Abstract
DNA sequence analysis and bioinformatic interpretations have identified two adjacent clusters of genes potentially involved in the formation of a bc1 complex and in the maturation of a cytochrome c-type protein in two strains (ATCC 19859 and ATCC 33020) of the acidophilic, chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans (formerly Thiobacillus ferrooxidans). Reverse transcriptase-PCR experiments suggest that the two clusters are organized as operons, and +1 start sites of transcription for the operons have been determined by primer extension experiments. Potential promoters have been identified. The presence of these operons lends support to a recent model of reverse electron flow and is consistent with previous reports of phenotypic switching in this bacterium.
Acidithiobacillus ferrooxidans, formerly Thiobacillus ferrooxidans (13), is a chemolithoautotrophic gram-negative bacterium that derives energy and electrons from the oxidation of ferrous iron and/or sulfur and various reduced sulfur compounds at pH 2 by using oxygen as the electron acceptor (11). It fixes CO2 by the Calvin-Bassham-Benson scheme and can also fix nitrogen under microaerophilic conditions. A. ferrooxidans has been reported to be able to grow on hydrogen as an energy source both aerobically (4) and anaerobically by the dissimilatory reduction of Fe(III) (14). It has been shown to be active in the solubilization of copper and in the processing of refractory gold ores in bioleaching operations and is also a contributor to acid mine drainage.
An intriguing problem is how A. ferrooxidans can generate the reduced pyridine nucleotides NADH and NADPH when using Fe(II) as the sole source of electrons and energy. The problem is that the standard reduction half-potential of the Fe2+/Fe3+ couple (pH 2) is +0.77 V while that of the NAD+/NADH couple (pH 7) is −0.32 V. Therefore, to generate NADH, it is necessary to “push” electrons “uphill” from Fe(II) to NAD+ against a thermodynamically favorable gradient.
It has been speculated that the energy required for this uphill electron flow could come primarily from the proton motive force resulting from an influx of protons across the A. ferrooxidans membrane since the exterior pH of the medium is 2 and that of the interior of the cell is 6.5 (12). It was further speculated that the pathway that the electrons take during uphill flow is from Fe(II) via a bc1 complex and a membrane-soluble quinone to a NAD+ reductase complex (12).
A downhill pathway of electrons in A. ferrooxidans has been described; in this pathway electrons derived from the oxidation of Fe(II) reduce oxygen to water via a series of electron carriers that remain inconclusively identified but that may include an outer membrane cytochrome (19), an iron-sulfur protein, blue copper protein rusticyanin, a cytochrome c4, and a terminal cytochrome aa3 oxidase (1). This is considered the downhill pathway because electrons flow from the reduction potential of Fe2+ (+0.77 V) to the more positive reduction potential of O2, which is thought to be about +0.82 V since the reduction of oxygen takes place in the cytoplasm at pH 6.5. It has been estimated that 95% of the electron flux during Fe(II) oxidation is via the downhill pathway (12). The major role of these electrons is thought to be the neutralization of the protons that enter through the ATP synthetase complex during the biosynthesis of ATP. The remaining 5% of the electrons are thought to pass uphill to reduce NADP+ to NADPH, which is required for fixing CO2 and for anabolic metabolism.
There is genetic evidence for the presence of genes encoding a candidate bc1 complex in A. ferrooxidans and for the role of the cytochrome c-type biogenesis proteins, ResB and ResC, in the maturation of a cytochrome c type (2). More recently, compelling biophysical evidence for the uphill flow of electrons in A. ferrooxidans via a bc1 complex and an NADH-Q oxidoreductase was presented (6). In this paper we describe the characterization of the petI operon of A. ferrooxidans, whose putative protein products appear to include the components of complex III, identified as being involved in uphill electron flow. In addition, we show that the petI operon encodes a short-chain dehydrogenase of the ribitol/glucose family and a diheme cytochrome c4. We also demonstrate that the resB and resC genes and hyp2, of unknown function, constitute an adjacent operon downstream from the petI operon.
Isolation and sequencing of the petI and res operons.
