Abstract
The bacterial cell division protein FtsW has been suggested to perform two functions: stabilize the FtsZ cytokinetic ring, and facilitate septal peptidoglycan synthesis by the transpeptidase FtsI (penicillin-binding protein 3). We show here that depleting Escherichia coli cells of FtsW had little effect on the abundance of FtsZ rings but abrogated recruitment of FtsI to potential division sites. Analysis of FtsW localization confirmed and extended these results; septal localization of FtsW required FtsZ, FtsA, FtsQ, and FtsL but not FtsI. Thus, FtsW is a late recruit to the division site and is essential for subsequent recruitment of its cognate transpeptidase FtsI but not for stabilization of FtsZ rings. We suggest that a primary function of FtsW homologues—which are found in almost all bacteria and appear to work in conjunction with dedicated transpeptidases involved in division, elongation, or sporulation—is to recruit their cognate transpeptidases to the correct subcellular location.
FtsW is an integral membrane protein required for cell division in Escherichia coli (6, 23, 26), and presumably most other bacteria as well, although its precise biochemical role in the process is not yet known. FtsW is one of nine essential cell division proteins in E. coli, all of which have been shown to localize to the division site (midcell) during septation.
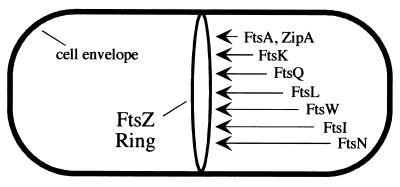
Studies of localization in various mutant backgrounds revealed a set of dependency relationships that are generally considered to reflect the order in which the division proteins are recruited to the division site (reviewed in reference 35). The first event in this pathway is assembly of FtsZ into a ring (Z-ring) at the future division site (Fig. 1). FtsA and ZipA each bind directly to FtsZ and localize next. After localization of FtsA, the proteins FtsK, FtsQ, FtsL, FtsI, and FtsN appear at the division site in that order. This recruitment sequence has been postulated to reflect the order of assembly of a multiprotein complex that mediates constriction of the cell envelope at the midcell (28, 30, 33, 35).
FIG. 1.
Recruitment of division proteins to the septal ring of E. coli. The first event is polymerization of FtsZ into the Z-ring. The other proteins localize in the order indicated. For a recent review, see reference 35. The position of FtsK is from reference 7. The position of FtsW is from this work.
Although FtsW is known to localize to the division site (43), its position in the pathway has not been established. The most pertinent study reported that depletion of FtsW severely destabilized Z-rings, implying that FtsW is an early recruit to the division site and has an important role in regulation of FtsZ dynamics (6). In contrast, inactivation of other division proteins has only a minor effect on Z-rings. For example, inactivation of FtsI delays Z-ring assembly and reduces the total number of rings per unit cell mass by about twofold compared to an unperturbed population of cells (34). Even inactivation of FtsA or ZipA, which localize immediately after FtsZ, has only a modest effect on the number of Z-rings observed per unit mass (2, 18, 27).
FtsW belongs to a large family of polytopic membrane proteins that appear to be present in all bacteria that have a peptidoglycan cell wall (21, 22, 24). This family has been named SEDS (21) for shape, elongation, division, and sporulation. The founding members of the SEDS family are the E. coli FtsW and RodA proteins and the Bacillus subtilis SpoVE protein, which are required for peptidoglycan synthesis during division, elongation, and sporulation, respectively.
Each SEDS protein appears to work in conjunction with a specific class B penicillin-binding protein (PBP) (31), transpeptidases that catalyze the formation of peptide cross-links in peptidoglycan (1, 15). One such pair is RodA and PBP2. These proteins are encoded in the same operon, and inactivation of either interferes with elongation, resulting in the production of large, spherical cells (32, 37, 39, 41). Likewise, the genes encoding FtsW and FtsI (also called PBP3) are cotranscribed (19), and inactivation of either protein blocks division without impairing elongation (26, 38). Interestingly, a recent analysis of 19 bacterial genomes for which a complete sequence is available showed that all organisms that contain ftsW also contain ftsI, and vice versa; FtsW and FtsI were the only pair of division proteins that showed perfect correlation (30). B. subtilis SpoVE is required for synthesis of spore cortex peptidoglycan (20); its partner has not yet been established but might be SpoVD (11).
Despite the large body of circumstantial evidence that SEDS proteins support the enzymatic activity of cognate transpeptidases, the mechanism by which they do so has remained obscure. We show here that FtsW is a late recruit to the division site of E. coli and is required for recruitment of its cognate transpeptidase FtsI (PBP3) to the septal ring. We did not find any evidence to suggest that FtsW has a major role in stabilization of Z-rings.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genetic markers or features | Construction or comment | Source or reference | |
|---|---|---|---|---|
| Miscellaneous strains | ||||
| MG1655 | Wild type | Lab collection | ||
