Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that may cause severe infections in humans and other vertebrates. In addition, a human clinical isolate of P. aeruginosa, strain PA14, also causes disease in a variety of nonvertebrate hosts, including plants, Caenorhabditis elegans, and the greater wax moth, Galleria mellonella. This has led to the development of a multihost pathogenesis system in which plants, nematodes, and insects have been used as adjuncts to animal models for the identification of P. aeruginosa virulence factors. Another approach to identifying virulence genes in bacteria is to take advantage of the natural differences in pathogenicity between isolates of the same species and to use a subtractive hybridization technique to recover relevant genomic differences. The sequenced strain of P. aeruginosa, strain PAO1, has substantial differences in virulence from strain PA14 in several of the multihost models of pathogenicity, and we have utilized the technique of representational difference analysis (RDA) to directly identify genomic differences between P. aeruginosa strains PA14 and PAO1. We have found that the pilC, pilA, and uvrD genes in strain PA14 differ substantially from their counterparts in strain PAO1. In addition, we have recovered a gene homologous to the ybtQ gene from Yersinia, which is specifically present in strain PA14 but absent in strain PAO1. Mutation of the ybtQ homolog in P. aeruginosa strain PA14 significantly attenuates the virulence of this strain in both G. mellonella and a burned mouse model of sepsis to levels comparable to those seen with PAO1. This suggests that the increased virulence of P. aeruginosa strain PA14 compared to PAO1 may relate to specific genomic differences identifiable by RDA.
Pseudomonas aeruginosa is an opportunistic pathogen that frequently causes severe systemic infections, particularly in patients with cystic fibrosis, burns, and immunosuppression (2, 3, 48, 52). The versatility of P. aeruginosa may be a consequence of its ability to produce a wide variety of both cell-associated and extracellular virulence factors. Cell-associated virulence factors include pili, flagellae, lipopolysaccharide, a type III secretion system, and alginate. Secreted products include low-molecular-weight toxins, such as phenazines, rhamnolipid, and cyanide, and numerous protein virulence factors, including ADP-ribosylating enyzmes, proteases, and phospholipases (12, 16, 42, 62). Additional virulence factors include proteins required for the expression or the secretion of these molecules, often in response to particular environmental stimuli (17, 18).
A soil inhabitant, P. aeruginosa is widely distributed in the natural environment and can also act as a plant pathogen. Recently, Rahme et al. (45) have exploited the broad host range of this pathogen and have shown that a clinical isolate of P. aeruginosa, strain PA14, is capable of causing disease both in an Arabidopsis thaliana leaf infiltration model and in a mouse full-thickness skin thermal burn model. Furthermore, mutations in a variety of PA14 genes reduced the virulence of this strain for both plants and mice, suggesting that at least some of the mechanisms of pathogenesis of P. aeruginosa infection may be conserved in evolutionarily divergent hosts (44).
These results have subsequently been extended to show that P. aeruginosa can also act as a pathogen for a variety of additional nonvertebrate hosts, including Caenorhabditis elegans (33, 58, 59), Drosophila melanogaster (13; Mahajan-Miklos et al., unpublished data), and the greater wax moth, Galleria mellonella (25). This has led to the development of a multihost pathogenesis system in which plants, nematodes, and insects have been used as adjuncts to animal models for the identification and study of bacterial virulence factors of P. aeruginosa. Indeed, random transposon mutagenesis has been used to create 8,000 individual mutants of strain PA14, and these clones have been screened for reduced virulence compared to the wild-type parent by using a plant leaf infection model (46), in fast- and slow-killing assays in C. elegans (33, 58, 59) and in the G. mellonella model system (25). The relevance to mammalian pathogenesis of virulence factors identified using these screens has been confirmed by using a mouse full-thickness burn model (54). Remarkably, of 20 genes in P. aeruginosa strain PA14 that are required for pathogenesis in at least one of the three different invertebrate hosts (a plant, a nematode, or an insect), 17 were also required for full pathogenicity in a mouse burn model. Of these 17 genes, eight encode novel proteins and three encode proteins not previously known to be involved in bacterial pathogenesis. Many classes of genes were identified, including genes encoding proteins involved in transcriptional and posttranscriptional regulation, efflux systems, biosynthetic enzymes involved in phenazine production, and proteins of unknown function (32). Since the 8,000 mutants screened so far represent approximately 25 to 33% of the total number that need to be screened to ensure a 95% probability of testing a mutation in each P. aeruginosa gene in these assays, many additional factors involved in the pathogenicity of P. aeruginosa may remain to be discovered.
Another approach to identifying virulence factors in bacteria is to take advantage of naturally occurring differences in pathogenicity between isolates of the same species, utilizing one of a variety of subtractive techniques to recover genes present in one isolate but not the other, such as those found on pathogenicity islands. One such technique is representational difference analysis (RDA), a procedure involving subtractive hybridization and kinetic enrichment that has been used previously to recover differences between two complex genomes, including identifying the genome of human herpesvirus 8 in the tissue of patients with Kaposi's sarcoma (29, 30). RDA has also been modified to utilize cDNA as the starting material, thereby allowing analysis of differential gene expression (24). Recently, RDA was adapted for use in detecting and cloning genomic differences between two closely related bacterial species or isolates of the same species (8, 41, 60).