The previously described resB and resC genes (2) served as starting points to walk up and down the A. ferrooxidans ATCC 19859 genome by using the Genome Walker kit (Clontech Laboratories Inc.) according to the manufacturer's recommendations. In addition, segments of genes and several intergenic regions of A. ferrooxidans ATCC 33020 were amplified by PCR using primers based on the sequence of the petI and res operons deduced from the partial genome of A. ferrooxidans ATCC 23270 (www.tigr.org/). The resulting amplified products were cloned and sequenced using standard procedures (15).
Five genes, cycA, sdrA, petA, petB, and petC, were identified upstream of resB; these genes potentially encoded a diheme cytochrome c4, a short-chain dehydrogenase of the ribitol/glucose family, a cytochrome b, an FeS reductase (Rieske), and a cytochrome c1, respectively (Table 1). We propose the term petI operon for this gene complex.
TABLE 1.
Predicted properties of the proteins encoded by the petI operon
| Gene, protein (acc. no.)a | Molecular mass (kDa) of precursor/mature protein | No. of TMb helices | Featuresc |
|---|---|---|---|
| cycA, diheme cytochrome c4 (AAF76296) | 27.6/21.9 | 0 | Signal peptide; two heme-binding cytochrome c family binding sites (PS000169); cytochrome class 1 signature (PF00034) |
| sdrA, short-chain dehydrogenase (AAF76297) | —d/28.9 | 1 (weak) | SDR family signature (PS00061); short-chain alcohol dehydrogenase signature (PF00106); glucose/ribitol SDR signature (PR00081) |
| petA, Rieske (AAF76298) | —/22.7 | 1 | Rieske 2Fe-2S signature (PS00200, PR00162, PF00355) |
| petB, cytochrome b (AAF76299) | —/45.5 | 9 | Cytochrome b/b6 signature (PS00192, PF00033). |
| petC, cytochrome c1 (AAF79300) | 27.6/24.9 | 2 | Signal peptide; cytochrome c family heme binding site (PS00190); cytochrome c1 (PR00603, PF02167) |
Acc. no., GenBank accession number.
TM, transmembrane.
PS, Prosite; PF, Pfam; PR, prints. PS, PR, and PF were accessed through InterPro (http://www.ebi.ac.uk/interpro/).
—, not applicable.
Cotranscription of the genes fo the res and petI operons.
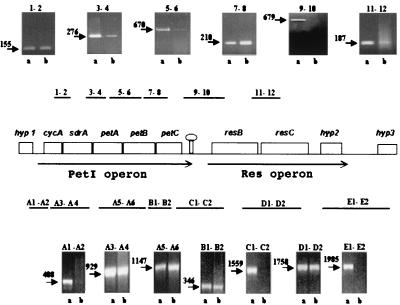
Reverse transcriptase-PCR (RT-PCR) was carried out in order to identify cotranscribed genes. A. ferrooxidans ATCC 19859 and ATCC 33020 were grown in Fe(II) at pH 2, 30°C (20), and RNA was isolated from cells in mid-log phase as previously described (9). RT-PCR was carried out as previously described and included various control reactions that accompanied each experiment (9). The locations, directions, and DNA sequences of the various primers used for both the RT and PCRs are available on request. RT-PCR products of the predicted sizes between the five genes of the proposed petI operon and between the three genes of the res operons were obtained (Fig. 1). In contrast, no RT-PCR products between the petI and res operons, between hypothetical gene hyp1 (upstream of the petI operon) and the petI operon, or between hypothetical gene hyp3 (downstream of the res operon) and the res operon could be detected. We conclude that the genes of the petI and res operons constitute two adjacent but separate transcriptional units in A. ferrooxidans ATCC 19859 and ATCC 33020. It has been suggested that A. ferrooxidans ATCC 33020 might not conform to the characteristics of an Acidithiobacillus sp. (13), but these results show that the organization and expression of its petI and res operons are similar to those of A. ferrooxidans ATCC 19859.
FIG. 1.