| KS272 | F− ΔlacX74 galE galK thi rpsL ΔphoA(PvuII) | 40 | ||
| MC4100 | F−araD139 ΔlacU169 ΔrelA1 rpsL150 thi mot flb5301 deoC7 ptsF25 rbsR | J. Beckwith | ||
| JP313 | MC4100 Δara-714 | J. Pogliano | ||
| DY330 | W3110 ΔlacU169 gal-490 λc1857 Δ(cro-bioA) | 46 | ||
| EC850 | DY330 ftsW::kan/pDSW406 | From pKD4 using Red recombinase | This study | |
| EC912 | W3110 ΔlacU169 gal-490 ftsW::kan/pDSW406 | MG1655 attB region into EC850 using Red recombinase | This study | |
| SM551 | F− Δlac(MS562) λ−λsmel NalArsupF58 (=suIII+) | S. Michaelis | ||
| DHB6521 | SM551 λInCh1 (Kanr) | 5 | ||
| DHB6504 | SM551 λInCh1 · pSX102 (Ampr) | D. Boyd | ||
| MM61 | F−araD139 ΔlacU169 StrrftsA12(Ts) leu::Tn10 | J. Beckwith | ||
| DRC14 | MC4100 ftsZ84(Ts) leu::Tn10 | D. RayChaudhuri | ||
| EC530 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-ftsI | 44 | ||
| EC442 | MC4100 Δ(λattL-lom)::bla lacIqP207-gfp-ftsQ | 8 | ||
| EC447 | MC4100 Δ(λattL-lom)::bla lacIqP209-ftsA-gfp | 44 | ||
| EC528 | JP313 Δ(λattL-lom)::bla lacIqP208-ftsZ-gfp | 14 | ||
| EC530 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-ftsI | 44 | ||
| EC531 | JP313 Δ(λattL-lom)::bla lacIqP207-gfp-ftsL | 14 | ||
| EC535 | JP313 Δ(λattL-lom)::bla lacIqP208-zipA-gfp | Lab collection | ||
| EC549 | KS272 ftsI::TnphoAI173 ΔIS 50R (Kanr)/pDSW262 | = EC548 | 44 | |
| JOE170 | KS272 ftsQ::TnphoA80 (Kanr)/pJC10 | 8 | ||
| JMG265 | KS272 ftsL::TnphoAL81 ΔIS 50R (Kanr)/pBAD33-LLL | = JM265 | J.-M. Ghigo | |
|
gfp-ftsW fusions in wild-type background
|
||||
| EC785 | MC4100 Δ(λattL-lom)::kan lacIqP209-gfp-ftsW | From pDSW360 using λnCh1 · pSX102 (Ampr) | This study | |
| EC791 | MC4100 Δ(λattL-lom)::bla lacIqP209-gfp-ftsW | From pDSW311 using λInCh1 (Kanr) | This study | |
|
gfp-ftsW fusions in fts background
|
||||
| EC787 | EC785 ftsA12(Ts) leu::Tn10 | P1 (MM61) × EC785 → select Tetr | This study | |
| EC788 | EC785 ftsZ84(Ts) leu::Tn10 | P1 (DRC14) × EC785 → select Tetr | This study | |
| EC798 | JMG265 Δ(λattL-lom)::bla lacIqP209-gfp-ftsW | P1 (EC791) × JM265 → select Ampr | This study | |
| EC799 | JOE170 Δ(λattL-lom)::bla lacIqP209-gfp-ftsW | P1 (EC791) × JOE170 → select Ampr | This study | |
| EC829 | EC549 Δ(λattL-lom)::bla lacIqP209-gfp-ftsW | P1 (EC791) × EC549 → select Ampr | This study | |
|
gfp fusions in FtsW depletion backgrounda
|
||||
| EC856 | EC850 Δ(λattL-lom)::bla lacIqP208-ftsZ-gfp | P1 (EC528) × EC850 → select Ampr | This study | |
| EC860 | EC850 Δ(λattL-lom)::bla lacIqP207-gfp-ftsQ | P1 (EC442) × EC850 → select Ampr | This study | |
| EC861 | EC850 Δ(λattL-lom)::bla lacIqP207-gfp-ftsI | P1 (EC530) × EC850 → select Ampr | This study | |
| EC881 | EC850Δ(λattL-lom)::bla lacIqP207-gfp-ftsL | P1 (EC531) × EC850 → select Ampr | This study | |
| EC892 | EC850 Δ(λattL-lom)::bla lacIqP208-zipA-gfp | P1 (EC535) × EC850 → select Ampr | This study | |
| EC904 | EC850 Δ(λattL-lom)::bla lacIqP210-ftsA-gfp | P1 (EC447) × EC850 → select Ampr | This study | |
| Plasmids | ||||
| pBAD33 | Arabinose regulation, Cmr | 16 | ||
| pJC10 | pBAD33-ftsQ | 8 | ||
| pJMG197 | pBAD33-ftsL | 14 | ||
| pDSW209 | pDSW206-gfp-MCS (fusion vector) | 44 | ||
| pKD4 | Kanr template plasmid | 12 | ||
| pHR485 | pSY309-Pmra-envA (a mini-F that carries ftsW) | 19 | ||
| pDSW311 | pDSW209-ftsW | This study | ||
| pDSW360 | Kanr derivative of pDSW311 | This study | ||
| pDSW406 | pBAD33-ftsW | This study | ||
These EC850 derivatives are temperature independent because the transduction crosses the prophage out of the recipient.
Media.
Cells were grown in Luria-Bertani (LB) medium or LB with 0% NaCl. l-Arabinose or d-glucose was added at 0.2% as indicated to modulate expression of genes under control of the PBAD promoter (16). For localization studies, IPTG (isopropyl-β-d-thiogalactopyranoside) was used at the following concentrations to induce expression of gfp fusions: 1 mM for gfp-ftsW, 2.5 μM for ftsZ-gfp, 100 μM for ftsA-gfp, 50 μM for zipA-gfp, 5 μM for gfp-ftsQ, 2.5 μM for gfp-ftsL, and 2.5 μM for gfp-ftsI. (The gfp-ftsW and ftsA-gfp fusions are under control of weaker promoters than are the other fusions.) Antibiotics were used at the following concentrations: ampicillin, 200 μg/ml for plasmids and 25 μg/ml for chromosomal alleles; kanamycin, 40 μg/ml; chloramphenicol, 30 μg/ml; and tetracycline, 10 μg/ml.
Molecular biological procedures.
Standard procedures for cloning and analysis of DNA, PCR, electroporation, and transformation were used (36). Enzymes used to manipulate DNA were from New England Biolabs (Beverly, Mass.) Oligonucleotides were from Integrated DNA Technologies (Coralville, Iowa). DNA sequencing was performed by the DNA Core Facility of the University of Iowa. All constructs made by the PCR were sequenced to verify their integrity.
Construction of a gfp-ftsW fusion.
The ftsW gene was amplified by PCR with pHR485 as the template and primers FtsW1 (5"GCAGAATTCAACAACAACAACATGCGTTTATCTCTCCCTCGCCTG) plus FtsW2 (5"CGTAAGCTTTCATCGTGAACCTCGTACAAACGC). The amplified product was digested with EcoRI and HindIII (sites underlined) and ligated into the same sites of the gfp fusion vector pDSW209 to create pDSW311. The fusion protein encoded by pDSW311 has the linker sequence YKEFNNNMR, where YK are the last two residues of GFP and MR are the first two residues of FtsW. pDSW311 confers resistance to ampicillin.
