclease β activity in D. radiodurans strain 302 (10), and it has been reported that each mutation affects a separate coding sequence, designated uvsC, uvsD, and uvsE. Evans and Moseley (9) have argued that endonuclease β is a multisubunit protein, but there is no definitive biochemical evidence to support this claim. A cell that is uvrA-1 and that carries any one of the three uvs mutations is unable to incise its DNA following UV irradiation and has lost the capacity to remove pyrimidine dimers from its chromosomal DNA (22).
Although this alternative excision repair pathway has not been thoroughly characterized, Gutman et al. (14) have proposed that endonuclease β may be a pyrimidine dimer DNA glycosylase (PD glycosylase), because expression of the denV gene of bacteriophage T4 partially restores UV resistance to a uvrA-1 uvs strain of D. radiodurans. Two lines of evidence, however, argue against this suggestion. (i) The annotation of the D. radiodurans R1 genome failed to identify a coding sequence similar to known PD glycosylases (19, 34). (ii) Endonuclease β does not cleave the N-glycosyl bond of the 5" base in a pyrimidine dimer as do known PD glycosylases. Instead, this enzyme appears to recognize pyrimidine dimers and cleave a phosphodiester bond immediately 5" to the lesion (8). In this respect, endonuclease β mimics the behavior of the UV damage endonucleases (UVDE proteins) of the eukaryotes Neurospora crassa (35) and Schizosaccharomyces pombe (6, 30). The eukaryotic proteins recognize a wide range of UV-induced DNA damage and introduce an incision into the DNA backbone 5" to the lesion, initiating a sequence of events that remove the damage (7, 15, 30, 36). The nick in the DNA results in exonucleolytic digestion of the strand carrying the damage, followed by DNA synthesis to fill in the resulting gap (2, 16, 17).
The annotation of the D. radiodurans genome did reveal the existence of a gene, dr1819, that encodes a protein that has approximately 30% amino acid sequence identity with the UVDE protein of S. pombe, suggesting that dr1819 encodes endonuclease β. (The gene designations used throughout this paper are based on those used in The Institute for Genomic Research [TIGR] Comprehensive Microbial Resource [http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gdr]. The prefix is given in lowercase letters, and loci are italicized.) In addition, a DNA sequence (accession number BAA85759), annotated as the uvs coding sequence, was deposited in the National Center for Biotechnology Information (NCBI) database in 1999 without evidence supporting the annotation. This sequence is identical to dr1819.
In this study, we have disrupted the coding sequence of dr1819 using an in vitro transposition system designed specifically for D. radiodurans and demonstrate that the resulting strain is sensitive to UV light in a uvrA-1 background. We also provide evidence that dr1819 is the uvsE gene described in earlier literature.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. All D. radiodurans strains were grown at 30°C in TGY broth (0.5% tryptone, 0.3% yeast extract, 0.1% glucose) or on TGY agar (1.5% agar). E. coli strains were grown in Luria-Bertani broth (28) or on Luria-Bertani plates at 37°C. Plasmids were propagated in E. coli strain DH5α MCR.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| D. radiodurans | ||
| R1 | ATCC 13939 | Anderson et al. (3) |
| 302 | As R1 but uvrA-1 | Moseley and Copland (21), Narumi et al. (26) |
| UVS78 | As 302 but uvsE | Moseley and Evans (22) |
| LSU1000 | As 302 but dr1819::TnDrCat | This study |
| LSU2000 | As R1 but dr1819::TnDrCat | This study |
| E. coli DH5α MCR | K-12 F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80d lacZΔ15 Δ(lacZYA-argF)U169 endA1 recA1 deoR thi-1 phoA supE44 λ−gyrA96 relA1 | Invitrogen Inc. |
| Plasmids | ||
| pGEM-T | Ampr | Promega, Madison, Wis. |
| pBC | Ampr | Stratagene, La Jolla, Calif. |
| pGPS3 | Ampr Kanr | New England Biolabs, Beverly, Mass. |
| pTTC101 | pGEM-T derivative with a TnDrCat (2,623-bp) insert; Catr Ampr | This study |
| pGTC101 | pGPS3 derivative with a TnDrCat insert; Catr Kanr Ampr | This study |
| pdr1771 | pGEM-T derivative with 3,441 bp of dr1771 (uvrA-1) and adjacent sequence, Ampr | This study |
| pdr1819 | pGEM-T derivative with 3,441 bp of dr1819 (putative uvsE) and adjacent sequence, Ampr | This study |
| pdr1819-54 | pdr1819 derivative with a TnDrCat insertion within the dr1819 coding sequence | This study |
| pdr1819-65 | pdr1819 derivative with a TnDrCat insertion within the dr1819 coding sequence | This study |
| pdr1819-77 | pdr1819 derivative with a TnDrCat insertion within the dr1819 coding sequence | This study |
| pdr1819-80 | pdr1819 derivative with a TnDrCat insertion within the dr1819 coding sequence | This study |
Transformation in liquid culture.
