Abstract
A new cellulosomal protein from Clostridium cellulolyticum Cel9M was characterized. The protein contains a catalytic domain belonging to family 9 and a dockerin domain. Cel9M is active on carboxymethyl cellulose, and the hydrolysis of this substrate is accompanied by a decrease in viscosity. Cel9M has a slight, albeit significant, activity on both Avicel and bacterial microcrystalline cellulose, and the main soluble sugar released is cellotetraose. Saccharification of bacterial microcrystalline cellulose by Cel9M in association with two other family 9 enzymes from C. cellulolyticum, namely, Cel9E and Cel9G, was measured, and it was found that Cel9M acts synergistically with Cel9E. Complexation of Cel9M with the mini-CipC1 containing the cellulose binding domain, the X2 domain, and the first cohesin domain of the scaffoldin CipC of the bacterium did not significantly increase the hydrolysis of Avicel and bacterial microcrystalline cellulose.
The mesophilic cellulolytic bacterium Clostridium cellulolyticum produces multienzyme complexes of about 600 kDa, termed cellulosomes (13), which are efficient for degradation of crystalline cellulose. The catalytic subunits of the cellulosome are bound to a noncatalytic scaffolding protein called CipC, which contains a cellulose binding domain (CBD), two X2 domains of unknown function, and eight reiterated domains called cohesins (24). The binding of each cellulosomal enzyme to the scaffoldin is accomplished by means of a dockerin domain, generally located at the C termini of the enzymes, which interacts with one of the eight cohesins of CipC (22). At least 13 proteins harboring a dockerin have been identified in rough cellulosome preparations (13).
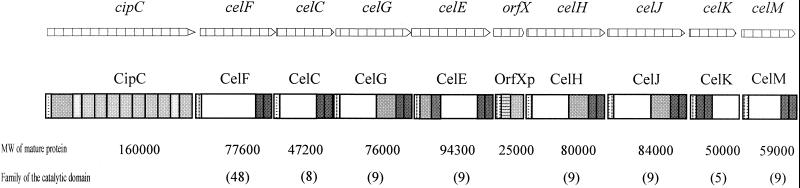
Six of these proteins, produced as cloned enzymes in Escherichia coli, have been characterized: Cel5A (9, 10, 11), Cel5D (30, 31), Cel8C (12), Cel9G (14), Cel9E (16), and Cel48F (26, 27, 28). The genes celF, celC, celG, and celE are located on the same cluster downstream from cipC (1, 27). Chromosome walking downstream of celE by successive inverse PCR experiments has allowed us to identify new genes coding for cellulases (15; H. P. Fierobe, unpublished data ), including celH and celJ coding for cellulases belonging to family 9 and, more recently, celM. This cluster is shown in Fig. 1. The enzymatic subunits of the cellulosome are made up of at least two domains, including a catalytic domain and a dockerin, although some of them are larger and contain other, extra domains of known or unknown function. Cel9G, Cel9H, and Cel9J are multidomain enzymes made up of a glycosyl hydrolase family 9 catalytic domain (GH9), a family IIIc CBD, and a dockerin. Cel9E is made up of a family IV CBD, an immunoglobulin (Ig)-like domain, a GH9 domain, and a dockerin. Both enzymes have been extensively studied, and it has been demonstrated that the CBDs of Cel9G and Cel9E are essential for catalysis. celM encodes a new family 9 cellulase containing only a GH9 catalytic domain and a dockerin. In this paper, the biochemical properties of the cloned enzyme are studied and compared to those of the corresponding multidomain enzymes Cel9G and Cel9E.
FIG. 1.
Cellulosomal gene cluster and corresponding proteins of C. cellulolyticum.
MATERIALS AND METHODS
Bacterial strains and plasmids.
C. cellulolyticum ATCC 35319 was used as the source of genomic DNA and for production of cellulosomes. E. coli DH5α (Bethesda Research Laboratory) was used as a host for pET-22b(+) (Novagen) derivatives. E. coli BL21(DE3) was used as the host for expression vectors. C. cellulolyticum was grown anaerobically at 32°C on basal medium supplemented with cellobiose (2g/liter) or cellulose MN300 (5 g/liter) as a carbon and energy source. E. coli was grown at 37°C in Luria-Bertani medium supplemented with ampicillin (100 or 200 μg/ml) when required.