A derivative, pDSW360, that confers kanamycin resistance was constructed as follows. The kan gene from pUC4K (Amersham Pharmacia Biotech, Piscataway, N.J.) was amplified by PCR with pUC4K1 and pUC4K2 as primers (5"-GCAGATATCGGCGCTGAGGTCTGCCTC-3" and 5"-TGGGATATCGGGAAAGCCACGTTGTCTC-3", respectively). The amplified product was digested with EcoRV (sites underlined) and then ligated into pDSW311 that had been digested with FspI and ScaI, which cut within the bla gene. gfp-ftsW fusions were moved from plasmids to the chromosome using λInCh (5).
Construction of an ftsW depletion strain.
To understand our approach for constructing a depletion strain, it is helpful to know that ftsW resides in a large operon in the 2-min region of the E. coli chromosome along with several other genes involved in division and/or peptidoglycan synthesis. The gene order in the relevant region is murD-ftsW-murG, all of which overlap and are essential. We disrupted ftsW by using the λ Red recombination system to replace ftsW with an ftsW::kan PCR fragment that had 50 nucleotides of flanking homology at either end (12, 46). The ftsW::kan replacement allele preserved the first six codons of ftsW (so the stop codon for murD was not affected), extended to the stop codon of ftsW, and provided a strong Shine-Dalgarno sequence and a start codon for murG to minimize polarity (12). In addition, the kan gene was oriented so that transcription proceeds towards murG and other downstream genes.
The depletion strain was constructed in several steps. First, ftsW was cloned under control of an arabinose-dependent promoter in pBAD33 (16). To do this, ftsW was amplified by PCR with pHR485 as the template and primers P300 (5"CACGAGCTCGGAGTGAAACGATGCGTTTATCTCTCCCTCGC) and P301 (5"GTGGCATGCTCATCGTGAACCTCGTACAAAC). The amplified product was digested with SacI and SphI (sites underlined) and ligated into the same sites of pBAD33 to create pDSW406.
Second, pDSW406 was transformed into DY330, a W3110 derivative with a defective λ prophage that expresses the Red recombinase upon heat induction (46).
Third, the ≈1.6-kb ftsW::kan fragment used to disrupt ftsW was obtained by PCR on the Kanr template plasmid pKD4 using P307 as the 5" primer (5"AGTTTGCCCGTCTGGCGAAGGAGTTAGGTTGATGCGTTTATCTCTCCCTCgtgtaggctggagctgcttc) and P308 as the 3" primer (5"TGTCCACCGGTTCCGCCTGCCATCACCATTAATCGCTTTCCTTGACCACTcatatgaatatcctcctta). Start and stop codons for ftsW are underlined. Italics indicate a start codon and Shine-Dalgarno sequence embedded in the 3" primer. Lowercase sequences anneal to the template plasmid, while uppercase sequences are homologous to the E. coli chromosome either within or immediately adjacent to ftsW and serve as sites for the homologous recombination event that replaces ftsW with ftsW::kan. In the case of the 3" primer, almost all of the flanking homology is within murG. Because recombination depended largely on primer-encoded homology in murD and murG, the PCR fragment was directed to the chromosome rather than the complementing plasmid.
Fourth, the ftsW::kan fragment was introduced by electroporation into DY330/pDSW406 cells that had been made proficient for recombination as described (46). Recombinants were obtained on LB supplemented with kanamycin to select for ftsW::kan, chloramphenicol to select for pDSW406, and arabinose to induce the complementing copy of ftsW on pDSW406. Presumptive recombinants were arabinose dependent.
Three PCR tests were used to confirm that ftsW had been replaced by ftsW::kan. Primer P320 (5"CCAGCCTTGATCAGTTCAAGA, binds in murD) and primer P321 (5"GCCATTAGATGGTGCGCAACC, binds in murG) are predicted to give a ≈1.66-kb fragment from a murD-ftsW::kan-murG recombinant, but will give a ≈1.41-kb product from the murD-ftsW-murG parent. Primers K1 and K2 (12) bind within the kan gene and should yield a 593-bp product in the recombinant but no product in the parent. Finally, PCR with P321 plus K2 should produce a 979-bp product in the recombinant but no product in the parent. We tested five putative recombinants by PCR and found all of them to be correct. One recombinant was saved and designated EC850. This strain is temperature sensitive owing to the presence of the defective λ prophage inserted at attB.
The fifth and final step in constructing our FtsW depletion strain was to remove the prophage. This was accomplished in a couple of ways. In several cases we used generalized transduction to move gfp fusion genes into EC850. The gfp fusions resided at the attB locus of the donors in these crosses, so the resulting transductants lost the prophage in the recipient. In addition, we constructed a derivative of EC850 in which the defective prophage was removed by homologous recombination (46). Primers P327 (5"CTCCGGTCTTAATCGACAGCAAC) and P355 (5"GAGGTACCAGGCGCGGTTTGATC) were used to amplify DNA from the attB region of MG1655, a nonlysogen. The ≈2.3-kb PCR product was used to electroporate EC850 cells that had been made recombination proficient. Recombinants were selected at 42°C on LB plates that contained kanamycin, chloramphenicol, and arabinose. Numerous temperature-independent isolates were obtained. We chose seven and demonstrated by PCR with primers P327 and P355 that all had lost the prophage; the product obtained was 2.3 kb, whereas no product was obtained from EC850 because the presence of the prophage places the priming sites ≈9.5 kb apart. One isolate obtained by this procedure was designated EC912.
Complementation by transduction.
A P1 lysate grown on strain EC912 (ftsW::kan/pDSW406) was used to transduce EC791 (gfp-ftsW Ampr) to kanamycin resistance on LB containing kanamycin and ampicillin, with and without 1 mM IPTG. Eight Kanr transductants were chosen at random, made phage free, and analyzed by PCR using primer pair P307 and P308. All isolates yielded the expected ≈1.6-kb PCR product indicative of ftsW::kan.
Microscopy.