Map of the petI and res operons of A. ferrooxidans. A rho-independent-like secondary structure element is schematically represented between the operons. Above and below the map are shown both the locations of the regions amplified by RT-PCR (numbers, regions primed using RNA from strain ATCC 33020; numbered letters, regions primed using RNA from strain ATCC 19859) and also an analysis of the respective RT-PCR-amplified products by agarose gel electrophoresis (a, PCR amplification of the DNA to determine the expected size of the fragment between the respective primer pairs; b, RT-PCR amplification of the RNA). Expected sizes (in base pairs) of RT-PCR products (arrows) are given.
Initiation and termination of transcription; ribosome binding sites.
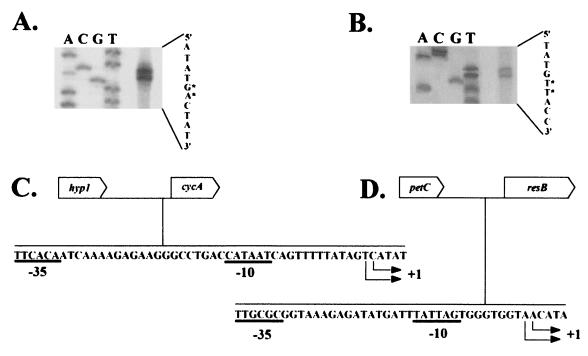
Primer extension analysis of RNA prepared from A. ferrooxidans ATCC 19859 was carried out (15) by using primers whose locations and sequences are available on request. Two equally likely adjacent possible start sites of transcription for the petI and res operons were detected (Fig. 2A and B). We have identified possible −10 and −35 regions in which the −10 sites are 12 and 8 bp upstream of the more upstream of the two possible start sites of the petI (Fig. 2C) and res (Fig. 2D) operons, respectively. It is difficult to experimentally verify the locations of the promoters in vivo in A. ferrooxidans due to the lack of reproducible experimental techniques for the genetic manipulation of the bacterium.
FIG. 2.
(A and B) Polyacrylamide gel electrophoresis analysis of the primer extension products resulting from primers specific for the pet and res operons, respectively. The primers used were A2 and C2 (see Fig. 1 for their genetic locations). Additional primers A4 and C1 (Fig. 1) were also used and confirm the location of the respective +1 start sites (data not shown). The four lanes of a DNA-sequencing gel are indicated with the appropriate nucleotide, A, C, G, or T. The deduced DNA sequence of the template strand is shown at the right. ∗, possible start sites of transcription. (C and D) DNA sequences of the coding strands upstream of cyc4 and resB. Possible +1 start sites of transcription and possible −10 and −35 promoter sites for the pet (C) and res (D) operons are indicated.
The mfold RNA secondary structure program (bioinfo.math.rpi.edu/∼mfold/rna/) identified a sequence between the petI and res operons that could potentially fold into an RNA secondary structure with a GC-rich stem and loop, followed by several Us, typical of a rho-independent-type transcription termination signal (see GenBank accession no. AF220499 for its location). No obvious rho-independent stop site could be detected in the intergenic space downstream of the res operon.
Potential ribosome binding sites for all the genes of the petI and res operons with the exceptions of resB and hyp2 have been identified and are described under GenBank accession no. AF220499.
Predicted properties of the putative gene products of the operon.
The deduced characteristics of the proteins encoded by the petI operon are summarized in Table 1. Deduced characteristics of the proteins encoded by the res operon have already been described (2). It is reasonable to propose that putative cytochrome c1, Rieske protein, and cytochrome b form part of a membrane-associated complex III electron transport system. In particular, the precursor cytochrome c1 exhibits a predicted molecular mass of 27.6 kDa (Table 1) similar to the 30-kDa cytochrome c1 proposed to be involved in uphill electron flow in A. ferrooxidans (5, 6). The role of ResB and ResC may be to aid in the maturation of the cytochrome c1 and/or the diheme cytochrome c4 encoded by the petI operon.