Expression of recombinant CelM protein.
The region of the celM gene that encodes the mature protein was amplified by PCR with the primers celMdir. (5" CCCCCATATGGCAGGAACACATGATTATT 3") and celMrev. (5" CCCCCTCGAGACCTAAGATAGCCTTCTTT 3"). These primers create an NdeI and an XhoI site (boldface) upstream and downstream of the coding sequence, respectively. After digestion by NdeI and XhoI, the PCR fragment was cloned into NdeI/XhoI-digested pET-22b(+). The coding sequence of CelM was fused in frame with a downstream sequence of the vector encoding six histidine residues (His tag). The resulting plasmid, pET-M, was used to transform E. coli BL21(DE3) cells for the production of the recombinant protein. This construction produces a recombinant protein which contains two additional residues (Leu and Glu) between the C terminus of the mature protein and the His tag.
Production and purification of CelM in E. coli.
Cells were grown in Luria-Bertani medium supplemented with ampicillin (200 μg/ml) at 37°C with shaking to an optical density of 2 at 600 nm. They were then cooled to 15°C. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 50 μM, and the culture was incubated with shaking at 15°C for 15 h. The final optical density of the culture was about 6. The cells were then cooled to 5°C, harvested by centrifugation, resuspended in cold 30 mM Tris-HCl (pH 8) buffer (RB), and broken twice in a French pressure cell. The crude extract was centrifuged at 26,000 × g for 15 min, and the supernatant was loaded onto a 3-ml Ni-nitrilotriacetic acid column previously equilibrated with RB. After washes with RB and RB supplemented with 5 and 10 mM imidazole, the protein was eluted with RB supplemented with 60 mM imidazole. The eluate was dialyzed against RB and concentrated in an Amicon concentrator. The protein purification was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis visualization. Small aliquots of the protein solution were stored at −20°C.
The protein concentration was calculated by measuring the absorbance at 280 nm in 6 M guanidium chloride using a molar extinction coefficient of 102,630 M−1 cm−1.
Substrates used.
Carboxymethyl cellulose, medium viscosity (CMC) (Sigma), was prepared as a 1% (wt/vol) solution in 20 mM Tris-maleate buffer (pH 6). Avicel (Merck) was used as a 0.8% suspension in the same buffer. Bacterial microcrystalline cellulose (BMCC) was prepared from cubes of commercial bacterial cellulose (Nata de coco; Fujikko Co., Kobe, Japan) according to the protocol described by Boisset (4). After the treatments, the cellulose was suspended in 20 mM Tris-maleate buffer (pH 6) at a final concentration of 3.3 g/liter and stored at 4°C. Phosphoric acid swollen cellulose (PASC) was prepared from Avicel as described by Walseth (34) and suitably adjusted to obtain a final concentration of 1% (wt/vol) in 20 mM Tris-maleate buffer (pH 6). In each case, the concentration was estimated by the phenol-sulfuric acid method (21) or by performing dry-weight measurement. Cellodextrins with degrees of polymerization ranging from 5 to 2 were purchased from Sigma.
Enzyme assays.
Carboxymethyl cellulase activity was assayed at 37°C by mixing 4 ml of CMC solution with 1 ml of enzyme solution at the appropriate concentration. Aliquots of 0.5 ml were removed at specific intervals and stored on ice. The reducing-sugar contents were determined by the ferricyanide method of Park and Johnson (25). One international unit corresponds to 1 μmol of d-glucose equivalent released per min.
To measure the hydrolysis of insoluble sugars, such as Avicel or BMCC, 3.5 ml of suspension was incubated at 37°C with slight shaking (60 rpm). The reaction was started with 50 μl of enzyme solution at the appropriate concentration. Samples (0.7 ml) were taken at specific intervals and centrifuged. The reducing-sugar content of 0.5 ml of the supernatant was estimated as described above. The activities on cellodextrins were measured as follows: cellodextrins ranging from cellotriose (Glc3) to cellopentaose (Glc5) were purchased from Sigma. Two hundred microliters of each cellodextrin (1 and 2.5 g/liter) in 20 mM Tris-maleate buffer (pH 6) was mixed at 37°C with 50μl of diluted enzyme; 75 μl was sampled at specific intervals and immediatly incubated at 75°C for 10 min to stop the reaction. The sugars were analyzed using a DX 500 chromatographic system (Dionex, Sunnyvale, Calif.).