Fluorescence micrographs were recorded on an Olympus BX60 microscope equipped with a 100× UPlanApo objective (numerical aperture, 1.35). Images were captured using a black-and-white Spot 2 cooled charge-coupled device camera (Diagnostic Instruments, Sterling Heights, Mich.) with a KAF1400E chip (class 2), a Uniblitz shutter, and a personal computer with Image-Pro software version 4.1 (Media Cybernetics, Silver Spring, Md.). Filter sets were from Chroma Technology Corp (Brattleburo, Vt.). For GFP and fluorescein, the filter set comprised a 450- to 490-nm excitation filter, 495-nm dichroic mirror (long pass), and a 500- to 550-nm emission filter. The DAPI (4′,6′-diamidino-2-phenylindole) filter set was a 340- to 380-nm excitation filter, 400-nm dichroic mirror (long pass), and a 435- to 485-nm emission filter. Typical exposure times were in the range of 2 to 8 s for GFP, 1 to 2 s for fluorescein, and 0.1 s for DAPI.
Image-Pro was also used to measure cells and to quantitate fluorescence signal in transects across the long axis of cells. Images were prepared for publication using Canvas 5 (Deneba Software, Miami, Fla.). The “levels” command was used to darken the background and make the fluorescent bands appear brighter, so that the printed images would match their appearance on a computer screen, where they are backlit and exhibit strong contrast. Because such processing raises issues regarding the quality of the original data, we include either direct quantitation of the fluorescence signal or appropriate fluorescence standards (control cells) in the relevant figures.
Growth and processing of cells for protein localization experiments.
Experiments for localization of GFP-FtsW were done essentially as described (44). For experiments involving depletion of FtsW, overnight cultures that had been grown in LB containing antibiotics and 0.2% arabinose were inoculated 1:10 or 1:100 into fresh medium containing chloramphenicol, 0.02% arabinose, and IPTG. Cultures were grown at 30°C to optical density at 600 nm (OD600) of ≈0.5. Cells were washed once with LB and inoculated 1:400 into medium containing either 0.2% arabinose or 0.2% glucose. Samples were fixed at various times; filaments of a useful length typically formed after about 4 to 5 h, at which time the OD600 was ≈0.5.
Immunofluorescence microscopy was performed on fixed cells that were processed essentially as described (34). The primary antibody was anti-FtsZ rabbit serum raised against a hexahistidine-tagged FtsZ protein (Covance, Denver, Pa.), used at a dilution of 1:20,000. The secondary antibody was fluorescein-conjugated goat anti-rabbit immunoglobulin G (IgG) (heavy and light chains) (Molecular Probes, Eugene, Oreg.) used at 0.01 mg/ml.
Western blotting.
Immunoblotting of GFP-FtsW and GFP-FtsI was performed essentially as described (44). Because FtsW aggregates upon heating, extracts were made by resuspending cells in BugBuster with benzonase (Novagen, Madison, Wis.) and incubating at room temperature for 10 min, at which time an equal volume of 2× sample buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was added and samples were loaded onto gels containing 8% polyacrylamide. The affinity-purified primary antibody against GFP (a gift of W. Margolin) was used at a dilution of 1:2,500 and incubated with blots for 16 h. The anti-GFP was incubated with an E. coli cell lysate before use (14) to reduce cross-reaction with cellular proteins. The secondary antibody was a 1:8,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit IgG (Pierce Chemical Co., Rockford, Ill.) and was incubated with blots for 4 h. Blots were developed with SuperSignal chemiluminescent reagent and exposed to film or quantified using a Typhoon 8600 imager.
RESULTS
Localization of Fts proteins after depletion of FtsW.
To determine which Fts proteins are dependent upon FtsW for localization to the division site, we constructed a strain in which ftsW expression was under control of an arabinose-dependent promoter (16). Thus, in the presence of arabinose the strain grew similarly to the wild type, but after several generations in the presence of glucose the strain became depleted of FtsW and formed filaments (Fig. 2). The filaments ultimately lysed (not shown).
FIG. 2.
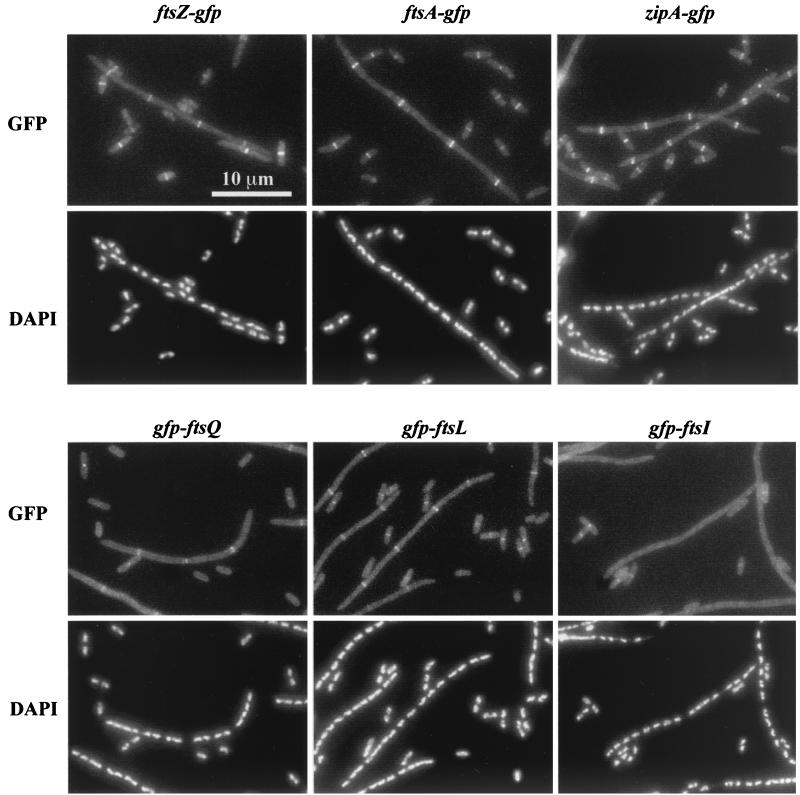
Localization of division proteins in an FtsW depletion background. Strains expressing the indicated GFP fusion were grown in the presence of arabinose or glucose to induce or prevent expression of ftsW, respectively. Cells were fixed and then mixed prior to spreading on slides for microscopy. The short (arabinose-grown) cells often display septal localization of the target division protein and serve as internal controls for microscopy and subsequent image processing. The strains shown are EC856, EC892, EC904, EC860, EC881, and EC861.