In this study, we have taken advantage of the fact that the sequenced strain of P. aeruginosa, strain PAO1, has substantial differences in virulence from strain PA14 in several of the multihost models of pathogenicity described above. Strain PAO1 has been used in numerous previous investigations related to the genetics and virulence of P. aeruginosa (21, 36, 37, 38). We utilized the technique of RDA to directly detect genomic differences between P. aeruginosa strains PA14 and PAO1 and examined whether any of the identified genomic differences relate to the differences in virulence between the two strains in the various models of pathogenesis. We found that the pilC, pilA, and uvrD genes in P. aeruginosa strain PA14 differ substantially from their counterparts in strain PAO1. We also recovered a ybtQ gene homolog that is specifically present in strain PA14 but absent in strain PAO1. We demonstrated that mutation of the ybtQ homolog in P. aeruginosa strain PA14 significantly attenuates the virulence of this strain in both G. mellonella and a burned mouse model of sepsis, to levels comparable to those seen with PAO1, suggesting that the increased virulence of P. aeruginosa strain PA14 compared to PAO1 may relate to specific genomic differences identifiable by RDA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are shown in Table 1. P. aeruginosa strain PA14 is a human clinical isolate used for identification of novel virulence-related genes (45). P. aeruginosa strain PAO1 has been studied extensively in many laboratories (21, 36, 37, 38), and the genomic sequence has been determined (55). All strains were maintained at −70°C in Luria-Bertani (LB) medium containing 15% glycerol. LB broth and agar were used for the growth of P. aeruginosa and Escherichia coli strains at 37°C. Chelex-100 treated syncase medium (15) was used for the low-iron growth conditions for the P. aeruginosa ybtQ mutant. Antibiotic concentrations were as follows: for E. coli, ampicillin at 100 μg ml−1; for P. aeruginosa, rifampin at 100 μg ml−1 and carbenicillin at 300 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotypea | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Wild-type laboratory strain | Lab collection |
| PA14 | Human clinical isolate; Rifr | 45 |
| JY2M | pilC mutant of PA14; Rifr Cbr | This study |
| JY5M | pilA mutant of PA14; Rifr Cbr | This study |
| JY11M | ybtQ mutant of PA14; Rifr Cbr | This study |
| JY15M | uvrD mutant of PA14; Rifr Cbr | This study |
| E. coli | ||
| DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 λ− φ80 dlacZΔM15 | Gibco BRL |
| SM10 | thi-I thi leu tonA lacY supE recA::RP4-2-Tc::Mu; Kmr | Lab collection |
| Plasmids | ||
| pUC19 | Cloning vector; Apr | Lab collection |
| pEX18 | Suicide vector for P. aeruginosa; oriT+sacB+, gene replacement vector with MCS from pUC18; Apr | 23 |
| pGEM-T Easy | PCR cloning vector; Apr | Promega |
| pJY1 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY2 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY3 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY4 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY5 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY6 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY8 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY10 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY11 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY12 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY13 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY15 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY16 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY17 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY19 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY20 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY22 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY23 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY24 | pEX18 derivative carrying RDA product; Apr | This study |
| pJY25 | pEX18 derivative carrying RDA product; Apr | This study |
| pJYP25 | pGEM-T Easy with a 3.0-kbp fragment of pilABC gene cluster; Apr | This study |
| pJY2A | pGEM-T Easy with a 1,160-bp IPCR product of pilC; Apr | This study |
| pJY5A | pGEM-T Easy with a 1,030-bp IPCR product of pilA; Apr | This study |
| pJY15A | pGEM-T Easy with a 1,150-bp IPCR product of uvrD; Apr | This study |
| pJY2-1 | pGEM-T Easy with a 600-bp internal fragment of pilC; Apr | This study |
| pJY5-1 | pGEM-T Easy with a 500-bp internal fragment of pilA; Apr | This study |
| pJY11-1 | pGEM-T Easy with a 1.3-kbp internal fragment of ybtQ; Apr | This study |
| pJY15-1 | pGEM-T Easy with a 1.1-kbp internal fragment of uvrD; Apr | This study |
| pJY2M | pEX18 derivative carrying a 600-bp internal fragment of pilC; Apr | This study |
| pJY5M | pEX18 derivative carrying a 500-bp internal fragment of pilA; Apr | This study |
| pJY11M | pEX18 derivative carrying a 1.3-kbp internal fragment of ybtQ; Apr | This study |
| pJY15M | pEX18 derivative carrying a 1.1-kb internal fragment of uvrD; Apr | This study |
| pJYYBT | 2.1-kb EcoRI fragment containing the intact ybtQ gene cloned into pUC19 | This study |
Apr, ampicillin resistance; Cbr, carbenicillin resistance; Kmr, kanamycin resistance; Rifr, rifampin resistance.
Molecular genetic techniques.
Isolation of plasmid DNA, restriction enzyme digests, and agarose gel electrophoresis were performed according to standard molecular biological techniques (50). Restriction enzyme-digested chromosomal and plasmid DNA fragments were separated on 0.8% agarose gels; fragments of interest were cut from the gel under UV illumination and purified with a QIAEX II gel extraction kit (Qiagen Inc., Valencia, Calif.).
DNA sequencing was performed at the Massachusetts General Hospital Department of Molecular Biology in the DNA Sequencing Core Facility using ABI Prism DiTerminator cycle sequencing with AmpliTaq DNA polymerase FS and an ABI377 DNA sequencer (Perkin-Elmer/Applied Biosystems, Foster City, Calif.). The sequences obtained were analyzed against the P. aeruginosa PAO1 genome sequence generated by the P. aeruginosa genome project (Cystic Fibrosis Foundation and Pathogenesis Corporation) and at the National Center for Biotechnology Information via the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST).
Representational difference analysis.
The procedure of RDA was originally described by Lisitsyn et al. (29) and adapted to the comparison of bacterial strains by Tinsley and Nassif (60) and Calia et al. (8). In the present study, 2 μg of DNA from P. aeruginosa strain PA14 was cleaved with Sau3AI, precipitated with ethanol-sodium acetate, and ligated for 18 h at 16°C with 5 nmol of the oligonucleotide adapter pair (RSau24 [5"-AGCACTCTCCAGCCTCTCACCGCA-3"] and RSau12 [5"-GATCTGCGGTGA-3"]). The mixture was gel purified on 2% low-melting-point agarose (taking fragments above 200 bp) to remove unincorporated primers, phenol purified, precipitated, and redissolved in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) at a DNA concentration of 0.1 μg μl−1. This procedure results in DNA fragments whose two 5" ends are covalently linked to the 24-base adapter (RSau24). To prepare the subtracting DNA, chromosomal DNA of P. aeruginosa strain PAO1 was sheared by repeated passage through a 30-gauge hypodermic needle to give fragments ranging from about 3 to 10 kbp. The DNA was repurified by phenol extraction, precipitated, and redissolved in TE buffer at a concentration of 0.1 μg μl−1. The first subtractive hybridization was performed with 40 μg of P. aeruginosa strain PAO1 subtracting DNA and 400 ng of Sau3AI-digested, RSau adapter-linked PA14 DNA fragments. The DNA was mixed, ethanol precipitated, and redissolved in 8 μl of EE buffer (10 mM N-[2-hydroxyethyl]piperazine-N"-[3-propanesulfonic acid], 1 mM EDTA [pH 8.0]). The liquid was overlaid with 30 μl of mineral oil, denatured at 100°C for 2 min, and then placed at 67°C. After the addition of 2 μl of 5 M NaCl to the aqueous phase, the mixture was left to hybridize at 67°C for 20 h. The reaction mixture was then diluted 10-fold with preheated EE buffer-NaCl and immediately placed on ice. A portion of the subtraction mixture (10μl) was diluted into 400 μl of PCR mix [67 mM Tris-HCl (pH 8.9)], 16 mM (NH4)2SO4, 4 mM MgCl2, 10 mM β-mercaptoethanol, 0.1 mg of bovine serum albumin ml−1, a 0.125 mM concentration of each deoxynucleoside triphosphate, 15 U of Taq polymerase per ml) to fill in the ends corresponding to the 24-base adapter (RSau24). The reaction mixture was diluted 10-fold further, and PCR amplification was performed on 400 μl of the dilution. After denaturation for 5 min at 94°C and addition of the appropriate 24-base oligonucleotide (RSau24 primer), the mixture was amplified by PCR (10 cycles of 1 min at 95°C and 3 min at 72°C, with the last cycle followed by an extension at 72°C for 10 min). Single-stranded DNA molecules present after amplification were degraded by a 30-min incubation with 20 U of mung bean nuclease, diluted (1:5) in 50 mM Tris-HCl (pH 8.9), and then heated to 95°C for 5 min to inactivate the enzyme. A portion (40 μl) of the solution was further amplified for 20 cycles under the conditions described above. The amplified P. aeruginosa DNA fragments were separated by agarose gel electrophoresis from the primers and high-molecular-weight subtracting DNA. The RSau adapters were cleaved from the PCR products by digestion with Sau3AI, and 2 nmol of the second-round adapters (JSau24 [5"-ACCGACGTCGACTATCCATGAACA-3"] and JSau12 [5"-GATCTGTTCATG-3"]) were ligated with 2 μg of the first-round difference products in a volume of 50 μl. The ligated fragments were gel purified, phenol extracted, and ethanol precipitated. A second round of subtractive hybridization and PCR enrichment was performed with 400 ng of first-round products religated to the JSau adapters and 40 μg of sheared DNA from P. aeruginosa strain PAO1 as described above. Fragments amplified from the second round were cleaved with Sau3AI, gel purified, and cloned into the pEX18 vector (23) digested with BamHI. The recombinant plasmids were maintained in E. coli strain DH5α. DNA sequences of the RDA products corresponding to the inserted DNA were determined with primers flanking the polylinker site of pEX18. The pool of fragments obtained after the second round of RDA was also tested by Southern hybridization to ensure that they were absent in PAO1 and present in PA14.