Viscosimetry assays.
Viscosimetry assays were performed by monitoring the flow time of a 0.8% (wt/vol) CMC solution incubated with the enzyme. The procedure was as previously described (12); 0.5-ml aliquots sampled at various times were boiled for 15 min, and the fluidity was measured at room temperature in a viscosimeter adapted for handling small volumes. The specific fluidity was estimated using the formula Fsp = T0/(T − T0), where T0 is the flow time of water and T is the flow time of the CMC solution after the reaction.
Analysis of degradation products.
The products of cellulose enzymatic hydrolysis were analyzed by high-performance anion-exchange chromatography coupled with a pulsed amperometric detector (HPAE-PAD) using a DX 500 system equipped with a GP 40 gradient pump and an ED 40 electrochemical detector. A Dionex eluent degas module was used to sparge and pressurize the mobile phase with helium C. Water used in HPAE-PAD was ultrapure 18-MΩ-cm deionized water. Sodium hydroxide aqueous solution (48%) was from Fisher Scientific (Illkirch, France). Sodium acetate was purchased from Merck.
The eluents were 100 mM sodium hydroxide-5 mM sodium acetate (eluent A) and 100 mM sodium hydroxide-500 mM sodium acetate (eluent B). Amperometric detection was performed using four pulses applied on a gold working electrode. The following pulse potentials and durations were used: step E1, +0.1 V (0 to 0.4 s); step E2, −2 V (0.4 to 0.42 s); step E3, +0.6 V (0.42 to 0.44 s); and step E4,−0.1 V (0.44 to 0.5 s). The system was piloted by a Peaknet chromatography workstation (Dionex). Glucose, cellobiose, cellotriose, cellotetraose, and cellopentaose were analyzed using a Carbopac PA (Dionex) guard column (25 by 4 mm) and a Carbopac PA-100 analytical anion-exchange column (250 by 4 mm). The elution was performed at a constant flow rate of 1 ml/min at room temperature using a 15-min linear gradient from 0 to 50% of eluent B. The injection volume was 10 μl in each case. The reaction products were identified and quantified by reference to known standards.
Nucleotide sequence accession number.
The updated sequence of the cluster including the celH, celJ, and celM genes was submitted to GenBank (accession no. AF316823).
RESULTS
Sequence analysis of Cel9M.
celM, encoding a family 9 cellulase, was identified in the large cluster of genes that includes several genes encoding cellulosomal proteins (1, 15, 27). It encodes a 526-amino-acid protein which begins with a putative peptide signal sequence characteristic of the signal sequence of C. cellulolyticum cellulases sequenced so far. The putative cleavage site may be located between the two Ala residues 30 and 31. The mature protein contains 496 amino acids with a calculated molecular mass of 54,618 Da. It is composed of a GH9 catalytic domain and a dockerin at the C terminus characteristic of C. cellulolyticum cellulases (23). Comparison of the Cel9M catalytic domain with those of Cel9G and Cel9E (Fig. 2) reveals quite a significant amino acid sequence conservation with respect to Cel9G (43% identity and 55% similarity) and a sequence conservation about two times lower with respect to Cel9E (24% identity and 34% similarity).
FIG. 2.
Alignment of the catalytic domains of cellulases Cel9M (1), Cel9G (2), and Cel9E (3) from C. cellulolyticum cellulosome. Identical amino acids are in boldface. The mutated Glu is underlined and indicated by an arrow.
Production and purification of recombinant Cel9M.
Cel9M was purified using a one-step process. This purification was performed as fast as possible in order to prevent the C-terminal cleavage of the dockerin domain. About 30 mg of pure protein was produced from 3 liters of culture. Small aliquots of proteins were stored at −20°C. The recombinant protein, made up of 505 amino acids, had an apparent mass of 56,000 Da, which is in good agreement with the theoretical mass (55,701 Da) of the recombinant protein.
Polyclonal antibodies against Cel9M were produced. The immunodetection of the protein in the cellulosome, prepared from cell cultures grown on cellulose medium, was performed using the Western blotting procedure previously described by Gal et al. (13). A protein with a molecular mass of about 55,000 Da was detected (not shown). This result confirms the identification of Cel9M as a component of the cellulosome, as was expected due to the presence of the dockerin signature sequence.