The petI operon also encodes a predicted diheme cytochrome c4 whose N-terminal sequence is identical to that of the N-terminal 50 amino acids of a 27-kDa diheme cytochrome c reported by Guidici-Orticoni et al. (8). However, it differs in amino acid sequence and molecular weight from a second hypothetical diheme cytochrome c4 in A. ferrooxidans (21 kDa) previously described (3, 7, 18). An inspection of the partial genome sequence of A. ferrooxidans (www.tigr.org/; 17) reveals genes for both the 21- and 27-kDa diheme cytochrome c4 proteins. This demonstrates that they are not merely variants of the same gene arising from the use of different strains of A. ferrooxidans. Giudici-Orticoni et al. (8) speculated that the 27-kDa diheme cytochrome c4 directly oxidizes Fe(II) and passes electrons to rusticyanin, but, based on its cotranscription with the genes for the bc1 complex, we propose that it is the cytochrome that feeds electrons to complex III during uphill electron flow. Simultaneous activation of the 27-kDa diheme cytochrome c4 with the proteins of complex III could promote the shunting of electrons from the downhill pathway to the uphill pathway of electron flow.
The petI operon also contains sdrA, which potentially encodes a short-chain oxidoreductase that has the characteristics of the family of ribitol/glucose oxidoreductases, including the Prosite signature for ribitol/glucose binding (Table 1). Ribitol dehydrogenase generates NADH from NAD+ via the oxidation of ribitol to ribulose. The latter could then be converted to ribulose-5-P, which could enter the Calvin cycle. This suggests a model in which activation of sdrA could help stimulate a quiescent Calvin cycle so that it is ready to receive the NADH (NADPH) generated by uphill electron flow.
The identification of the petI and res operons also supports a model for phenotypic switching (10) in which the reversible insertion of ISAfe1 (formerly IST1) into resB results in a mutant phenotype in which cells are unable to utilize Fe(II) as a sole energy and electron source but can still grow in the presence of reduced sulfur (2, 16). Insertional inactivation of resB by ISAfe1 could lead to an incorrectly matured cytochrome c, possibly the diheme cytochrome c4 and/or the cytochrome c1 encoded by the petI operon, effectively knocking out uphill electron flow through complex III.
Nucleotide sequence accession numbers.
The sequence of the DNA resulting from walking up and down the A. ferrooxidans ATCC 19859 genome and the deduced amino acid sequences of the putative proteins were deposited in GenBank under accession no. AF220499. The sequences resulting from the amplification of A. ferrooxidans ATCC 33020 gene segments and intergenic regions were assigned GenBank accession no. AJ318500 to AJ318506.
Acknowledgments
This work was supported by Fondecyt grants 1980665 and 1010623 and a grant from Ecos/Conicyt, C99B05.
We thank The Institute for Genomic Research and Integrated Genomics for making available the partial genome sequence of A. ferrooxidans.
REFERENCES
- 1.Blake, R. C., and E. A. Shute. 1994. Respiratory enzymes of Thiobacillus ferrooxidans. Kinetic properties of an acid-stable iron:rusticyanin oxidoreductase. Biochemistry 33:9220-9228. [DOI] [PubMed] [Google Scholar]
- 2.Cabrejos, M. E., H.-L. Zhao, M. Guacucano, S. Bueno, G. Levican, E. Garcia, E. Jedlicki, and D. S. Holmes. 1999. IST1 insertional inactivation of the resB gene: implications for phenotypic switching in Thiobacillus ferrooxidans. FEMS Microbiol. Lett. 175:223-229. [DOI] [PubMed] [Google Scholar]
- 3.Cavazza, C., M. T. Giudici-Orticoni, W. Nitschke, C. Appia, V. Bonnefoy, and M. Bruschi. 1996. Characterization of a soluble cytochrome c4 isolated from T. ferrooxidans. Eur. J. Biochem 242:308-314. [DOI] [PubMed] [Google Scholar]