Localization experiments were done using derivatives of the FtsW depletion strain that expressed gfp fusions to various division genes (8, 14, 44). These strains were merodiploids in that the gfp fusions were expressed from a defective prophage at the lambda attachment site, while a wild-type copy of the corresponding fts gene was expressed from the normal chromosomal locus. These strains were grown in LB with IPTG to induce expression of the gfp fusion and either arabinose or glucose to induce or repress ftsW, respectively. Cells from both growth conditions were fixed with cross-linking agents, mixed together, and examined by fluorescence microscopy. Mixing resulted in normal-length cells containing FtsW lying next to filaments that were depleted of FtsW. Because the normal-length cells often displayed localization of the GFP-Fts fusion protein, they served as internal controls for microscopy and subsequent image processing. The results are summarized in Fig. 2 and Table 2. Depletion of FtsW had only a minimal effect (about twofold) on localization of FtsZ, FtsA, ZipA, FtsQ, and FtsL. In contrast, GFP-FtsI essentially failed to localize in FtsW depletion filaments. These findings suggest that FtsW localizes after FtsL and before FtsI.
TABLE 2.
Localization of Fts proteins in FtsW depletion backgrounds
| GFP fusion | Sugar | No. of cells scored | Average cell length (μm) | % of cells with the indicated no. of rings
|
Spacing of ringsa (μm/ring) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | ≥5 | |||||
| FtsZ-GFP | Arabinose | 175 | 2.7 | 30 | 70 | 0 | 0 | 0 | 0 | 3.8 |
| Glucose | 146 | 22 | 0 | 26 | 23 | 11 | 15 | 21 | 7.3 | |
| ZipA-GFP | Arabinose | 257 | 2.9 | 39 | 61 | 0 | 0 | 0 | 0 | 4.8 |
| Glucose | 190 | 30 | 0 | 3 | 26 | 16 | 24 | 31 | 5.8 | |
| FtsA-GFP | Arabinose | 244 | 3.6 | 14 | 86 | 0 | 0 | 0 | 0 | 3.6 |
| Glucose | 177 | 28 | 0 | 12 | 31 | 11 | 17 | 29 | 7.3 | |
| GFP-FtsQ | Arabinose | 307 | 3.3 | 44 | 56 | 0 | 0 | 0 | 0 | 5.9 |
| Glucose | 216 | 23 | 4 | 28 | 33 | 17 | 13 | 6 | 10.3 | |
| GFP-FtsL | Arabinose | 338 | 2.9 | 48 | 52 | 0 | 0 | 0 | 0 | 6.7 |
| Glucose | 176 | 24 | 3 | 22 | 43 | 17 | 7 | 7 | 11 | |
| GFP-FtsI | Arabinose | 298 | 3.3 | 44 | 56 | 0 | 0 | 0 | 0 | 6.0 |
| Glucose | 124 | 22 | 95 | 5 | 0 | 0 | 0 | 0 | 460 | |
The spacing is a measure of the frequency of rings per unit cell mass and is calculated by dividing the total number of rings (not shown) into the total length of cells or filaments scored (column 3 multiplied by column 4).
We confirmed the FtsZ localization results using immunofluorescence microscopy to visualize FtsZ in EC912, an FtsW depletion strain that did not harbor a gfp-ftsW fusion gene (not shown). About 95% of the filaments, which averaged 26 μm in length, had a Z-ring. The longest filament that we observed, 60 μm, had 11 Z-rings. When averaged over the population, 1 Z-ring was observed per 9.5 μm of filament length, similar to the value of 7.3 μm/ring obtained with FtsZ-GFP (last column of Table 2).
Fusion of gfp to ftsW.
FtsW of E. coli is 415 amino acids long and is predicted by TopPred II to span the cytoplasmic membrane 10 times, with both the N and C termini located in the cytoplasm (9). We attempted to fuse a bright variant of GFP, gfpmt2 (10), at either the N or C terminus of FtsW. The gfp-ftsW fusion was readily obtained, but the ftsW-gfp fusion was not, despite multiple attempts, suggesting that it might be toxic. The relative ease with which we recovered an N-terminal fusion is consistent with the finding that a short form of FtsW lacking the first 30 amino acids complements both temperature-sensitive and null alleles of ftsW (6, 25).
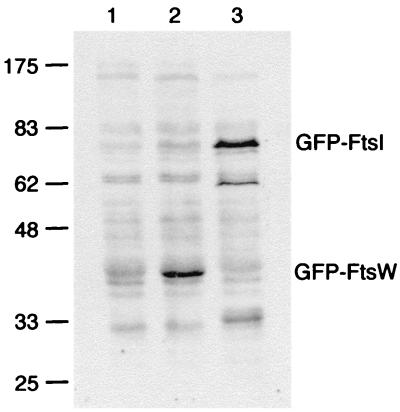
The gfp-ftsW fusion was integrated into the chromosome in single-copy at the λ attachment site using the shuttle vector λ InCh (5), creating an ftsW/gfp-ftsW merodiploid. Expression of gfp-ftsW was under control of a weak IPTG-regulated promoter (44), while expression of the wild-type copy of ftsW remained under control of its native promoter(s). Since we lack antibody to FtsW, we were not able to assess the level of gfp-ftsW expression relative to ftsW. But by using anti-GFP, we could compare the expression of gfp-ftsW to gfp-ftsI under conditions in which GFP-FtsI is present at a level of ≈200 molecules per cell (44). As expected, the amount of GFP-FtsW increased with increasing IPTG, ranging from <30 molecules/cell in the absence of IPTG to ≈200 molecules per cell with 1 mM IPTG (Fig. 3).
FIG. 3.
Steady-state levels of GFP-FtsW fusion protein as determined by Western blotting with an anti-GFP antibody. The positions of molecular mass standards (in kilodaltons) and the positions of GFP-FtsW and GFP-FtsI are indicated. Strain EC791 (P209-gfp-ftsW) was grown with 0 mM IPTG (lane 1) or 1 mM IPTG (lane 2). Strain EC436 (P207-gfp-ftsI) was grown with 2.5 μM IPTG (lane 3), which results in production of ≈200 molecules of GFP-FtsI per cell (44). Promoter P209 is much weaker than P207 (44). Chemiluminescence signal was recorded on film, which was then scanned to prepare this figure. In addition, chemiluminescence signal from bands corresponding to the GFP fusion proteins was quantified directly on a Typhoon 8600 imager. The values were 480 for GFP-FtsW uninduced, 3,300 for GFP-FtsW induced, and 3,500 for GFP-FtsI (arbitrary units, corrected for background).