Catalytic properties of Cel9M.
The specific activity of Cel9M toward specific substrates was tested and compared to those of Cel9G and Cel9E. The results are summarized in Table 1. Cel9M is very active on CMC (twice as active as Cel9G), whereas the activity on PASC is of the same order as those of Cel9E and Cel9G. Cel9M is able to hydrolyze Avicel and BMCC, although with less efficiency than the other two cellulases. As for Cel9G, no activity on p-nitrophenyl-cellobiose (pNPC) was detected.
TABLE 1.
Cel9M activity on various substrates and comparison with activities of Cel9G and Cel9E characterized in cellulosome of C. cellulolyticum (proteins with dockerins)
| Substrate | Activity (IU/μmol)
|
||
|---|---|---|---|
| Cel9M | Cel9G | Cel9E | |
| CMC | 2,500 | 1,300 | 13.5 |
| PASC | 57 | 38 | 42.5 |
| Avicel | 2.5 | 5.2 | 5.8 |
| BMCC | 2 | 10.3 | 5.5 |
| pNPCa | NDb | ND | 34.8 |
pNPC, p-nitrophenyl-cellobiose.
ND, no detectable activity.
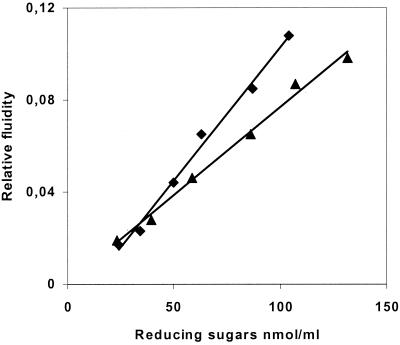
Information on the hydrolytic mechanism was obtained by studying the increase in CMC fluidity versus the release of reducing sugars and by comparing the results obtained with Cel9G. The degradation of CMC is concomitant with a decrease in viscosity (Fig. 3), thus revealing that Cel9M has an endo mode of action, like Cel9G.
FIG. 3.
Relative fluidity of CMC versus the liberation of reducing extremities by 0.01 μM Cel9M (▴) and 0.01 μM Cel9G (⧫).
The soluble sugars produced by Cel9M during the hydrolysis of BMCC were analyzed after 2, 6, and 22 h. For each sampling time, cellotetraose was found to be the major product (83%), with small amounts of cellotriose (11%) and cellobiose (4%) also being detected. In comparison, in the case of Cel9G, the major soluble products from BMCC after 2 h of hydrolysis were cellotetraose (16%), cellotriose (50%), and cellobiose (32%); after 22 h of hydrolysis, the composition of the soluble-sugar mixture was 2% cellotriose, 60% cellobiose, and 38% glucose. In the case of Cel9E, the main soluble product on the same substrate was cellobiose (96%) for each sampling time.
The activity of Cel9M on the natural cellodextrines Glc5 to Glc3 was also determined. Cellopentaose was cleaved into cellotetraose and glucose. Cellotetraose and cellotriose were not cleaved.
Production of a catalytic mutant.
Sequence alignments of GH9 catalytic domains led us to propose Glu410 as a catalytic residue. This residue was replaced by glutamine by using the two mutated oligonucleotides 5" CAGTATCAGTATACACAGGTAGCTCTAGACTACAA 3" and 5" TTGTAGTCTAGAGCTACCTGTGTATACTGATACTG 3" (the mutated codon is in boldface). Cel9ME410Q was purified by the same protocol used for the purification of the native protein. This mutation totally inactivated the carboxymethyl cellulase activity of the protein.
Cellulolytic activity of CelM complexed with the mini-CipC1.
To determine a possible influence of CBDIIIa on the activity of Cel9M toward Avicel and BMCC, a Cel9M-mini-CipC1 complex was engineered as follows: the mini-CipC1 containing the CBDIIIa as well as the first X2 domain and the cohesin1 domain from CipC was prepared as previously described (22). The complex was obtained due to the strong interaction that occurred between the cohesin domain of mini-CipC1 and the dockerin domain of Cel9M in the presence of 10 mM CaCl2. Nondenaturing gel electrophoresis indicated that mini-CipC1 and Cel9M formed a complex (mini-CipC1-Cel9M) that was stable for at least 24 h. After 22 h of hydrolysis of Avicel, the reducing-sugar concentration of the soluble fraction was slightly higher for mini-CipC1-Cel9M than for Cel9M alone, while no such increase was detected following the hydrolysis of BMCC.