Southern hybridization.
Bacteria from 10 ml of LB broth were resuspended in 1 ml of 10 mM Tris-HCl (pH 8.0)-10 mM EDTA-100 mM NaCl containing 2 μg of RNase A. After addition of 50μl of 20% sodium dodecyl sulfate and incubation at 65°C for 30 min, the mixture was digested for 2 h at 37°C with proteinase K (100 μg). The solution was then extracted once with an equal volume of phenol (pH 8.0), twice with phenol-chloroform-isopropanol (25:24:1), and once with chloroform-isopropanol (24:1). The solution was overlaid with an equal volume of ethanol and cooled to 0°C, and the DNA was spooled from the interface by mixing with a glass Pasteur pipette. DNA was washed in 70% ethanol, partially dried, and redissolved in TE buffer. The concentration of DNA was determined by UV spectrophotometry.
After digestion of purified DNA with Sau3AI and separation by agarose gel electrophoresis, Southern blotting was performed by capillary transfer onto Hybond-N+ positively charged nylon membranes (Amersham Pharmacia Biotech., Piscataway, N.J.). Hybridization of labeled probes and detection were performed with an enhanced chemiluminescence kit (Amersham) as described by the manufacturer.
IPCR.
To obtain chromosomal sequences flanking RDA fragments from pilC, pilA, and uvrD, we performed partial inverse PCR (IPCR) (39). P. aeruginosa strain PA14 chromosomal DNA (10 μg) was partially digested with Sau3AI at 2 U μg−1 for 1 h. The reactions were stopped by heating at 65°C for 20 min, and an aliquot of the reaction mixture was run on a 0.8% agarose gel to check the extent of cutting. The rest of the DNA was ethanol precipitated and resuspended in 1× ligation buffer to a concentration of 5 ng μl−1. T4 DNA ligase was added, and chromosomal fragments were allowed to self-ligate at 22°C for 4 h. Ligation was stopped by heating to 65°C for 20 min followed by phenol extraction, ethanol precipitation, and resuspension in TE buffer at a DNA concentration of 100 ng μl−1. Two primers (5"-CATTTAGGGAAGCTCATCA-3" and 5"-GAACTGTGGGACCACTTTTATC-3" [pilC], 5"-CTAGTGAAAGGGCAGGCCT-3" and 5"-GGCATGCAAGATGCTTTA-3" [pilA], 5"-ACTCTTCTTCAAGTTCGGA-3" and 5"-CAGATGCAGGGCAAGTTCT-3" [uvrD]) facing outwards from the RDA sequences of the pilC, pilA, and uvrD genes were used to carry out each IPCR. The PCR reaction mixture (20 μl) contained 200 ng of religated DNA, 0.2 mM concentrations of each deoxynucleoside triphosphate, 2 mM magnesium, 1 μM concentrations of each primer, and 1 U of Taq DNA polymerase in 1× buffer supplied by the manufacturer. PCR was performed for 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min. The largest IPCR products (1,160 bp, pilC; 1,030 bp, pilA; and 1,150 bp, uvrD) were excised from a gel, purified with the QIAEX II gel extraction kit, and cloned into the PCR cloning vector pGEM-T Easy, to generate pJY2A, pJY5A, and pJY15A. DNA sequencing confirmed plasmids carrying the correct insert corresponding to each RDA product.
To confirm that the pilC fragment in pJY2A and the pilA fragment in pJY5A were located near each other in the pilABC gene cluster, a 3.0-kbp fragment of pilABC was generated by PCR using the JY2F primer (5"-TGATGAGCTTCCCTAAATG-3"; 5" sequence of pJY2) in combination with the JY5G primer (5"-ACTGGACATAGGGGGTAAG-3"; 3" sequence of pJY5). The amplified product was cloned into the PCR cloning vector pGEM-T Easy and designated pJYP25.
Cloning of the ybtQ gene from a plasmid library.
Chromosomal DNA of P. aeruginosa strain PA14 was fully digested with various restriction enzymes, electrophoresed, and transferred to Hybond-N+ positively charged nylon membranes (Amersham). Southern blot analysis was performed as described above, using the insert of plasmid pJY11 as a probe; digestion with EcoRI gave a single hybridizing fragment of 2.1 kbp. We digested chromosomal DNA of PA14 with EcoRI, recovered 2.0 to 3.0-kbp fragments from the agarose gel after electrophoresis and extracted these with the QIAEX II gel extraction kit. The recovered DNA fragments were ligated into the EcoRI site of pUC19, and the resulting plasmids were transformed into ultracompetent E. coli DH5α. This library of plasmids was screened by colony blot hybridization (50) using the insert DNA of plasmid pJY11 as the probe. Individual positive plasmid clones with appropriate insert sizes were verified by Southern blot hybridization, and the DNA sequence of the 2,126-bp insert in plasmid pJYYBT was determined. DNA and deduced amino acid sequences were analyzed with CloneMap version 2.11 (CGC Scientific, Inc., Ballwin, Mo.) and the WU-Blast2 program in EMBL (European Bioinformatics Institute) (http://www2.ebi.ac.uk/blast2/). Motif analysis of deduced amino acid sequences was performed with the Expert Protein Analysis System (ExPASy) Molecular Biology Server (http://www.expasy.ch).
Strain constructions.
P. aeruginosa strain PA14 pilC, pilA, ybtQ, and uvrD mutants JY2M, JY5M, JY11M, and JY15M were constructed by inserting a suicide vector, containing an internal fragment of each gene, into the chromosome of PA14. A 700-bp internal fragment of pilC from PA14 was amplified by PCR (94°C for 3 min, 25 cycles at 94°C for 1 min, 50°C for 2 min, 72°C for 5 min, 72°C for 10 min), using primers JY2R (5"-GCAGCAAGGTCAAAGGAGAG-3") and JY2L (5"-TGAGCTTCCCTAAATGCAAAAG-3"), and cloned into the PCR cloning vector pGEM-T Easy vector to create plasmid pJY2-1. The 500-bp, 1.3-kbp, and 1.1-kbp internal fragments of pilA, ybtQ, and uvrD genes were similarly amplified with primer pairs JY5R (5"-GAAAGGCTTTACCTTGAT-3") and JY5L (5"-AGGAGCGAAACGAGCCG-3"), JY11R (5"-CTACGCAATCATGGCAGTA-3") and JY11L (5"-CGATTCCATGCAGCCTGTGT-3"), and JY15R (5"-CACGCATGCATTGTAGCGA-3") and JY15L (5"-GATCGGTAGCGCAAAACT-3"), respectively, and cloned into pGEM-T Easy to generate plasmids pJY5-1, pJY11-1, and pJY15-1. The SacI-SphI fragments from pJY2-1, pJY5-1, pJY11-1, and pJY15-1 were cloned into the SacI and SphI sites in the polylinker of pEX18 to generate plasmids pJY2M, pJY5M, pJY11M, and pJY15M. Plasmid constructions were verified by DNA sequencing.