Two unusual aspects of the Western blot are worth noting. GFP-FtsW ran at an apparent molecular mass of about 43 kDa, considerably smaller than its predicted mass of 73.5 kDa. In addition, we observed the highest levels of GFP-FtsW when samples were maintained at room temperature before loading the gels. Even mild heating (37°C for 5 min) caused the protein to aggregate and prevented entry into the gel (data not shown). Both the aberrant mobility and heat sensitivity of FtsW have been reported previously (26). Despite the problem with aggregation, we think our Western blots provide realistic estimates of the amount of GFP-FtsW because exposure times of several seconds were needed to photograph GFP-FtsW in fixed cells. These exposure times are similar to those needed to visualize other GFP fusions present at a few hundred molecules per cell (8, 14, 44).
To determine whether the gfp-ftsW fusion was functional, we determined whether it could complement a null mutation in the native ftsW gene. This was done by transducing the ftsW/gfp-ftsW merodiploid to kanamycin resistance with P1 phage grown on an ftsW::kan donor strain that carried a complementing copy of ftsW on a plasmid. Several hundred transductants were obtained, whereas no colonies resulted from control crosses into an ftsW/gfp-ftsI recipient. The transductants were shown via PCR to have acquired the expected ftsW::kan insertion. Interestingly, equal numbers of transductants were obtained in the presence and absence of IPTG, which induces expression of the gfp-ftsW fusion, implying that low levels of GFP-FtsW (Fig. 3) are sufficient to support division. Nevertheless, in the absence of IPTG, colonies were small and contained numerous filamentous cells, while colonies from plates with IPTG appeared normal and contained normal-length cells. We conclude that our gfp-ftsW fusion functions in cell division.
Localization of GFP-FtsW in wild-type and fts mutant backgrounds.
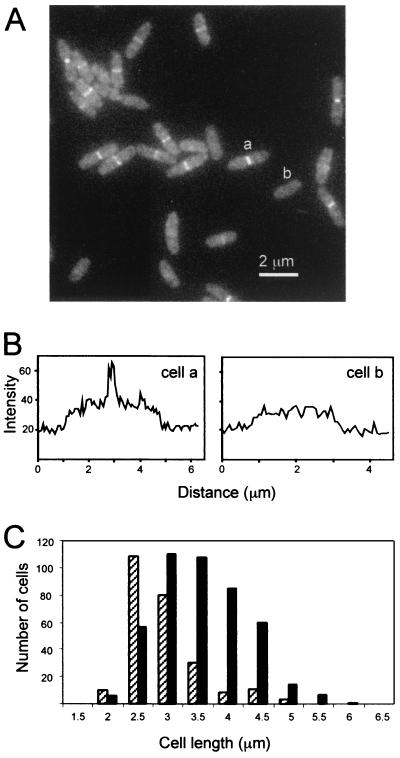
To determine the subcellular location of GFP-FtsW, an ftsW/gfp-ftsW merodiploid strain was grown in LB containing 1 mM IPTG to an OD600 of ≈0.3, fixed with cross-linking agents, and examined by fluorescence microscopy (Fig. 4). These cells often had a bright band of fluorescence at the midcell, confirming a previous report of septal localization based on immunofluorescence microscopy (43). We did not observe localization to sites other than the midcell. About 65% of the cells (n = 696) in this experiment exhibited septal localization of GFP-FtsW. In some experiments ≈50% of the cells displayed septal localization. The cells with FtsW at the division site were on average longer than those without, indicating that FtsW, like other Fts proteins, is recruited to the division site during the later stages of cell growth and remains at that site until division is complete.
FIG. 4.
Localization of GFP-FtsW during exponential growth. (A) GFP imaged in a field of cells of strain EC791. (B) Quantitation of the fluorescence in cells that do (a) and do not (b) display septal localization of GFP-FtsW. Transects run lengthwise through the cells indicated in panel A, starting about 1 μm before (lower left) and extending about 1 μm beyond (upper right) each cell. (C) Size distribution of cells that display septal localization of GFP-FtsW. A total of 696 cells were measured and scored for the presence (black bar) or absence (hatched bar) of a fluorescent band extending across the midcell.
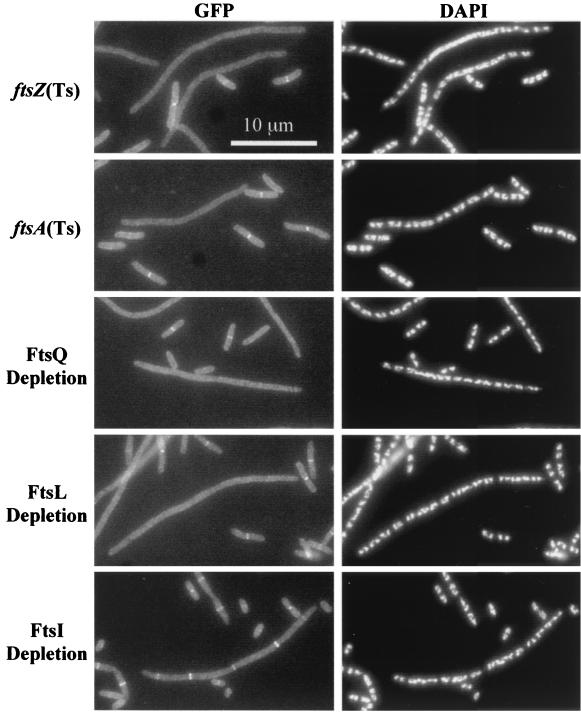
We introduced our gfp-ftsW fusion into strains that had conditional alleles of ftsZ, ftsA, ftsQ, ftsL, or ftsI and assayed localization of the fusion protein in filaments that formed upon inactivation or depletion of the indicated essential division proteins. These results are summarized in Fig. 5 and Table 3. GFP-FtsW did not localize in filaments formed upon shift of an ftsZ84(Ts) strain to 42°C for 1 h, whereas it did localize when this strain was grown at 30°C (compare filamentous and short cells in top panel of Fig. 5). DAPI staining verified that the filaments exhibited proper nucleoid segregation even though GFP-FtsW failed to localize, so we infer that the filamentous cells were healthy. Inactivation of ftsA [using the ftsA12(Ts) allele] also prevented localization of GFP-FtsW. In addition, GFP-FtsW localization was nearly eliminated upon depletion of FtsQ or FtsL in strains that expressed the respective genes under control of an arabinose-dependent promoter. In contrast, filaments formed by depleting cells of FtsI showed only about a twofold decrease in GFP-FtsW localization. Taken together, these results imply that FtsW localizes after FtsL but before FtsI.