Synergism among cellulases belonging to family 9.
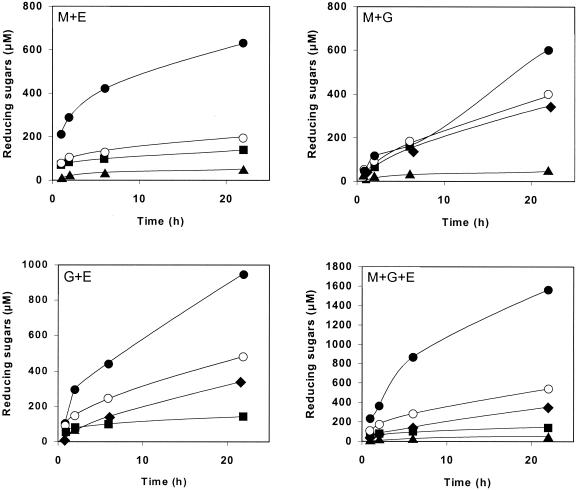
In order to identify possible synergistic interactions among the three family 9 cellulases belonging to the cellulosome of C. cellulolyticum, we studied the kinetics of cellulose hydrolysis generated by the following equimolar enzyme mixtures: Cel9M plus -G, Cel9M plus -E, Cel9G plus -E, and Cel9M plus -G plus -E. in these experiments, the hydrolysis of BMCC was monitored for 22 h. The reducing sugars released for each association of enzymes are given in Fig. 4. For the mixtures containing only two enzymes, the best stimulation (threefold increase) was observed with Cel9M and Cel9E. A very good stimulation (twofold increase) was also obtained with the Cel9G-Cel9E mixture, whereas no effect was detected during the first 6 h with the Cel9M-Cel9G mixture. In that case, only a 1.5-fold increase was monitored after 22 h. The mixture containing three enzymes was also three times more active than the sum of the activities of each protein considered separately.
FIG. 4.
Synergies among family 9 cellulases CelM (▴), CelG (⧫), and CelE (▪) in BMCC hydrolysis. Theoretical values (○) and experimental values (•) are shown. The concentration of each enzyme was 0.1 μM. The theoretical value was obtained by adding the values for the enzymes alone and was compared to the experimental value.
The soluble sugars produced during BMCC hydrolyses with the different mixtures were analyzed. In all of the mixtures, including Cel9E, cellobiose was the main product (accounting for over 80% of the products); the other sugar detected was cellotriose. The association Cel9M plus -G gave cellotetraose (37%), cellotriose (38%), and cellobiose (44%) after 2 h of hydrolysis; after 22 h, cellotetraose had disappeared and the remaining products were cellotriose (37%), cellobiose (36%), and glucose (23%).
DISCUSSION
To date, 129 cellulases belonging to family 9 of glycosyl hydrolases have been identified, and almost all of them contain additional domains (6). Family 9 cellulases conform to one of the four thematic modular arrangements (2, 7). Three of them contain additional helper modules. Group A has a family IIIc CBD immediately downstream from its GH9 catalytic module. Endoglucanase E4 from Thermobifida fusca (formerly Thermomonospora fusca) (19), the three-dimensional (3-D) structure of which has been resolved (29), belongs to this group. Group B1 bears an Ig-like domain upstream of the catalytic module. Cel9D from Clostridium thermocellum is a member of this group (17, 18). Its 3-D structure was obtained 9 years ago (20). A third thematic type, B2, also contains an Ig-like domain, but in addition, it has an N-terminal family IV CBD. Finally, about 30 enzymes belong to the simplest GH9 theme (group C), which consists of a simple catalytic module alone. This group is typical of many plant cellulases. Recently, the GH9 module located at the N-terminal position of the cellulosomal scaffoldin CipV from Acetivibrio cellulolyticus was placed in a position adjacent to the plant enzyme group (7).