Plasmids pJY2M, pJY5M, pJY11M, and pJY15M were transformed into E. coli SM10 and subsequently transferred to P. aeruginosa strain PA14 by conjugation. Carbenicillin- and rifampin-resistant transconjugants contained the mobilized plasmid integrated into the genome by homologous recombination. Insertional mutation was confirmed by Southern hybridization of chromosomal DNA of each mutant strain compared with PA14, using the inserts of plasmids pJY2, pJY5, pJY11, and pJY15 as probes.
Virulence testing.
We determined the virulence of various strains of P. aeruginosa for a number of nonvertebrate hosts, including G. mellonella (caterpillars) and C. elegans. To examine virulence in G. mellonella, overnight cultures were grown in LB broth, diluted 1:100 in the same medium, and grown to an optical density at 600 nm of 0.3 to 0.4. Cultures were pelleted and resuspended in 10 mM MgSO4. After dilution to an optical density at 600 nm of 0.1 with 10 mM MgSO4, serial 10-fold dilutions were made in 10 mM MgSO4 with 2 mg of rifampin per ml for P. aeruginosa strain PA14 (and derivatives) and 0.5 μg of ampicillin per ml for strain PAO1. A 10-μl Hamilton syringe was used to inject 5-μl aliquots into individual fifth-instar G. mellonella larvae (Van der Horst Wholesale, St. Marys, Ohio), via the hindmost left proleg. A series of 10-fold serial dilutions containing from 106 to 0 bacteria were injected into the G. mellonella larvae. Ten larvae were injected at each dilution, and larvae were scored as live or dead after 60 h at 25°C. Data from three independent experiments were combined. The Systat computer program was used to fit a curve to the infection data using the following formula: Y = A + (1 − A)/{1 + exp[B − G · ln(X)]}, where X is the number of bacteria injected, Y is the fraction of larvae killed by the infection, A is the fraction of larvae killed by control injections, and B and G are parameters which are varied for optimal fit of the curve to the data points. The 50% lethal dose (LD50) is calculated from the curve. Statistical significance of differences was determined by using Fisher's exact test. Slow killing assays in C. elegans were performed as described previously (58, 59).
To examine virulence of various strains in mice, a 5% total surface area burn was fashioned on the outstretched abdominal skin of 6-week-old male AKR/J mice weighing between 25 and 30 g (The Jackson Laboratories, Bar Harbor, Maine). Immediately following the burn, mice were injected subcutaneously with 5 × 105 CFU of the P. aeruginosa strain being analyzed, and the number of animals that died as a result of sepsis was monitored each day for 10 days. For each strain, data from two independent experiments (eight mice per experiment) were combined (except that P. aeruginosa strain PAO1 was tested only once). Animal study protocols were reviewed and approved by the Institutional Animal Care and Use Committee.
Nucleotide sequence accession numbers.
The sequences of the P. aeruginosa PA14 ybtQ, uvrD, pilC, and pilA genes have been submitted to the GenBank database; accession numbers are AY049068, AY049069, AY049070, and AY049071.
RESULTS
Construction of a library of fragments of P. aeruginosa PA14 DNA not found in the genome of PAO1.
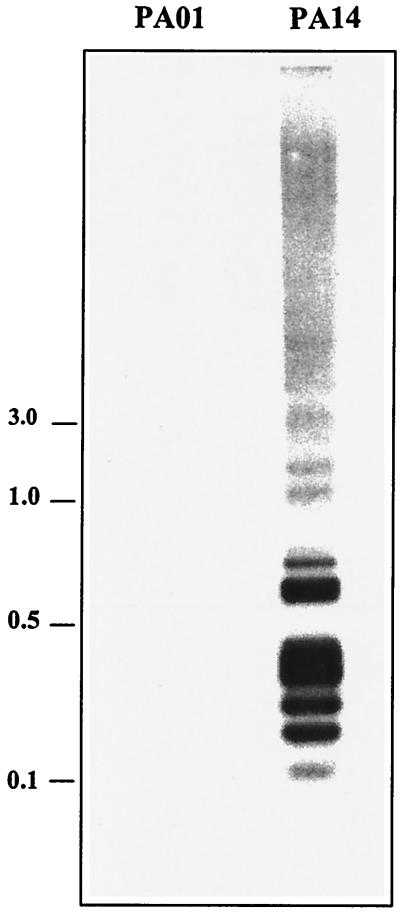
Using RDA, we subtracted total genomic DNA of the sequenced strain, P. aeruginosa PAO1, from strain PA14. Following two rounds of subtractive and kinetic enrichment, agarose gel electrophoresis of the amplified difference products revealed a small number of DNA fragments between 100 and 800 bp, with bands in the 200- to 500-bp range present in the greatest abundance (Fig. 1). We confirmed that the sequences amplified by RDA were specific to P. aeruginosa PA14 by using the pool of second-round difference products to probe genomic DNA of P. aeruginosa strains PAO1 and PA14 (Fig. 2).
FIG. 1.
Agarose gel electrophoresis (2% gel) of RDA products between P. aeruginosa PA14 and PAO1. The tester DNA amplicons before DNA hybridization and amplification (a) and the difference products after the first (b) and second (c) DNA hybridization-PCR amplification steps are shown. M, Molecular size markers; sizes are on the left, in base pairs.
FIG. 2.
Southern blot analysis confirms that the second-round RDA products are unique to PA14. Chromosomal DNA was isolated from P. aeruginosa strains PAO1 and PA14, digested with Sau3AI, separated on a 0.8% agarose gel, transferred to a membrane, and hybridized with a labeled pool of the second-round products. Molecular sizes are on the left, in kilobase pairs.
We ligated the pool of second-round difference products into pEX18 and recovered a total of 57 clones. Of these, the insert sequences of 20 representative clones, designated pJY, were determined using primers flanking the polylinker site of pEX18 (Table 2). When the sequences of these 20 inserts were compared to the P. aeruginosa PAO1 genome sequence (http://www.pseudomonas.com/GenomeSearch.asp), none showed a match, as expected from the Southern analysis results. Of the 20 clones chosen for further analysis, 14 showed homology to known genes, while 5 had no significant matches by BLAST search (http://www.ncbi.nlm.nih.goy/blast) (Table 2). One clone, pJY25, was a sibling of another (pJY13).
TABLE 2.