FIG. 5.
Dependency of GFP-FtsW localization on other Fts proteins. In each case the indicated fts mutant was grown in parallel under permissive and nonpermissive conditions (60 min for temperature-sensitive mutants). Cells were fixed and then mixed prior to spreading on slides for microscopy. The short (permissive condition) cells often display septal localization of GFP-FtsW and serve as internal controls for microscopy and subsequent image processing. The strains shown are EC788, EC787, EC799, EC798, and EC829.
TABLE 3.
Localization of GFP-FtsW in fts mutant backgroundsa
| Strain | Growth temp (°C) or sugar | No. of cells scored | Average cell length (μm) | Total no. of FtsW rings | % of cells with a ring(s) | Spacing of rings (μm/ring) |
|---|---|---|---|---|---|---|
| Wild type | 30 | 205 | 4.2 | 101 | 49 | 8.5 |
| 42 | 320 | 3.7 | 154 | 48 | 7.5 | |
| Temperature-sensitive mutants | ||||||
| ftsZ(Ts) | 30 | 182 | 6.0 | 73 | 40 | 15 |
| 42 (60 min) | 197 | 20 | 0 | 0 | >3,900 | |
| ftsA(Ts) | 30 | 151 | 4.9 | 75 | 47 | 9.9 |
| 42 (60 min) | 197 | 20 | 2 | 1 | 2,000 | |
| Depletion strains | ||||||
| FtsQ depletion | Arabinose | 199 | 4.2 | 106 | 53 | 7.9 |
| Glucose | 156 | 32 | 5 | 3 | 1,000 | |
| FtsL depletion | Arabinose | 232 | 5.7 | 146 | 63 | 9.1 |
| Glucose | 127 | 32 | 7 | 5 | 580 | |
| FtsI depletion | Arabinose | 188 | 3.1 | 93 | 50 | 6.3 |
| Glucose | 131 | 27 | 355 | 95 | 9.8 |
See Table 2, footnote a.
DISCUSSION
Our findings demonstrate that FtsW is a late recruit to the division site of E. coli, localizing after FtsL and before FtsI (Fig. 1). Three lines of evidence support this conclusion. First, 50 to 65% of cells in a population in exponential growth (doubling time, ≈30 min) displayed septal localization of GFP-FtsW. This value is similar to what we reported for the late recruit FtsI (44), but much less than the ≈90% typically observed for early recruits such as FtsZ, FtsA, and ZipA (2, 3, 17, 29). Second, GFP-FtsW did not localize in filaments that lack functional FtsZ, FtsA, FtsQ, or FtsL, but the fusion protein localized well in filaments depleted of FtsI. Finally, although filaments formed upon depletion of FtsW exhibited fairly normal localization of FtsZ, FtsA, ZipA, FtsQ, and FtsL, the septum-specific transpeptidase FtsI did not localize in such filaments.
An important implication of FtsW's being a late recruit to the division site is that FtsW probably has little if any role in regulating the assembly and/or stability of FtsZ rings. This was an issue because of a previous report that depletion of FtsW resulted in the production of filaments with few if any Z-rings (6). In our hands, >90% of FtsW depletion filaments had at least one Z-ring, as determined by immunofluorescence microscopy, and 100% had at least one Z-ring, as determined using GFP. Overall, depletion of FtsW reduced the frequency of Z-rings per unit cell mass only about twofold.
A similar modest decrease has been observed in several laboratories upon inactivation of many other Fts proteins (2, 8, 14, 18, 27, 34, 44), so FtsW is not special in this regard. Although the basis of this twofold reduction has yet to be determined, we would have expected to see a more profound defect if FtsW were uniquely important among the Fts proteins in stabilization of Z-rings. Finally, it is worth noting that four Z-ring-dependent proteins localized well in the FtsW depletion filaments, FtsA (3, 29), ZipA (18, 27), FtsQ (8), and FtsL (14). This observation provides strong evidence for the presence of Z-rings that is independent of the methods used to detect Z-rings directly.
Why our FtsW depletion filaments contain many more Z-rings than those studied previously (6) is a matter of conjecture. We doubt that the difference originates in the methods used to detect Z-rings because we obtained similar results using two techniques, GFP and immunofluorescence. One potential basis for the discrepancy is the extent of depletion, which ultimately leads to cell death, making it difficult to separate primary phenotypes from secondary effects of declining health. We deal with this problem in two ways. First, we always stain nucleoids with DAPI and only score protein localization in filaments that exhibit normal segregation (7, 44). In general, we find that filaments with aberrant nucleoids, either highly compacted or extremely diffuse, become common about 1.5 h after onset of filamentation. Such filaments tend not to exhibit localization of Fts proteins, and we presume that they are unhealthy. Second, we take comfort when independent experimental approaches produce compatible results, such as demonstrated here with requirements for localization of GFP-FtsW on the one hand and the localization of several Fts proteins in an FtsW depletion strain on the other.
FtsW belongs to a large family of bacterial membrane proteins, the SEDS family (21), that appear to work together with specific class B penicillin-binding proteins during the final stages of peptidoglycan assembly (31). FtsW's partner appears to be FtsI (PBP3). Whereas it is well established that FtsI is a transpeptidase involved in cross-linking peptidoglycan in the division septum (4, 38), no specific function has previously been demonstrated for FtsW.
Our results show that FtsW is required to recruit FtsI to its site of action, the division site. To our knowledge, this is the first demonstration of a well-defined role by which a SEDS protein supports its cognate transpeptidase and raises the possibility that other SEDS proteins are also required for proper localization of their cognate transpeptidases. At least in E. coli, peptidoglycan synthesis during elongation is diffuse, whereas during division most synthesis occurs in a zone at the midcell (13, 45). Thus, sites of active peptidoglycan synthesis are harder to define during elongation (and presumably sporulation) than during septum assembly, so our hypothesis may prove difficult to test.