In the large cluster of genes (26 kb) from C. cellulolyticum, which contains almost all the cellulase genes coding for cellulosomal proteins, five genes, celG, celE, celH, celJ, and celM, code for GH9 enzymes. The proteins Cel9G, Cel9H, and Cel9J are multidomain proteins, and they belong to group A. Cel9G has an endo mode of action. It is active on CMC, and the decrease in viscosity on this substrate is concomitant with the release of reducing sugars. However, this enzyme is very efficient in the hydrolysis of crystalline cellulose, particularly BMCC (14). This property is due to the presence of the CBDIIIc domain, which extends the active site cleft. Cel9G is not one of the major enzymes of the cellulosome (13); however, two other proteins very similar to Cel9G are also synthesized by the bacterium: Cel9H is a hypothetical cellulosome protein, and Cel9J has been identified as a cellulosomal protein (15). Both proteins show a high degree of similarity to Cel9G (55% for Cel9H and 46% for cel9J), and it seems reasonable to hypothesize that they act on crystalline cellulose in the same way as Cel9G does. The reason why C. cellulolyticum synthesizes three almost identical enzymes is not known. Cel9E, a protein typical of group B2, is one of the major proteins of the cellulosome. It acts first in a random mode of attack on cellulose and then in a processive mode of action, releasing mainly cellobiose (more than 90% of the total soluble sugars produced) (16). No protein equivalent to the C. thermocellum Cel9D (group B1) has ever been found in the C. cellulolyticum cellulosome.
Cel9M is a new protein possessing only a GH9 catalytic domain and a dockerin. The celM gene was found to be located in the cluster downstream from the manK gene coding for the mannanase Man5K. To date, no protein like Cel9M has been found in the cellulosome of C. thermocellum, although it contains various GH9 proteins (32). In Clostridium cellulovorans, however, a very close topological context has been reported (8, 33). The engL gene, found in the large cluster identified in the genome of the bacterium and located upstream of the manA gene (coding for Man5A), codes for a hypothetical cellulosomal protein, Cel9L, which shows 49% identity and 67% similarity with Cel9M. A higher degree of identity (51%) was found between Cel9M and the GH9 module of CipV from A. cellulolyticus. Cel9M and Cel9L could therefore be classified in the group containing this module and be positioned adjacent to group C, containing plant enzymes.
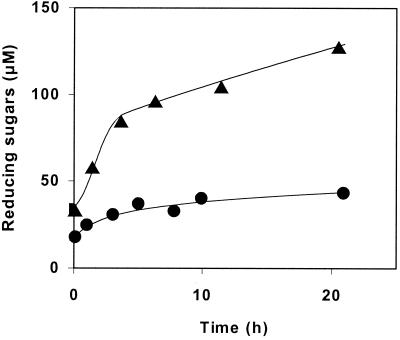
Cel9 M is as active on CMC as the two endoglucanases, Cel5A and Cel8C, from C. cellulolyticum and is twice as active as Cel9G. Viscosimetry assays of CMC with Cel9M and Cel9G yielded similar profiles, indicating that both enzymes have the same endo mode of action on this substrate. Cel9M is able to hydrolyze BMCC less efficiently than Cel9G or Cel9E but much more efficiently than the endoglucanases Cel5A and Cel8C. The activity of Cel9M on BMCC was compared to that of Cel5A (Fig. 5). The rate of degradation of the substrate is lower for Cel5A than for Cel9M during the first 4 h. After that, the degradation of BMCC by Cel5A reached a plateau while the degradation of this substrate by Cel9M continued at a low but constant rate until the 21-h point. Another difference from the other family 9 enzymes is the composition of the soluble-sugar mixture released by Cel9M. Cellotetraose was found to be the main product (more than 80%), and it was demonstrated that cellotetraose and cellotriose were not cleaved. As previously shown for Cel9G, Cel9M did not cleave pNPC. Crystals of the protein have been obtained by G. Parsiegla, and the structure of the protein has recently been determined (G. Parsiegla, A. Belaich, J. P. Belaich, and R. Haser, unpublished results). Structural studies will provide some information regarding the mode of action of this enzyme.
FIG. 5.
Comparison between reducing sugars released by Cel9M (▴) and Cel5A (•) on BMCC (3.3 mg/ml). The concentration of each enzyme was 0.2 μM.