Calcium chloride from a 1 M stock solution was added to D. radiodurans cultures in exponential growth until a final concentration of 30 mM was achieved. This mixture was incubated at 30°C for 80 min (32). Either 1 μg of plasmid DNA or 10 μg of chromosomal DNA was added to 1 ml of TGY containing 2 × 107 cells and incubated on ice for 30 min. The transformation mixture was diluted 10-fold with TGY broth and incubated for another 18 h at 30°C. When this transformation protocol was used to identify uvrA+ cells, transformants were selected on TGY plates containing 60 ng of mitomycin C per ml (32).
Chromosomal DNA isolation.
TGY broth (200 ml) was inoculated with a 2-ml overnight culture (approximately 2 × 108 CFU/ml) of D. radiodurans R1. After 48 h, the 200-ml culture was harvested by centrifugation at 4°C at 3,000 × g for 15 min. Pellets were resuspended in 20 ml of 95% ethanol and held at room temperature for 10 min to remove the D. radiodurans outer membrane. The ethanol-stripped cells were collected by centrifugation at 4°C at 3,000 × g for 15 min, and the resulting pellet was resuspended in 9 ml of TE buffer (10 mM Tris-HCl, 0.1 mM EDTA [pH 8.0]). Two milligrams of lysozyme (Sigma Chemical, St. Louis, Mo.) was added to the stripped cells, and this mixture was incubated at 37°C for 30 min. Then 0.5 ml of 10% sodium dodecyl sulfate (SDS) and 50 μl of 20-mg/ml proteinase K (Sigma Chemical) were added to lysozyme-treated cells and incubated for 3 h at 56°C.
Lysed cells were transferred to a centrifuge tube and extracted once with an equal volume of phenol-chloroform (1:1) and twice with equal volumes of chloroform-isoamyl alcohol (24:1). The DNA was precipitated from the extracted material with 1 ml of 3 M sodium acetate (pH 7.0) and 20 ml of ice-cold 100% ethanol. The DNA was spooled out with a curved glass rod and washed twice with 70% ethanol. The DNA was air dried, dissolved in 5 ml of TE buffer (pH 8.0), and stored at 4°C.
Survival curves.
Only D. radiodurans cultures in exponential growth (2 × 106 to 4.5 × 107 CFU/ml) were evaluated for their ability to survive UV or ionizing radiation. All D. radiodurans cultures were treated at 25°C. UV irradiation was conducted with a germicidal lamp with a calibrated dose rate of 25 J/m2/s generated UV light. Gamma irradiation was conducted with a model 484R 60Co irradiator (J. L. Shepherd & Associates, San Fernando, Calif.) at a rate of 30.8 Gy/min. Irradiated cultures were diluted, plated in triplicate on TGY agar plates, and incubated for 3 days at 30°C before scoring for survivors.
Construction of pdr1819 and pdr1771.
A PCR fragment encoding the putative uvs gene (dr1819) of D. radiodurans R1 was amplified directly from purified chromosomal DNA using an appropriate pair of primers derived from the published sequence of the R1 genome. The primers, designated uvsup and uvsdwn (Table 2), generate a 1,348-bp fragment that includes the coding sequence plus 180 bp of upstream and 190 bp of downstream sequence. PCRs were carried out using Ready-To-Go PCR Beads (Amersham Pharmacia, Piscataway, N.J.) and were supplemented with betaine and dimethyl sulfoxide at final concentrations of 1.3 M and 1.3%, respectively. The PCR products were isolated using the Prep-A-Gene DNA purification systems (Bio-Rad, Hercules, Calif.). The dr1819-containing PCR fragments were inserted directly into the vector pGEM-T (Promega, Madison, Wis.) to generate the construct pdr1819.
TABLE 2.
Primers used for construction of pGTC101, pdr1819, and pdr1771
| Primer | Sequence |
|---|---|
| uvsup | 5"-CGAATCCATCGGAACCTCCTCAGAGTAAGC |
| uvsdwn | 5"-GTACTTCTGGCAAAACGCCGACTGCGTGAC |
| uvrAFP | 5"-TCGAAGACCGGCAGCTTATCG A |
| uvrADP | 5"-TCCATCTCCCGCAGGACGTAT T |
| Tu1 | 5"-AGCTTTGTTTAAACACGTGTACAACGCCTCCAAGGAC |
| Tu2-1 | 5"-TGATTTTTTTCTCCATTGTCTTACTCCCTCCAAGCGGTG |
| Cat1-1 | 5"-GGAGGGAGTAAGACAATGGAGAAAAAAATCACTGGATATACCAC |
| Cat2-2 | 5"-ACTTATTCAGGCATAGCAACCAGGC |
| N | 5"-ACTTTATTGTCATAGTTTAGATCTATTTTG |
| S | 5"-ATAATCCTTAAAAACTCCATTTCCACCCCT |
Plasmid pdr1771 was constructed in a similar manner. A PCR fragment encoding the uvrA-1 gene of D. radiodurans R1 was amplified using a pair of primers derived from the published sequence of the R1 genome (Table 2). The primers, designated uvrAFP and uvrADP, generate a 3,320-bp fragment that includes the uvrA-1 coding sequence plus 306 bp of upstream and 87 bp of downstream sequence. The PCR product was ligated in pGEM-T as described above. The inserts were sequenced to confirm that the clones were the D. radiodurans R1 genes designated dr1819 and dr1771. The sequences obtained were identical to that reported at the TIGR web site (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gdr).