Homologies of the P. aeruginosa PA14 strain-specific RDA clones
| Clone | Length (bp) | Homology with BLASTX | GenBank accession no. |
|---|---|---|---|
| pJY1 | 195 | Ubiquitin-specific protease Ubp7p of Saccharomyces cerevisiae | NC001141 |
| pJY2 | 175 | PilC of P. aeruginosa | M32066 |
| pJY3 | 138 | Chlorophyll b synthetase of Dunaliella salina | AB021312 |
| pJY4 | 338 | KIAA0054 gene product; helicase of Homo sapiens | XP008261 |
| pJY5 | 348 | Type 4 pilin of P. aeruginosa | L37109 |
| pJY6 | 180 | Unknown | |
| pJY8 | 397 | Hypothetical protein Y68A4A.10 of C. elegans | AL021503 |
| pJY10 | 117 | Unknown | |
| pJY11 | 196 | YbtQ, ABC transporter, ATP-binding component of Y. pestis | AF091251 |
| pJY12 | 234 | Unknown | |
| pJY13 | 215 | Alpha-1 tubulin of C. elegans | D16439 |
| pJY15 | 368 | UvrD, DNA helicase of C. trachomatis | AE001331 |
| pJY16 | 116 | Unknown | |
| pJY17 | 230 | Patched-related proteins of C. elegans | AC006670 |
| pJY19 | 274 | Unknown protein of Pasteurella multocida | AE006141 |
| pJY20 | 128 | PilC of N. gonorrhoeae | AJ00121 |
| pJY22 | 217 | Unknown | |
| pJY23 | 264 | Nuclear receptor NHR-18 of C. elegans | AF083232 |
| pJY24 | 366 | Hypothetical 119.5K protein of Micrococcus luteus | JQ0405 |
| pJY25 | 215 | Alpha-1 tubulin of C. elegans | D16439 |
We focused our attention on the inserts in pJY2, pJY5, pJY11, and pJY15, which BLAST searches revealed to be homologs, respectively, of the type IV fimbrial assembly protein, PilC, in P. aeruginosa, the type IV pilin, PilA, in P. aeruginosa, the ABC transporter protein, YbtQ, in Yersinia pestis, and the DNA helicase, UvrD, in Chlamydia trachomatis.
Cloning and sequencing of the pilA, pilC, and uvrD genes from P. aeruginosa strain PA14.
We recovered 1,160-, 1,030-, and 1,150-bp fragments containing P. aeruginosa strain PA14 pilC, pilA, and uvrD genes by inverse PCR and cloned these into pGEM-T Easy to construct plasmids pJY2A, pJY5A, and pJY15A, respectively.
The deduced amino acid sequence encoded by bp 1 to 1,160 in the insert of pJY2A showed 72% identity and 79% similarity by BLAST search to the type IV fimbrial assembly protein PilC in P. aeruginosa strain PAO1. A pairwise BLAST alignment (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html) of the PilC amino acid sequences from P. aeruginosa strains PAO1 and PA14 showed that these genes shared a number of variable regions separated by conserved domains; the RDA product originally isolated in pJY2 was derived from one of the variable regions that differ substantially between PA14 and PAO1.
A BLAST search using the insert in pJY5A showed 91% identity over 179 amino acids to the type IV pilin from the P. aeruginosa G7 and G9 strains (53). The N-terminal 30 amino acids showed significant homology to various other members of the type IV group A prepilins, including PilA of P. aeruginosa strain PAO1 (26), FimA of Dichelobacter nodosus (6), and PilE of Neisseria gonorrhoeae (5). The C-terminal regions of these proteins were more variable, and the RDA product originally isolated in pJY5 was from a region quite variable in the pilA genes of PA14 and PAO1. Overall, pilA genes from PA14 and PAO1 were only 53% similar to each other.
We recovered a 3.0-kbp fragment of the pilABC gene cluster from PA14 in plasmid pJYP25, and sequence analysis demonstrated that the pilC gene in pJY2A and the pilA gene in pJY5A were linked in a pilABC gene cluster that was otherwise nearly identical to the same cluster in PAO1.
A BLAST search of the insert in pJY15A showed high levels of similarity to the DNA helicase (UvrD) of C. trachomatis (27% identity and 45% similarity over 902 bp), the DNA helicase of Borrelia burgdorferi (22% identity and 42% similarity over 758 bp), and other DNA helicase family members from other organisms. P. aeruginosa strain PAO1 also has uvrD in its genome (http://www.pseudomonas.com/GenomeSearch.asp), but there is only 45% similarity between the uvrD genes found in P. aeruginosa strains PA14 and PAO1. The insert from pJY15A came from an area that was particularly divergent between the sequences of the single uvrD genes in each of these two strains.
Analysis of the ybtQ gene homolog in P. aeruginosa strain PA14.
DNA sequence analysis of pJY11 showed that the insert was 196 nucleotides in length and had homology to the ybtQ gene of Y. pestis. This insert was used to recover a 2.1-kbp fragment of P. aeruginosa strain PA14 chromosomal DNA overlapping the insert in pJY11, which was cloned into plasmid pJYYBT. Sequencing of pJYYBT revealed a 2,126-bp complete open reading frame, encoding a protein of 585 amino acids with a predicted molecular mass of 63,549 Da and pI of 9.57 (Fig. 3). BLAST search revealed that the highest homologies of this open reading frame were with the ABC transporter protein, YbtQ, of Y. pestis (24% identity and 38% similarity in a 1,061-bp overlap), the inner membrane ABC transporter, Irp7, of Yersinia enterocolitica (24% identity and 38% similarity over 962 bp), the ABC transporter protein, YbtP, of Y. pestis (21% identity and 36% similarity over 1,061 bp), and the ABC transporter protein, Irp6, of Y. enterocolitica (21% identity and 36% similarity over 1,061 bp). The genes encoding the Ybt systems of Y. pestis and Y. enterocolitica show >97% sequence identity (7, 19, 20, 47, 51), and ybtP and ybtQ in Y. pestis are orthologs of irp6 and irp7 in Y. enterocolitica. Similarity analyses of deduced amino acid sequences by the WU-Blast2 program in EMBL also revealed that the amino acid sequence of the protein encoded in pJYYBT showed 30.7, 31.2, 28.6, and 28.8% similarity to YbtQ, Irp7, YbtP, and Irp6, respectively.
FIG. 3.
Map and nucleotide sequence of ybtQ gene from P. aeruginosa strain PA14. The deduced protein sequence is shown below the coding region of ybtQ. Numbering refers to the letter of the nucleotide sequence. The 196-bp of the RDA product recovered in pJY11 is underlined. The asterisks bracket the ABC transporter domain, and the four boxed regions are conserved motifs of the ABC transporter domain (Walker A, ABC signature, Walker B, and an unnamed fourth motif), as defined by Linton and Higgins (28).