Complexes associating mini-CipC1 and Cel9M did not significantly increase the release of soluble sugars from Avicel and BMCC. Similar experiments performed recently by Carrard et al. (5), who complexed Cel9D from C. thermocellum with various chimeric CBD-cohesin proteins, generated different results. Among the CBDs tested, CBDIIIa promoted the largest rise in BMCC hydrolysis, which was approximately fivefold greater than with the enzyme alone. Similar results were obtained with the chimeric protein Cel5A-CBDIIIa, in which the CBD of CipC was grafted downstream of the catalytic domain of C. cellulolyticum Cel5A. Using this hybrid protein, the hydrolysis of Avicel was three times higher than with Cel5A alone (3). It has also been observed that the presence of CBDIIIa leads to macroscopic changes in cellulose particle flocculation (3). The mechanism responsible for this phenomenon remains unknown. The assumption that the action of CBD concerns the crystalline parts of BMCC would explain its strong beneficial effect for the hydrolysis of the substrate when it is added to enzymes that are not efficient on crystalline cellulose, such as Cel9D or Cel5A. It would also explain its quite minor effect on cellulases that are able to hydrolyze crystalline cellulose, like Cel9M or Cel9G (14).
The synergism in the hydrolysis of BMCC can be interpreted from the intrinsic properties of the enzyme reported in Table 1. The greater effect obtained with the Cel9M plus -E mixture could be explained by the fact that Cel9M is a very good endoglucanase while Cel9E is a cellobiohydrolase. On the other hand, the lesser effect found with the Cel9M plus -G mixture could be due to the endoglucanase activity of both enzymes. Finally, the intermediate effect observed with the Cel9E plus -G mixture could be due to the endo mode of attack of Cel9G.
Comparison of the three enzymes Cel9M, Cel9G, and Cel9E shows an apparent increase in the complexity of the structures, resulting from the addition of a CBDIIIc domain to the catalytic domain to form Cel9G and of two extra domains (CBDIV and an Ig-like domain) to form Cel9E, with a correlative increase of the molecular mass (59, 76, and 94 kDa, respectively). Table 1 shows that this evolution is accompanied by a deep evolution from an endo activity of Cel9M to the endo-processive activity of Cel9G and finally to an exo activity or, more precisely, a total loss of endo activity of Cel9E. Such an evolution suggests that the bacterium was able to construct, by fusing separate extra domains to the same type 9 catalytic domain, a kit of three enzymes acting synergistically. The resolution of the 3-D structures of these three enzymes is under way and will provide new information on the putative contribution of each domain to the specific activities of these enzymes and on the molecular evolution of these proteins.
Acknowledgments
Protein sequences were obtained via the Carbohydrate-Active Enzyme server (CAZy and CAZyModO websites (http://almb.cnrs-mrs.fr/∼pedro/CAZY/db.htlm and http://afmb.cnrs-mrs.fr/∼pedro/DB/db.htlm, respectively) designed by P. M. Couthino and B. Henrissat. We are grateful to O. Valette and S. Bruno for technical assistance and to M. Johnson for proofreading the manuscript. We are indebted to C. Gaudin for helpful discussions.
This work was supported by contracts from the European Commission (Biotech BIO4-97-2000), from the Centre National de la Recherche Scientifique and Université de Provence, and from Conseil Général des Bouches du Rhône and Région Provence-Alpes-Côte d'Azur.
REFERENCES
- 1.Bagnara-Tardif, C., C. Gaudin, A. Bélaich, P. Hoest, T. Citard, and J.-P. Bélaich. 1992. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene 119:17-28. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., L. J. W. Shimon, Y. Shoham, and R. Lamed. 1998. Cellulosomes—structure and ultrastructure. J. Struct. Biol. 124:221-234. [DOI] [PubMed] [Google Scholar]
- 3.Belaich, A., J. P. Belaich, H. P. Fierobe, L. Gal, C. Gaudin, S. Pagès, C. Reverbel-Leroy, and C. Tardif. 1998. Cellulosome analysis and cellulases CelF and CelG from Clostridium cellulolyticum, p. 73-86. In M. Claeyssens, W. Nerinckx, and K. Piens (ed.), Carbohydrases from Trichoderma reesei and other microorganisms: structure, biochemistry, genetics and applications. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 4.Boisset, C. 1996. Modification de matériaux cellulosiques par des enzymes cellulolytiques. Ph.D. thesis. Université Joseph Fourier, Grenoble, France.