Construction of pGTC101.
The tuf1 (dr0309) gene product, elongation factor Tu, is the only protein known that is constitutively expressed at high level in D. radiodurans (31). For this reason, we chose to fuse the 1,907-bp sequence immediately upstream of tuf1 (here designated pTu) to the cat gene of Tn9 in a three-step process. (i) Primers were designed (Table 2) that permitted efficient amplification of pTu. Prior to amplification, D. radiodurans R1 chromosomal DNA (200 ng) was denatured in the presence of the primers (150 ng of each) by heating to 95°C for 5 min. Four units of Pfu polymerase (Stratagene, La Jolla, Calif.) were added to the denatured template to initiate PCR. The conditions for the first 15 cycles of amplification were denaturation at 95°C for 1 min, annealing at 64°C for 1 min, and extension at 72°C for 4 min. For the next 10 cycles, the length of the extension step was increased by 10 s per cycle. After 25 cycles, the reaction was held at 72°C for 10 min. The resulting PCR product duplicated pTu, beginning 1,907 bp upstream of the initiation codon of the tuf1 gene and ending with the base pair adjacent to the initiation codon. Primer Tu1-1 included a tail that, when amplified, resulted in formation of a PmeI restriction site. Primer Tu2-1 included a tail that overlapped the cat gene found on the vector pBC (Stratagene, La Jolla, Calif.).
(ii) The 716 bp of the cat gene from pBC was also amplified as described using the primers Cat1-1 and Cat1-2 (Table 2). The Cat1-1 primer included a tail that overlapped the pTu promoter region. The Cat1-2 primer included a tail that, when amplified, resulted in formation of a NotI restriction site.
(iii) The purified PCR products from steps i and ii were combined with primers Tu1-1 and Cat1-2 in a final PCR. The overlap between the amplified pTu fragment and the amplified cat gene resulted in the formation of a fusion product with a PmeI site upstream of pTu and a NotI site downstream of cat. For undetermined reasons, it was not possible to efficiently cut the spliced fragment with either PmeI or NotI. The spliced PCR fragment was therefore inserted into pGEM-T (Promega), a vector that provides a 3"-compatible overhang for PCR products. The resulting construct, designated pTTC101, was propagated in the E. coli strain DH5α MCR (Invitrogen, Inc., Gaithersburg, Md.).
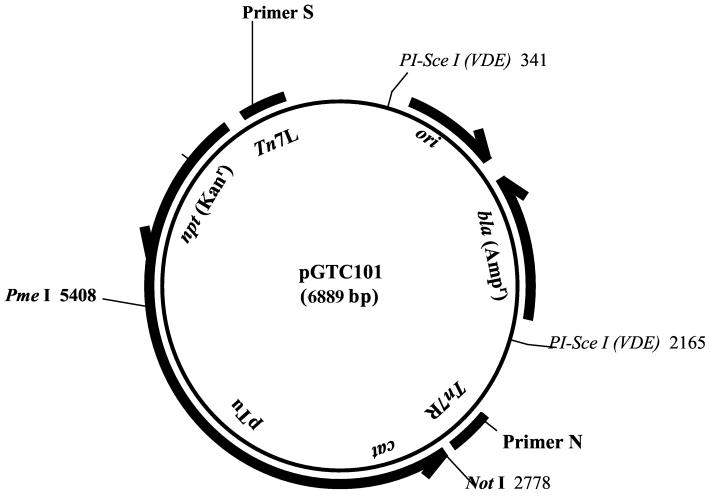
Purified pTTC101 was digested with PmeI and NotI restriction enzymes, and the pTu-cat fusion was isolated. This fragment was inserted into PmeI- and NotI-digested pGPS3 (New England Biolabs, Beverly, Mass.), creating a construct that could be used in an in vitro transposition reaction. The resulting construct, designated pGTC101 (Fig. 1), was propagated in DH5α MCR.
FIG.1.
Plasmid pGTC101. A PmeI- and NotI-digested TuCat fragment from pTTC101 was cloned into pGPS3. PI-Sce I(VDE), PI-SceI (intein from VDE [VMAI-derived endonuclease of S. cerevisiae]).
RESULTS
In vitro transposition using pGTC101.
In vitro transposition was performed using the protocol developed by New England Biolabs for the GPS-M mutagenesis system. Twenty nanograms of purified, circular pGTC101 was combined with the TnsABC* transposase supplied with the system (4) and target DNA (pdr1819). The target-to-donor molar ratio was maintained at 4:1. Following the transposition reaction, unreacted pGTC101 molecules were destroyed by digestion with 8 U of PI-SceI (a rare-cutting site-specific DNA endonuclease derived from an intein in Saccharomyces cerevisiae; New England BioLabs) for 4 h at 37°C. The parent of pGTC101, pGPS3, was constructed to contain a pair of PI-SceI restriction sites. The transposon generated by the action of the TnsABC* transposase on pGTC101 was designated TnDrCat.