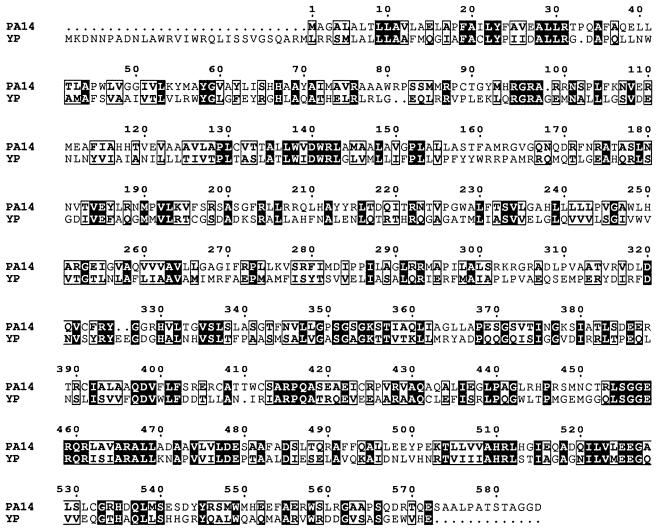
The N-terminal region of the YbtQ protein in P. aeruginosa PA14 was less homologous to YbtQ in Y. pestis than was the C-terminal region, which is hypothesized to act as a signal sensor (Fig. 4). Analysis of the deduced amino acid sequence of the open reading frame in pJYYBT by ExPASy suggested an amino-terminal hydrophobic region with six possible transmembrane segments, as well as an ABC transporter signature motif in the carboxy-terminal portion of the protein (Fig. 3) (1, 22, 28).
FIG. 4.
Alignment of YbtQ homolog in P. aeruginosa strain PA14 with YbtQ of Y. pestis. An alignment of the YbtQ homolog of P. aeruginosa PA14 with YbtQ of Y. pestis (YP) was generated with Clustal W. Dark boxes indicate identical residues and light boxes enclose similar residues. Numbers above each pair of sequences reflect the deduced protein sequence of the YbtQ homolog in P. aeruginosa PA14.
Nucleotide sequence comparison with the genome sequence database of PAO1 (http://www.pseudomonas.com/GenomeSearch.asp) revealed no sequences corresponding to the ybtQ homolog in PA14, suggesting that the entire ybtQ gene (and possibly surrounding sequences) is uniquely present in P. aeruginosa strain PA14 and absent from PAO1. We compared the ybtQ gene sequence from PA14 with the sequence of a recently described pathogenicity island, PAGI, present in the majority of pathogenic isolates of P. aeruginosa (27) but did not find any sequences homologous to ybtQ in PAGI.
Construction of mutations in the pilC, pilA, uvrD, and ybtQ genes of PA14 and determination of phenotypes.
We used standard methods to insertionally inactivate the pilC, pilA, uvrD, and ybtQ genes of P. aeruginosa strain PA14. Plasmids pJY2M, pJY5M, pJY11M, and pJY15M, encoding 5"- and 3"-truncated pilC, pilA, uvrD, and ybtQ, respectively, were integrated into the genome of P. aeruginosa strain PA14 by single homologous recombination events. The insertional disruption of the appropriate gene in the corresponding mutant was confirmed by Southern blot analysis.
Virulence testing of the four mutants in both nonvertebrate and mouse models of infection.
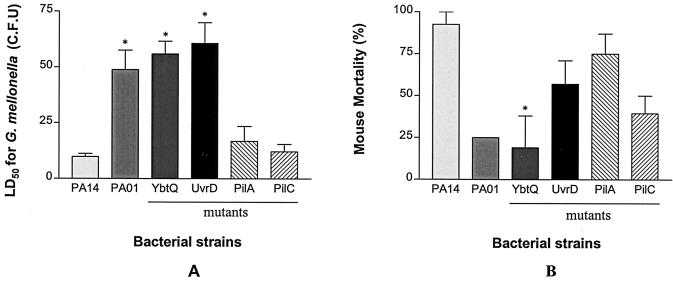
The strains PA14 and PAO1 show differences in a number of models of virulence, including the slow killing of C. elegans, killing of wax moth caterpillars, and a burned mouse model of sepsis; we wished to determine if the differences in virulence of these two P. aeruginosa strains in these various models related to the genetic differences uncovered by RDA. We therefore compared strains PA14, PAO1, and PA14 mutated in pilC, pilA, uvrD, and ybtQ in each of these virulence assays. In the C. elegans slow-killing model, there were no differences in virulence between strain PA14 and the mutants JY2M, JY5M, JY11M, and JY15M (data not shown). However, when we determined the LD50 of PA14, PAO1, and the four mutants in G. mellonella, the uvrD and ybtQ mutants of PA14 exhibited attenuated virulence, very similar to that of PAO1 (Fig. 5A). The LD50s for G. mellonella for these three strains were all approximately five-fold higher than for PA14 or the pilA or pilC mutants (P < 0.05).
FIG. 5.
Evaluation of mutants for virulence defects in wax moths and mice. (A) LD50s of P. aeruginosa strains PA14 and PAO1 and four mutants in fifth-instar G. mellonella larvae. Ten larvae were injected at each dilution (containing 0 to 106 bacteria), and larvae were scored as live or dead after 60 h at 25°C. The data are the means and standard deviations of three independent experiments. Asterisks indicate statistically significant differences from P. aeruginosa strain PA14. (B) Mortality in a burned mouse model for P. aeruginosa strains PA14 and PAO1 and for four mutants. Eight mice per experiment were injected subcutaneously with 5 × 105 CFU of each P. aeruginosa strain, and the number of animals that died as a result of sepsis was monitored each day for 10 days. The data are the means and standard deviations of two independent experiments for PA14 and the four mutants; PAO1 was tested once. The asterisk indicates a statistically significant difference from PA14.
Rahme et al. (44) reported that P. aeruginosa strain PA14 is more virulent in a burned mouse model of sepsis than PAO1, and we confirmed this observation (Fig. 5B). When we examined virulence of the four mutants of PA14 in this model, the ybtQ mutant was significantly attenuated, comparable to the attenuation seen with PAO1. Thus, a mutation of ybtQ, a gene missing in strain PAO1 (and identified by RDA), in P. aeruginosa strain PA14 significantly attenuates the virulence of strain PA14 in two different model systems of infection, to levels comparable to that seen with PAO1.
In Yersinia, YbtP and YbtQ are involved in iron acquisition from the environment. We examined the ability of the ybtQ mutant of PA14 (strain JY11), to grow in iron-chelated syncase medium. However, we detected no differences between growth of the mutant in this medium and that of wild-type strain PA14, perhaps reflecting the many other iron acquisition systems present in P. aeruginosa (data not shown).
DISCUSSION
In this study, we examined the hypothesis that differences in pathogenesis between specific strains of P. aeruginosa may reflect discrete genomic differences identifiable by RDA. Recently, RDA has been used to detect differences between virulent and avirulent Mycobacterium bovis strains (31) and between N. gonorrhoeae and Neisseria meningitidis (60). Previous studies have demonstrated that P. aeruginosa strain PA14 is significantly more virulent in a slow killing assay with C. elegans (58) and in a burned mouse model of infection (45) than the well-characterized and sequenced strain PAO1. We utilized these differences in pathogenesis, in combination with RDA, to examine specific genes in strain PA14 that might underlie differences in pathogenesis from strain PAO1. We identified differences in the pilC, pilA, and uvrD genes between P. aeruginosa strains PA14 and PAO1 and identified a ybtQ homolog in strain PA14 that was entirely missing in strain PAO1. We constructed mutations in each of these four genes and compared the pathogenesis of these mutants with that of the parent, PA14, and the reference strain, PAO1, in both nonvertebrate model systems and a mouse model of burn infection.