Whether FtsW recruits FtsI via a protein-protein interaction or some less direct mechanism remains to be established. Likewise, FtsW might have functions in addition to its role in localization of FtsI to the septal ring. The final stages of peptidoglycan synthesis involve flipping of a lipid-linked precursor from the cytoplasm to the periplasm and its incorporation into the cell wall via transglycosylation and transpeptidation reactions (42). Interestingly, of these reactions, FtsI apparently only catalyzes transpeptidation (1).
Acknowledgments
We thank J. Chen for sharing results prior to publication. We thank J. Chen, D. Court, J.-M. Ghigo, H. Hara, and B. Wanner for strains and plasmids and W. Margolin for affinity-purified antibody against GFP. We thank D. Boyd, J. Chen, H. Kaback, and members of the Weiss laboratory for helpful discussions.
This work was supported by a grant from the National Institutes of Health (GM59893).
REFERENCES
- 1.Adam, M., C. Fraipont, N. Rhazi, M. Nguyen-Distèche, B. Lakaye, J. M. Frère, B. Devreese, J. Van Beeumen, Y. van Heijenoort, J. van Heijenoort, and J. M. Ghuysen. 1997. The bimodular G57-V577 polypeptide chain of the class B penicillin-binding protein 3 of Escherichia coli catalyzes peptide bond formation from thiolesters and does not catalyze glycan chain polymerization from the lipid II intermediate. J. Bacteriol. 179:6005-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall, S. G., E. Bi, and J. Lutkenhaus. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Addinall, S. G., and J. Lutkenhaus. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178:7167-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botta, G. A., and J. T. Park. 1981. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J. Bacteriol. 145:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, D. S., M. M. Khattar, S. G. Addinall, J. Lutkenhaus, and W. D. Donachie. 1997. ftsW is an essential cell-division gene in Escherichia coli. Mol. Microbiol. 24:1263-1273. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK but not FtsN for colocalization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed]
- 8.Chen, J. C., D. S. Weiss, J. M. Ghigo, and J. Beckwith. 1999. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol. 181:521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 10.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 11.Daniel, R. A., S. Drake, C. E. Buchanan, R. Scholle, and J. Errington. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J. Mol. Biol. 235:209-220. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Pedro, M. A., J. C. Quintela, J. V. Höltje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghigo, J. M., D. S. Weiss, J. C. Chen, J. C. Yarrow, and J. Beckwith. 1999. Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol. 31:725-737. [DOI] [PubMed] [Google Scholar]
- 15.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale, C. A., and P. A. de Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175-185. [DOI] [PubMed] [Google Scholar]
- 18.Hale, C. A., and P. A. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara, H., S. Yasuda, K. Horiuchi, and J. T. Park. 1997. A promoter for the first nine genes of the Escherichia coli mra cluster of cell division and cell envelope biosynthesis genes, including ftsI and ftsW. J. Bacteriol. 179:5802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henriques, A. O., H. de Lencastre, and P. J. Piggot. 1992. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie 74:735-748. [DOI] [PubMed] [Google Scholar]
- 21.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda, M., T. Sato, M. Wachi, H. K. Jung, F. Ishino, Y. Kobayashi, and M. Matsuhashi. 1989. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 171:6375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishino, F., H. K. Jung, M. Ikeda, M. Doi, M. Wachi, and M. Matsuhashi. 1989. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J. Bacteriol. 171:5523-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joris, B., G. Dive, A. Henriques, P. J. Piggot, and J. M. Ghuysen. 1990. The life-cycle proteins RodA of Escherichia coli and SpoVE of Bacillus subtilis have very similar primary structures. Mol. Microbiol. 4:513-517. [DOI] [PubMed] [Google Scholar]
- 25.Khattar, M. M., S. G. Addinall, K. H. Stedul, D. S. Boyle, J. Lutkenhaus, and W. D. Donachie. 1997. Two polypeptide products of the Escherichia coli cell division gene ftsW and a possible role for FtsW in FtsZ function. J. Bacteriol. 179:784-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khattar, M. M., K. J. Begg, W. D. Donachie, K. Ehlert, and J. V. Höltje. 1994. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J. Bacteriol. 176:7140-7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Z., A. Mukherjee, and J. Lutkenhaus. 1999. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 31:1853-1861. [DOI] [PubMed] [Google Scholar]
- 28.Lutkenhaus, J., and S. G. Addinall. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93-116. [DOI] [PubMed] [Google Scholar]
- 29.Ma, X., D. W. Ehrhardt, and W. Margolin. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93:12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 31.Matsuhashi, M., M. Wachi, and F. Ishino. 1990. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res. Microbiol. 141:89-103. [DOI] [PubMed] [Google Scholar]
- 32.Matsuzawa, H., S. Asoh, K. Kunai, K. Muraiso, A. Takasuga, and T. Ohta. 1989. Nucleotide sequence of the rodA gene, responsible for the rod shape of Escherichia coli: rodA and the pbpA gene, encoding penicillin-binding protein 2, constitute the rodA operon. J. Bacteriol. 171:558-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pogliano, J., K. Pogliano, D. S. Weiss, R. Losick, and J. Beckwith. 1997. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. USA 94:559-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothfield, L., S. Justice, and J. Garcia-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spratt, B. G. 1977. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J. Bacteriol. 131:293-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spratt, B. G., A. Boyd, and N. Stoker. 1980. Defective and plaque-forming lambda transducing bacteriophage carrying penicillin-binding protein-cell shape genes: genetic and physical mapping and identification of gene products from the lip-dacA-rodA-pbpA-leuS region of the Escherichia coli chromosome. J. Bacteriol. 143:569-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauch, K. L., and J. Beckwith. 1988. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc. Natl. Acad. Sci. USA 85:1576-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamaki, S., H. Matsuzawa, and M. Matsuhashi. 1980. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J. Bacteriol. 141:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Heijenoort, J. 1996. Murein synthesis, p. 1025-1034. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 43.Wang, L., M. M. Khattar, W. D. Donachie, and J. Lutkenhaus. 1998. FtsI and FtsW are localized to the septum in Escherichia coli. J. Bacteriol. 180:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wientjes, F. B., and N. Nanninga. 1989. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J. Bacteriol. 171:3412-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]