The transposition reaction mixture was transformed by electroporation into approximately 5 × 105 CFU of electrocompetent DH5α MCR. Successful transposon insertions into the target were selected by plating the electroporated cells onto LB medium containing 25 μg of chloramphenicol per ml. Approximately 200 Catr colonies were recovered following in vitro transposition and electroporation. Thirty of the Catr colonies were picked, and the plasmids they carried were isolated. These plasmids were digested with a combination of ApaI and PstI to release the gene of interest from the vector. Digests were separated on 1% agarose and stained with ethidium bromide to determine whether the transposon had inserted into the putative uvs gene. Four of the first 80 pdr1819::TnDrCat plasmids examined appeared to have inserts in the putative uvs sequence. DNA sequencing using primer-binding sites (primer N and primer S; Table 2) within TnDrCat confirmed the position of insertion. The position of each insertion is identified in Table 3.
TABLE 3.
Position of insertions within the dr1819 coding sequence after in vitro transposition with TnDrCat
| Plasmid | Transposon inserted after base: |
|---|---|
| pdr1819-54 | 2 |
| pdr1819-65 | 1030 |
| pdr1819-77 | 1203 |
| pdr1819-80 | 726 |
A PstI-ApaI fragment was isolated and purified from pdr1819-80::TnDrCat. One microgram of this fragment was added to competent cultures of D. radiodurans strain 302 (approximately 107CFU/ml). After an 8-h incubation, 300 μl of the transformation mixture was plated onto TGY agar plates containing 5 μg of chloramphenicol per ml. Following this protocol, 20 to 50 Catr colonies were recovered from each transformation reaction. Individual colonies were used to inoculate TGY broth containing 5 μg of chloramphenicol per ml, and cultures were grown to stationary phase. One hundred microliters of this broth culture was used to inoculate TGY broth containing 10 μg of chloramphenicol per ml, and cultures were grown to stationary phase. This culture was diluted (1:106) and plated on TGY agar containing 10 μg of chloramphenicol per ml.
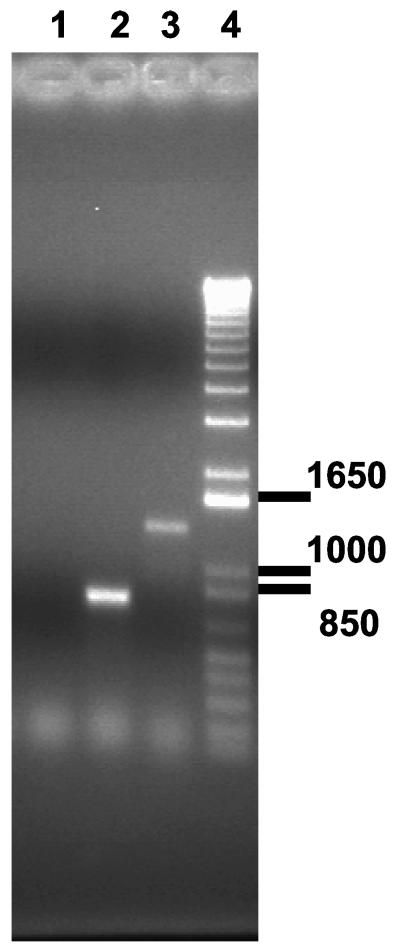
Since D. radiodurans is multigenomic, individual colonies had to be screened to determine if they were homozygous for the disruption. TnDrCat insertions into dr1819 of strain 302 were verified by PCR. The set of primers designed to amplify dr1819, uvsup and uvsdwn, were combined with primer S, which anneals within the transposon (Table 2). A 1,300-bp fragment corresponding to intact dr1819 was the only product produced when the three primers were combined with strain 302 genomic DNA in the reaction (Fig. 2, lane 3). In contrast, an approximately 750-bp product was obtained using genomic DNA isolated from the strain carrying the transposon (Fig. 2, lane 2). The 750-bp fragment was sequenced, and the sequence obtained was that of the 5" end of dr1819, signifying that the 750-bp product is the result of an amplification involving uvsup and primer S. We did not observe the 1,340-bp fragment corresponding to intact dr1819 in the strain carrying TnDrCat, indicating that the strain was homozygous for the transposon insertion. The strain containing the disruption was designated LSU1000.
FIG. 2.
Ethidium bromide-stained agarose gel illustrating that LSU1000 carries a homozygous insertion of dr1819-80::TnDrCat. Chloramphenicol-resistant colonies were isolated, and their genomic DNA was screened using appropriate primers and PCR to establish whether intact dr1819 remained in LSU1000. Lane 1, attempt to amplify dr1819 from LSU1000 genomic DNA with primers uvsup and uvsdwn. Lane 2, amplification of a 750-bp product when primers uvsup, uvsdwn, and S are combined with LSU1000 genomic DNA. Lane 3, amplification of a 1,300-bp product when primers uvsup, uvsdwn, and S are combined with R1 genomic DNA. Lane 4, 1-kbp ladder (Invitrogen, Gaithersburg, Md.). Sizes are shown in base pairs.