Mutations of the pilC and pilA genes in strain PA14 did not alter the virulence of the strains in the various model systems tested. The pilC and pilA genes in P. aeruginosa strain PA14 were located in the same pilABC cluster as in strain PAO1 but were divergent at the sequence level; additional homologs of pilA and pilC were not identified in P. aeruginosa strain PA14 by Southern blotting (data not shown). When the deduced amino acid sequence of PilC from P. aeruginosa strain PA14 was compared with that from strain PAO1, the proteins were shown to have very high homology in a number of conserved domains, separated by a number of variable regions. This structure is reminiscent of the PilE protein in N. gonorrhoeae, which has been shown to undergo pilus antigenic variation. This antigenic variation occurs by the high-frequency, unidirectional transfer of DNA sequences from one of several silent pilin loci (pilS genes) into the expressed pilin gene locus (pilE), resulting in changes in the primary pilin protein sequence (5, 35, 56, 57). Since we did not detect any additional pilC homologs in P. aeruginosa by Southern blot, the mechanism of divergence between the pilC genes in P. aeruginosa strains PA14 and PAO1 is currently uncertain.
The PA14 uvrD mutant was attenuated in virulence in G. mellonella but not in C. elegans or the burned mouse model, whereas the ybtQ mutant was attenuated in virulence in both G. mellonella and the burned mouse model (but not in C. elegans). Previous studies of P. aeruginosa mutants that were tested in both C. elegans and G. mellonella have shown that several mutants exhibit attenuated pathogenesis specifically in one host but not the other (25, 58, 59). This suggests that screening for pathogenesis in a variety of different hosts may lead to the identification of subsets of virulence factors that are critical for infections in specific hosts, as well as others important across hosts. For example, Mahajan-Miklos et al. (32) have suggested that screening for mutants of P. aeruginosa attenuated in a fast-killing assay of C. elegans under hyperosmolar and low-pH conditions may identify genes that are also important for virulence in a cystic fibrosis lung infection model.
P. aeruginosa utilizes several siderophores for high-affinity iron uptake, including pyoverdin and pyochelin (10, 11, 43). Pyoverdin production has been previously shown to be required for bacterial colonization of the lung in a rat infection model (43) and to correlate with lethality in a burned mouse infection model (34). Pyochelin has been shown to promote bacterial growth and lethality when injected into the peritoneal cavities of mice simultaneously with an avirulent mutant of P. aeruginosa strain PAO1, isolated by repetitive passage in mice (9).
Yersiniabactin, a phenolate-thiazole siderophore, was first purified from Y. enterocolitica and subsequently from Y. pestis. In Y. pestis, yersiniabactin may play a role in establishing infection at the site of a flea bite, as mutants that are unable to produce or transport yersiniabactin are avirulent in mice by peripheral routes of infection but fully virulent when infected intravenously (4, 61). Several of the genes needed for the biosynthesis and utilization of yersiniabactin are clustered in a pathogenicity island that is part of an unstable region on the Y. pestis chromosome, the pgm locus (4, 14, 19, 20, 40). All highly pathogenic species of Yersinia have similar genes clustered on a high-pathogenicity island.
Our results suggests that the ybtQ homolog that is present in strain PA14 but absent in strain PAO1 may at least partially explain the differences in pathogenesis of these two strains for the burned mouse model as well as for G. mellonella. One possibility is that P. aeruginosa strain PA14 contains a pathogenicity island encoding the ybtQ homolog and other genes absent from strain PAO1 and acquired by horizontal gene transfer. Extensive genomic rearrangements, as well as acquisition and loss of large blocks of DNA, have been previously demonstrated in different P. aeruginosa isolates (49). We have not yet recovered sufficient DNA flanking the ybtQ homolog to further examine the possibility of a pathogenicity island; such experiments are under way.
The role of the ybtQ homolog in the pathogenesis of P. aeruginosa strain PA14 for G. mellonella and mice is also uncertain. We were not able to demonstrate impaired growth under iron-limited conditions by the ybtQ mutant, but this may relate to the multiple siderophore systems available for use by P. aeruginosa during in vitro growth. Further experiments to examine the relationship between this gene, iron uptake, and pathogenesis of P. aeruginosa strain PA14 in various model systems are also under way.
Acknowledgments
We thank members of the Calderwood, Ausubel, and Rahme laboratories for helpful advice and Sachiko Miyata for helpful comments on the wax moth experiments.
This work was supported by a postdoctoral fellowship from the Korean Science and Engineering Foundation (KOSEF) to J.Y.C., by a Post-doctoral Research Fellowship for Physicians from the Howard Hughes Medical Institute to C.D.S., and by a research grant from Aventis Pharmaceuticals.
REFERENCES
- 1.Ames, G. F., C. S. Mimura, S. R. Holbrook, and V. Shyamala. 1992. Traffic ATPases: a superfamily of transport proteins operating from Escherichia coli to humans. Adv. Enzymol. Relat. Areas Mol. Biol. 65:1-47. [DOI] [PubMed] [Google Scholar]
- 2.Baltch, A. L. 1994. Pseudomonas bacteremia, p. 73-128. In R. P. Smith and A. L. Baltch (ed.), Pseudomonas aeruginosa infections and treatment. Marcel Dekker, New York, N.Y.