- 4.Drobner, E., H. Huber, and K. O. Stetter. 1990. Thiobacillus ferrooxidans, a facultative hydrogen oxidizer. Appl. Environ. Microbiol. 56:2922-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbehti, A., W. Nitschke, P. Tron, C. Michel, and D. Lemesle-Meunier. 1999. Characterization of cytochrome bc1-type complex of the acidophilic ferrous ion-oxidizing bacterium Thiobacillus ferrooxidans. J. Biol. Chem. 274:16760-16765. [DOI] [PubMed] [Google Scholar]
- 6.Elbehti, A., G. Brasseur, and D. Lemesle-Meunier. 2000. First evidence for existence of an uphill electron transfer through the bc1 and NADH-Q oxidoreductase complexes of the acidophilic obligate chemolithotrophic ferrous iron-oxidizing bacterium Thiobacillus ferrooxidans. J. Bacteriol. 182:3602-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giudici-Orticoni, M. T., F. Guerlesquin, M. Bruschi, and W. Nitschke. 1999. Interaction-induced redox switch in the electron transfer complex rusticyanin-cytochrome c(4). J. Biol. Chem. 274:30365-30369. [DOI] [PubMed] [Google Scholar]
- 8.Giudici-Orticoni, M. T., G. Leroy, W. Nitschke, and M. Bruschi. 2000. Characterization of a new dihemic c(4)-type cytochrome isolated from Thiobacillus ferrooxidans. Biochemistry 39:7205-7211. [DOI] [PubMed] [Google Scholar]
- 9.Guacucano, M., G. Levican, D. S. Holmes, and E. Jedlicki. 15December2000, posting date. An RT-PCR artifact in the characterization of bacterial operons. Electronic J. Bio/Technol. 3. [Online.] http://www.ejb.org/content/vol3/issue3/full/5/index.html.
- 10.Holmes, D. S., H.-L. Zhao, G. Levican, J. Ratouchniak, V. Bonnefoy, P. Varela, and E. Jedlicki. 2001. ISAfe1, a family ISL3 insertion sequence from Acidithiobacillus ferrooxidans ATCC 19859. J. Bacteriol. 183:4323-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holt, J. G. (ed.). 1993. Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, Md.
- 12.Ingledew, W. J. 1982. T. ferrooxidans, the bioenergetics of an acidophilic chemolithotrophic bacteria. Biochim. Biophys. Acta 683:89-117. [DOI] [PubMed] [Google Scholar]
- 13.Kelly, D. P., and A. P. Wood. 2000. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. E vol. Microbiol. 50:511-516. [DOI] [PubMed] [Google Scholar]
- 14.Ohmura, N., N. Matsumoto, K. Sasaki, T. Nagaoka, and H. Saiki. 1999. Growth of Thiobacillus ferrooxidans on hydrogen by the dissimilatory reduction of ferric iron under anaerobic conditions, p. 767-775. In R. Amils and A. Ballester (ed.), Biohydrometallurgy and the environment: toward the mining of the 21st century. Elsevier, Amsterdam, The Netherlands.
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Schrader, J., and D. Holmes. 1988. Phenotypic switching of Thiobacillus ferrooxidans. J. Bacteriol. 170:3915-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selkov, E., R. Overbeek, Y. Kogan, L. Chu, V. Vonstein, D. Holmes, S. Silver, R. Haselkorn, and M. Fonstein. 2000. Functional analysis of gapped microbial genomes: amino acid metabolism of Thiobacillus ferrooxidans. Proc. Natl. Acad. Sci. USA 97:3509-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamanaka, T., and Y. Fukumori. 1995. Molecular aspects of the electron transfer system which participates in the oxidation of ferrous ion by Thiobacillus ferrooxidans. FEMS Microbiol. Rev. 17:401-413. [DOI] [PubMed] [Google Scholar]
- 19.Yarzábal, A., G. Brasseur, J. Ratouchniak, K. Lund, D. Lemesle-Meunier, J. A. DeMoss, and V. Bonnefoy. The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J. Bacteriol. 184:313-317. [DOI] [PMC free article] [PubMed]
- 20.Yates, J. R., and D. S. Holmes. 1987. Two families of repeated DNA sequences in Thiobacillus ferrooxidans. J. Bacteriol. 169:1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]