Genomic DNA from LSU1000 was isolated and used to transform D. radiodurans R1 to chloramphenicol resistance. Resulting colonies were screened as described above to establish that they were homozygous for the disruption. This strain, LSU2000, was uvrA-1+ (mitomycin C resistant) and dr1819::TnDrCat.
Disruption of dr1819 in D. radiodurans 302 sensitizes this strain to the lethal effects of UV light.
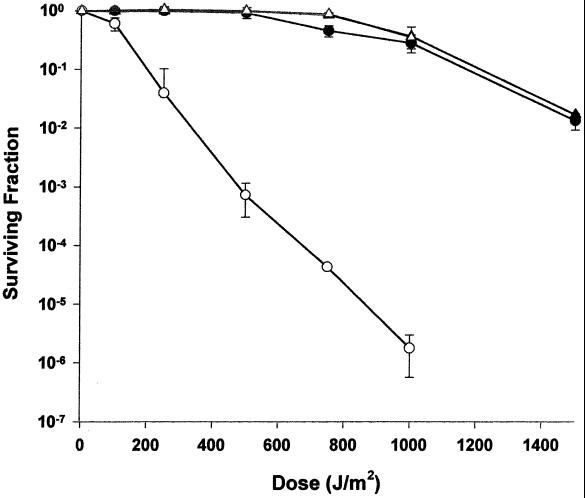
LSU1000 (uvrA-1 dr1819::TnDrCat) is very sensitive to UV light (Fig. 3), and the enhanced sensitivity was observed at every UV dose examined. For example, there is an approximately 100-fold increase in sensitivity at the 250-J/m2 dose relative to the parent strain. The shoulder that typifies the UV resistance of strains R1 and 302 disappears in LSU1000, indicating that the gene product of dr1819 is necessary for the UV resistance observed in a uvrA-1 background. Disruption of dr1819 in a uvrA-1+ background (LSU2000) had no effect on survival following UV treatment, demonstrating a complete overlap of function in the repair pathways involving DR1819 and nucleotide excision repair.
FIG. 3.
Representative survival curves for D. radiodurans strain LSU1000 uvrA-1 dr1819::TnDrCat (open circles) following exposure to UV radiation. Survival of strains R1 (triangles), 302 uvrA-1 (circles), and LSU2000 dr1819::TnDrCat (open triangles) is also shown. Values are the means ± standard deviations of triplicate experiments (n = 9).
LSU1000 and LSU2000 are fully resistant to ionizing radiation. The disruption of dr1819 did not alter the resistance of strain 302 or R1 to ionizing radiation at any of the doses examined (data not shown).
pdr1819 restores UV resistance to a uvrA-1 uvs strain of D. radiodurans
The pattern of UV sensitivity observed in LSU1000 duplicates that reported for UVS78, a uvrA-1 uvsE strain of D. radiodurans, suggesting that dr1819 encodes the uvsE gene product. To test this possibility, an attempt was made to transform UVS78 to UV resistance with pdr1819. UVS78 cultures were grown to a density of approximately 106 CFU/ml before 1 μg of transforming DNA was added to the culture. Following the transformation protocol, cells were diluted 1:100 in TGY broth and allowed to grow for 14 h at 30°C. The resulting cultures grew to a density of 4.8 × 107 ± 0.6 × 107 CFU/ml.
Aliquots of these cultures were exposed to 500 J of UV light m−2, and the surviving fraction was determined (Table 4). Genomic DNA isolated from UVS78 could not transform UVS78 to UV resistance, as evidenced by the failure to increase survival of the irradiated transformed culture versus the culture that did not receive transforming DNA. In contrast, UVS78 cultures transformed with genomic DNA isolated from strains R1 and 302 exhibited a 20-fold increase in survival after exposure to 500 J m−2 relative to the nontransformed control. Plasmids pdr1771 and pdr1819 were equally effective in restoring UV resistance to UVS78, yielding a 50-fold-higher number of survivors following UV irradiation. Since incorporation of the wild-type sequence of dr1819 is sufficient to restore UV resistance to UVS78, we assume that dr1819 is the uvsE gene described earlier (9, 10, 22).
TABLE 4.
Transformation of UVS78 to UV resistance with isolated DNA encoding the dr1771 (uvrA-1) and the dr1819 (putative uvsE) gene productsa
| Transforming DNA | Mean no. of cells survivingb (104) ± SD | Fold increasec |
|---|---|---|
| None | 3.8 ± 0.01 | |
| Genomic DNA from UVS78 | 3.2 ± 0.01 | 0.84 |
| Genomic DNA from R1 | 100 ± 8 | 26 |
| Genomic DNA from 302 | 75 ± 4.7 | 20 |
| pdr1771 | 190 ± 12 | 50 |
| pdr1819 | 200 ± 46 | 53 |
Cultures of UVS78 were exposed to UV light (500 J m−2) after transformation (see description in text).
Mean ± standard deviation (n = 54).
Fold increase relative to the nontransformed UVS78 culture.