- 3.Bayer, A. S., and D. C. Norman. 1990. Valve site-specific pathogenetic differences between right-sided and left-sided bacterial endocarditis. Chest 98:200-205. [DOI] [PubMed] [Google Scholar]
- 4.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergstrom, S., K. Robbins, J. M. Koomey, and J. Swanson. 1986. Piliation control mechanisms in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 83:3890-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billington, S. J., and J. I. Rood. 1991. Sequence of fimbrial subunit-encoding genes from virulent and benign isolates of Dichelobacter (Bacteroides) nodosus. Gene 99:115-119. [DOI] [PubMed] [Google Scholar]
- 7.Buchrieser, C., C. Rusniok, L. Frangeul, E. Couve, A. Billault, F. Kunst, E. Carniel, and P. Glaser. 1999. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 67:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calia, K. E., M. K. Waldor, and S. B. Calderwood. 1998. Use of representational difference analysis to identify genomic differences between pathogenic strains of Vibrio cholerae. Infect. Immun. 66:849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, C. D. 1982. Effect of pyochelin on the virulence of Pseudomonas aeruginosa. Infect. Immun. 36:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, C. D., and P. Adams. 1985. Siderophore activity of pyoverdin for Pseudomonas aeruginosa. Infect. Immun. 48:130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, C. D., and R. Graham. 1979. Isolation of an iron-binding compound from Pseudomonas aeruginosa. J. Bacteriol. 137:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darby, C., C. L. Cosma, J. H. Thomas, and C. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fetherston, J. D., V. J. Bertolino, and R. D. Perry. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32:289-299. [DOI] [PubMed] [Google Scholar]
- 15.Finkelstein, R. A., P. Atthasampunna, M. Chulasamaya, and P. Charunmethee. 1966. Pathogenesis of experimental cholera: biologic activities of purified procholeragen A. J. Immunol. 96:440-449. [PubMed] [Google Scholar]
- 16.Galloway, D. R. 1991. Pseudomonas aeruginosa elastase and elastolysis revisited: recent developments. Mol. Microbiol. 5:2315-2321. [DOI] [PubMed] [Google Scholar]
- 17.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambello, M. J., S. Kaye, and B. H. Iglewski. 1993. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 61:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehring, A. M., E. DeMoll, J. D. Fetherston, I. Mori, G. F. Mayhew, F. R. Blattner, C. T. Walsh, and R. D. Perry. 1998. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 20.Gehring, A. M., I. Mori, R. D. Perry, and C. T. Walsh. 1998. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry 37:11637-11650. [DOI] [PubMed] [Google Scholar]
- 21.Hassett, D. J. 1996. Anaerobic production of alginate by Pseudomonas aeruginosa: alginate restricts diffusion of oxygen. J. Bacteriol. 178:7322-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 23.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 24.Hubank, M., and D. G. Schatz. 1994. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 22:5640-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jander, G., L. G. Rahme, and F. M. Ausubel. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182:3843-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, K., M. L. Parker, and S. Lory. 1986. Nucleotide sequence and transcriptional initiation site of two Pseudomonas aeruginosa pilin genes. J. Biol. Chem. 261:15703-15708. [PubMed] [Google Scholar]
- 27.Liang, X., X. Q. Pham, M. V. Olson, and S. Lory. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 29.Lisitsyn, N., N. Lisitsyn, and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 30.Lisitsyn, N., and M. Wigler. 1995. Representational difference analysis in detection of genetic lesions in cancer. Methods Enzymol. 254:291-304. [DOI] [PubMed] [Google Scholar]
- 31.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahajan-Miklos, S., L. R. Rahme, and F. M. Ausubel. 2000. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol. Microbiol. 37:981-988. [DOI] [PubMed] [Google Scholar]
- 33.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 34.Meyer, J. M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer, T. F., C. P. Gibbs, and R. Haas. 1990. Variation and control of protein expression in Neisseria. Annu. Rev. Microbiol. 44:451-477. [DOI] [PubMed] [Google Scholar]
- 36.Nicas, T. I., and B. H. Iglewski. 1985. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can. J. Microbiol. 31:387-392. [DOI] [PubMed] [Google Scholar]
- 37.Ohman, D. E., R. P. Burns, and B. H. Iglewski. 1980. Corneal infections in mice with toxin A and elastase mutants of Pseudomonas aeruginosa. J. Infect. Dis. 142:547-555. [DOI] [PubMed] [Google Scholar]
- 38.Ostroff, R. M., B. Wretlind, and M. L. Vasil. 1989. Mutations in the hemolytic-phospholipase C operon result in decreased virulence of Pseudomonas aeruginosa PAO1 grown under phosphate-limiting conditions. Infect. Immun. 57:1369-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pang, K. M., and, D. A. Knecht. 1997. Partial inverse PCR: a technique for cloning flanking sequences. BioTechniques 22:1046-1048. [DOI] [PubMed] [Google Scholar]
- 40.Pelludat, C., A. Rakin, C. A. Jacobi, S. Schubert, and J. Heesemann. 1998. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol. 180:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrin, A., X. Nassif, and C. Tinsley. 1999. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to the pathogenic Neisseria species. Infect. Immun. 67:6119-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters, J. E., and D. R. Galloway. 1990. Purification and characterization of an active fragment of the LasA protein from Pseudomonas aeruginosa: enhancement of elastase activity. J. Bacteriol. 172:2236-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poole, K., C. Dean, D. Heinrichs, S. Neshat, K. Krebes, L. Young, and L. Kilburn. 1996. Siderophore-mediated iron transport in Pseudomonas aeruginosa, p. 371-383. In T. Nakazawa, K. Furukawa, D. Haas, and S. Silver (ed.), Molecular biology of pseudomonads. American Society for Microbiology, Washington, D.C.
- 44.Rahme, L. G., F. M. Ausubel, H. Cao, E. Drenkard, B. C. Goumnerov, G. W. Lau, S. Mahajan-Miklos, J. Plotnikova, M. W. Tan, J. Tsongalis, C. L. Walendziewicz, and R. G. Tompkins. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97:8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 46.Rahme, L. G., M. W. Tan, L. Le, S. M. Wong, R. G. Tompkins, S. B. Calderwood, and F. M. Ausubel. 1997. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 94:13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rakin, A., C. Noelting, S. Schubert, and J. Heesemann. 1999. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun. 67:5265-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reyes, M. P., and A. M. Lerner. 1983. Current problems in the treatment of infective endocarditis due to Pseudomonas aeruginosa. Rev. Infect. Dis. 5:314-321. [DOI] [PubMed] [Google Scholar]
- 49.Romling, U., K. D. Schmidt, and B. Tummler. 1997. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J. Mol. Biol. 271:386-404. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, (2nd ed.) Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 51.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heesemann. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, J. J., S. M. Travis, E. P. Greenberg, and M. J. Welsh. 1996. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell 85:229-236. [DOI] [PubMed] [Google Scholar]
- 53.Spangenberg, C., R. Fislage, W. Sierralta, B. Tummler, and U. Romling. 1995. Comparison of type IV-pilin genes of Pseudomonas aeruginosa of various habitats has uncovered a novel unusual sequence. FEMS Microbiol. Lett. 125:265-274. [DOI] [PubMed] [Google Scholar]
- 54.Stevens, E. J., C. M. Ryan, J. S. Friedberg, R. L. Barnhill, M. L. Yarmush, and R. G. Tompkins. 1994. A quantitative model of invasive Pseudomonas infection in burn injury. J. Burn Care Rehabil. 15:232-235. [DOI] [PubMed] [Google Scholar]
- 55.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 56.Swanson, J., S. Bergstrom, O. Barrera, K. Robbins, and D. Corwin. 1985. Pilus-gonococcal variants. Evidence for multiple forms of piliation control. J. Exp. Med. 162:729-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swanson, J., S. Bergstrom, K. Robbins, O. Barrera, D. Corwin, and J. M. Koomey. 1986. Gene conversion involving the pilin structural gene correlates with pilus+ in equilibrium with pilus− changes in Neisseria gonorrhoeae. Cell 47:267-276. [DOI] [PubMed] [Google Scholar]
- 58.Tan, M. W., S. Mahajan-Miklos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tinsley, C. R., and X. Nassif. 1996. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc. Natl. Acad. Sci. USA 93:11109-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Une, T., and R. R. Brubaker. 1984. In vivo comparison of avirulent Vwa- and Pgm- or PstR phenotypes of yersiniae. Infect. Immun. 43:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wick, M. J., D. W. Frank, D. G. Storey, and B. H. Iglewski. 1990. Structure, function, and regulation of Pseudomonas aeruginosa exotoxin A. Annu. Rev. Microbiol. 44:335-363. [DOI] [PubMed] [Google Scholar]