DISCUSSION
Although perhaps better known for its resistance to ionizing radiation, D. radiodurans also displays a startling ability to tolerate UV light (22-24). The UV radiation survival curves of log-phase cultures of D. radiodurans R1 exhibit a characteristic shoulder of resistance in which there is no loss of viability to approximately 500 J m−2. The D37 dose for D. radiodurans R1 is typically reported to be between 600 and 700 J m−2, at least 20-fold higher than the D37 dose of E. coli cultures UV irradiated under the same conditions (29). The energy deposited by a 500-J m−2 dose of UV is sufficient to convert approximately 1% of the thymine in the irradiated cell's DNA to pyrimidine dimers (5, 33). Since the D. radiodurans genome is 3.26 Mbp with a 67% GC content (34), as many as 5,000 thymine-containing pyrimidine dimers could form per genome following exposure to UV at 500 J m−2.
When D. radiodurans cultures are UV irradiated, there is an immediate and characteristic burst of chromosomal DNA degradation (25, 33), and it has been assumed that this phenomenon is a manifestation of necessary DNA repair processes, since strains that do not exhibit this DNA degradation are highly sensitive to UV light. Moseley and Evans (22) established that the process of UV-induced DNA degradation in D. radiodurans was initiated by either of two nucleases, which they named endonuclease α and endonuclease β. Each of these enzymes appears to be capable of recognizing UV-induced DNA damage and incising the genome in response to that damage. Furthermore, both endonucleases appear to be equally adept at protecting the cell from UV-induced lethality. Mutations inactivating each endonuclease were identified, and it was established that loss of either nuclease alone failed to affect the UV resistance of this species (10).
Although endonuclease α was subsequently identified as a homologue of the UvrABC endonuclease of E. coli, (1, 26) little is known about the function of endonuclease β. Like the characterized PD glycosylases of M. luteus (13) and bacteriophage T4 (12, 27), endonuclease β catalyzes an incision adjacent to a pyrimidine dimer, facilitating its removal, but the D. radiodurans protein appears to incise the DNA by a different mechanism (8). The PD glycosylases cleave the N-glycosyl bond of the 5" base in the dimer, generating a structure that will release free thymine on photoreversal. The action of endonuclease β does not produce this structure. Free thymine is not released when photoreversal is attempted on UV-irradiated DNA exposed to partially purified endonuclease β, and from this result it has been inferred that endonuclease β cleaves a phosphodiester bond adjacent to the lesion (8).
This study has determined that inactivation of dr1819 by insertional mutagenesis sensitizes a uvrA-1 strain of D. radiodurans to UV light, even though the same disruption has no effect on the UV resistance of a uvrA-1+ strain (Fig. 3). The dr1819 gene product apparently catalyzes a process that compensates for the loss of nucleotide excision repair in this species. Since the dr1819 gene product of D. radiodurans has 30% amino acid sequence identity and 40% similarity with the uve1+ gene product (Uve1p) of S. pombe (19, 34), we suggest that DR1819 is a UV DNA damage endonuclease that catalyzes repair of UV-induced DNA damage by a mechanism similar to that of Uve1p. However, understanding DR1819's specific role in DNA damage repair will require further biochemical investigation.
Uve1p is an endonuclease that binds to a wide spectrum of DNA lesions, including UV-induced photoproducts (6, 11), apurinic/apyrimidinic sites (15), and base-base mismatches (18). This protein introduces a nick immediately 5" to a lesion, leaving a 5" phosphate and a 3"-OH (2). The nicks become the focus of other repair proteins, which first digest the strand containing the damage and then fill in the resulting gapped heteroduplex. Like endonuclease β, the activity of Uve1p overlaps the nucleotide excision repair pathway in S. pombe. Strains of S. pombe that are nucleotide excision repair deficient remain proficient in the excision of UV photoproducts and are largely UV resistant (7, 15, 30, 36). Double mutants, lacking nucleotide excision repair and Uve1p activities, become hypersensitive to UV light.
UVS78 (10, 22) was used to establish whether the coding sequence dr1819 was the uvsE gene described in earlier literature. The dr1819 sequence was cloned, and that clone was used as donor DNA in a transformation protocol in which UVS78 was the recipient. As shown in Table 4, the dr1819 sequence is sufficient but not necessary to restore UVS78 to UV resistance. The coding sequence of dr1771 (uvrA-1) will also restore UV resistance to this strain. We conclude that the open reading frame dr1819 is the uvsE gene and that dr1819 encodes endonuclease β.
In addition to UVS78, Evans and Moseley isolated UVS9 and UVS25 while screening mutagenized strain 302 cultures for UV-sensitive strains (10, 22). As with UVS78, the UV sensitivity of these strains was manifest only in uvrA-1 backgrounds. Genomic DNA isolated from each of these strains could transform UVS78 to UV resistance, indicating that each strain carried a different mutation. Evans and Moseley concluded that the individual mutations were found in separate genes, which they designated uvsC and uvsD, and that these genes were different from uvsE. They reasoned that the mutations found in UVS9, UVS25, and UVS78 could not be located within the same gene because the frequency with which each mutant's genomic DNA restored the others to UV resistance did not differ significantly from that observed when R1 genomic DNA was used in the transformation protocol.
Apparently, Evans and Moseley assumed that if all three mutations were located within the same gene, the efficiency of transformation to UV resistance would be greatly reduced, because one mutation would simply replace the other during recombination. However, this is not a valid assumption when considering natural transformation in D. radiodurans. Since the publication of Evans and Moseley's manuscript (10), it has been established that the average size of the DNA fragment inserted during transformation is less than 950 bp, substantially smaller than that reported for other transformable species (20). Genomic DNA from deinococcal strains carrying different mutations within the same gene can transform each other to DNA damage resistance with efficiencies indistinguishable from those observed when R1 genomic DNA is used as the donor. It is therefore possible that uvsC and uvsD are also alleles of dr1819. We could not obtain strains UVS9 and UVS25 to test this possibility.
As part of this study, a method was developed for genetically manipulating D. radiodurans R1 that makes use of the commercially available GPS-M in vitro transposition system. A transposon was created that is capable of expressing chloramphenicol resistance in D. radiodurans R1 by fusing a 1,900-bp sequence upstream of the tuf1 gene of R1 to a cat gene and inserting that fusion into a vector containing the minimal set of sequences required for transposition. The resulting construct, pGTC101, was used to disrupt the cloned coding sequence of dr1819. The vector carrying the disrupted gene was used as transforming DNA, and the chloramphenicol resistance marker was integrated into the R1 genome. The resulting Catr colonies were screened for cells homozygous for the marker. The result was a stable loss-of-function mutation that is easily moved from one strain to another. Since the D. radiodurans R1 genome has been sequenced in its entirety and since any segment of that genome can be cloned after amplification in a PCR, it should be possible to rapidly inactivate any nonessential gene in this species using this approach.
Acknowledgments
This work was supported by National Science Foundation grant MCB-9728404.
We gratefully acknowledge F. A. Rainey and his laboratory at Louisiana State University for assistance in DNA sequencing.
EFERENCES
- 1.Agostini, H. J., J. D. Carroll, and K. W. Minton. 1996. Identification and characterization of uvrA, a DNA repair gene of Deinococcus radiodurans. J. Bacteriol. 178:6759-6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alleva, J. L., S. Zuo, J. Hurwitz, and P. W. Doetsch. 2000. In vitro reconstitution of the Schizosaccharomyces pombe alternative excision repair pathway. Biochemistry 39:2659-2666. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, A. W., H. C. Nordon, R. F. Cain, G. Parrish, and D. Duggan. 1956. Studies on a radio-resistant micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 10:575-578. [Google Scholar]
- 4.Bainton, R. J., K. M. Kubo, J. N. Feng, and N. L. Craig. 1993. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell 72:931-943. [DOI] [PubMed] [Google Scholar]
- 5.Boling, M. E., and J. K. Setlow. 1966. The resistance of Micrococcus radiodurans to ultraviolet radiation. III. A repair mechanism. Biochim. Biophys. Acta 123:26-33. [DOI] [PubMed] [Google Scholar]
- 6.Bowman, K. K., K. Sidik, C. A. Smith, J. S. Taylor, P. W. Doetsch, and G. A. Freyer. 1994. A new ATP-independent DNA endonuclease from Schizosaccharomyces pombe that recognizes cyclobutane pyrimidine dimers and 6-4 photoproducts. Nucleic Acids Res. 22:3026-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davey, S., M. L. Nass, J. V. Ferrer, K. Sidik, A. Eisenberger, D. L. Mitchell, and G. A. Freyer. 1997. The fission yeast UVDR DNA repair pathway is inducible. Nucleic Acids Res. 25:1002-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, D. M., and B. E. Moseley. 1988. Deinococcus radiodurans UV endonuclease β DNA incisions do not generate photoreversible thymine residues. Mutat. Res. 207:117-119. [DOI] [PubMed] [Google Scholar]
- 9.Evans, D. M., and B. E. Moseley. 1985. Identification and initial characterisation of a pyrimidine dimer UV endonuclease (UV endonuclease beta) from Deinococcus radiodurans; a DNA-repair enzyme that requires manganese ions. Mutat. Res. 145:119-128. [DOI] [PubMed] [Google Scholar]
- 10.Evans, D. M., and B. E. Moseley. 1983. Roles of the uvsC, uvsD, uvsE, and mtcA genes in the two pyrimidine dimer excision repair pathways of Deinococcus radiodurans. J. Bacteriol. 156:576-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freyer, G. A., S. Davey, J. V. Ferrer, A. M. Martin, D. Beach, and P. W. Doetsch. 1995. An alternative eukaryotic DNA excision repair pathway. Mol. Cell. Biol. 15:4572-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedberg, E. C., and I. R. Lehman. 1974. Excision of thymine dimers by proteolytic and amber fragments of E. coli DNA polymerase I. Biochem. Biophys. Res. Commun. 58:132-139. [DOI] [PubMed] [Google Scholar]
- 13.Grafstrom, R. H., L. Park, and L. Grossman. 1982. Enzymatic repair of pyrimidine dimer-containing DNA. A 5" dimer DNA glycosylase: 3"-apyrimidinic endonuclease mechanism from Micrococcus luteus. J. Biol. Chem. 257:13465-13474. [PubMed] [Google Scholar]
- 14.Gutman, P. D., H. L. Yao, and K. W. Minton. 1991. Partial complementation of the UV sensitivity of Deinococcus radiodurans excision repair mutants by the cloned denV gene of bacteriophage T4. Mutat. Res. 254:207-215. [DOI] [PubMed] [Google Scholar]
- 15.Kanno, S., S. Iwai, M. Takao, and A. Yasui. 1999. Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage. Nucleic Acids Res. 27:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaur, B., A. M. Avery, and P. W. Doetsch. 1998. Expression, purification, and characterization of ultraviolet DNA endonuclease from Schizosaccharomyces pombe. Biochemistry 37:11599-11604. [DOI] [PubMed] [Google Scholar]
- 17.Kaur, B., and P. W. Doetsch. 2000. Ultraviolet damage endonuclease (Uve1p): a structure and strand-specific DNA endonuclease. Biochemistry 39:5788-5796. [DOI] [PubMed] [Google Scholar]
- 18.Kaur, B., J. L. Fraser, G. A. Freyer, S. Davey, and P. W. Doetsch. 1999. A Uve1p-mediated mismatch repair pathway in Schizosaccharomyces pombe. Mol. Cell. Biol. 19:4703-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton, E. V. Koonin, and M. J. Daly. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattimore, V., K. S. Udupa, G. A. Berne, and J. R. Battista. 1995. Genetic characterization of forty ionizing radiation-sensitive strains of Deinococcus radiodurans: linkage information from transformation. J. Bacteriol. 177:5232-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moseley, B. E., and H. F. Copland. 1978. Four mutants of Micrococcus radiodurans defective in the ability to repair DNA damaged by mitomycin-C, two of which have wild-type resistance to ultraviolet radiation. Mol. Gen. Genet. 160:331-337. [DOI] [PubMed] [Google Scholar]
- 22.Moseley, B. E., and D. M. Evans. 1983. Isolation and properties of strains of Micrococcus (Deinococcus) radiodurans unable to excise ultraviolet light-induced pyrimidine dimers from DNA: evidence for two excision pathways. J. Gen. Microbiol. 129:2437-2445. [DOI] [PubMed] [Google Scholar]
- 23.Moseley, B. E., and A. Mattingly. 1971. Repair of irradiation transforming deoxyribonucleic acid in wild type and a radiation-sensitive mutant of Micrococcus radiodurans. J. Bacteriol. 105:976-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moseley, B. E. B. 1983. Photobiology and radiobiology of Micrococcus (Deinococcus) radiodurans. Photochem. Photobiol. Rev. 7:223-275. [Google Scholar]
- 25.Moseley, B. E. B. 1967. The repair of DNA in Micrococcus radiodurans following ultraviolet irradiation. J. Gen. Microbiol. 48:4-24. [Google Scholar]
- 26.Narumi, I., K. Cherdchu, S. Kitayama, and H. Watanabe. 1997. The Deinococcus radiodurans uvrA gene: identification of mutation sites in two mitomycin-sensitive strains and the first discovery of insertion sequence element from deinobacteria. Gene 198:115-126. [DOI] [PubMed] [Google Scholar]
- 27.Radany, E. H., and E. C. Friedberg. 1982. Demonstration of pyrimidine dimer-DNA glycosylase activity in vivo: bacteriophage T4-infected Escherichia coli as a model system. J. Virol. 41:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, p. 1.25-1.28, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sweet, D. M., and B. E. Moseley. 1974. Accurate repair of ultraviolet-induced damage in Micrococcus radiodurans. Mutat. Res. 23:311-318. [DOI] [PubMed] [Google Scholar]
- 30.Takao, M., R. Yonemasu, K. Yamamoto, and A. Yasui. 1996. Characterization of a UV endonuclease gene from the fission yeast Schizosaccharomyces pombe and its bacterial homolog. Nucleic Acids Res. 24:1267-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka, A., H. Hirano, M. Kikuchi, S. Kitayama, and H. Watanabe. 1996. Changes in cellular proteins of Deinococcus radiodurans following gamma-irradiation. Radiat. Environ. Biophys. 35:95-99. [DOI] [PubMed] [Google Scholar]
- 32.Udupa, K. S., P. A. O'Cain, V. Mattimore, and J. R. Battista. 1994. Novel ionizing radiation-sensitive mutants of Deinococcus radiodurans. J. Bacteriol. 176:7439-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varghese, A. J., and R. S. Day. 1970. Excision of cytosine-thymine adduct from the DNA of ultraviolet-irradiated Micrococcus radiodurans. Photochem. Photobiol. 11:511-517. [DOI] [PubMed] [Google Scholar]
- 34.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yajima, H., M. Takao, S. Yasuhira, J. H. Zhao, C. Ishii, H. Inoue, and A. Yasui. 1995. A eukaryotic gene encoding an endonuclease that specifically repairs DNA damaged by ultraviolet light. EMBO J. 14:2393-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yonemasu, R., S. J. McCready, J. M. Murray, F. Osman, M. Takao, K. Yamamoto, A. R. Lehmann, and A. Yasui. 1997. Characterization of the alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe. Nucleic Acids Res. 25:1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]