- 5.Carrard, G., A. Koivula, H. Söderlund, and P. Beguin. 2000. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. USA 97:10342-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coutinho, P. M., and B. Henrissat. 1999. The modular structure of cellulases and other carbohydrate-active enzymes: an integrated database approach, p. 15-23. In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimuna (ed.), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers Co., Ltd., Tokyo, Japan.
- 7.Ding, S. Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 1999. A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family 9 glycosyl hydrolase. J. Bacteriol. 181:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doi, R. H., and Y. Tamaru. 2000. The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity. Chem. Rec. 1:24-32. [DOI] [PubMed]
- 9.Ducros, V., M. Czjzek, A. Belaich, C. Gaudin, H. P. Fierobe, J. P. Belaich, G. J. Davies, and R. Haser. 1995. Crystal structure of the catalytic domain of bacterial cellulase belonging to family 5. Structure 3:939-949. [DOI] [PubMed] [Google Scholar]
- 10.Faure, E., A. Bélaich, C. Bagnara, C. Gaudin, and J.-P. Bélaich. 1989. Sequence analysis of the Clostridium cellulolyticum endoglucanase-A-encoding gene, celCCA. Gene 84:39-46. [DOI] [PubMed] [Google Scholar]
- 11.Fierobe, H.-P., C. Gaudin, A. Bélaich, M. Loutfi, E. Faure, C. Bagnara, D. Baty, and J.-P. Bélaich. 1991. Characterization of endoglucanase A from Clostridium cellulolyticum. J. Bacteriol. 173:7956-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fierobe, H.-P., C. Bagnara-Tardif, C. Gaudin, F. Guerlesquin, P. Sauve, A. Bélaich, and J.-P. Bélaich. 1993. Purification and characterization of endoglucanase C from Clostridium cellulolyticum. Catalytic comparison with endoglucanase A. Eur. J. Biochem. 217:557-565. [DOI] [PubMed] [Google Scholar]
- 13.Gal, L., S. Pagès, C. Gaudin, A. Bélaich, C. Reverbel-Leroy, C. Tardif, and J.-P. Bélaich. 1997. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ. Microbiol. 63:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gal, L., C. Gaudin, A. Belaich, S. Pagès, C. Tardif and J.-P. Belaich. 1997. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J. Bacteriol. 179:6595-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gal, L. 1997. Etude du cellulosome de Clostridium cellulolyticum et de l'un de ses composants la cellulase CelG. Ph.D. thesis. Université de Provence, Marseille, France.
- 16.Gaudin, C., A. Belaich, S. Champ, and J. P. Belaich. 2000. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J. Bacteriol. 182:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joliff, G., P. Béguin, and J.-P. Aubert. 1986. Nucleotide sequence of the cellulase gene celD encoding endoglucanase D of Clostridium thermocellum overproduced in Escherichia coli. Bio/Technology 4:896-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joliff, G., P. Béguin, M. Juy, J. Millet, A. Ryter, R. Poljak, and J.-P. Aubert. 1986. Isolation, crystallisation and properties of a new cellulase of Clostridium thermocellum. Nucleic Acids Res. 14:8605-8613. [DOI] [PMC free article] [PubMed]
- 19.Jung, E. D., G. Lao, D. Irwin, B. K. Barr, A. Benjamin, and D. B. Wilson. 1993. DNA sequences and expression in Streptomyces lividans of an exoglucanase gene and an endoglucanase gene from Thermonospora fusca. Appl. Environ. Microbiol. 59:3032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juy, M., A. G. Amit, P. M. Alzari, R. J. Poljak, M. Claeyssens, P. Béguin, and J.-P. Aubert. 1992. Three-dimensional structure of a thermostable bacterial cellulase. Nature 357:89-91. [Google Scholar]
- 21.Leray, C., J. Nicoli, and P. Audiffren. 1966. Microdosage colorimétrique des oses neutres par l'acide sulfurique et le phénol. Influence des sels et des protéines. Méd. Trop. 26:382-390. [PubMed] [Google Scholar]
- 22.Pagès, S., A. Bélaich, C. Tardif, C. Reverbel-Leroy, C. Gaudin, and J.-P. Bélaich. 1996. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 178:2279-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagès, S., A. Belaich, J. P. Belaich, E. Morag, R. Lamed, Y. Shoham, and E. A. Bayer. 1997. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29:517-527. [PubMed] [Google Scholar]
- 24.Pagès, S., A. Belaich, H.-P. Fierobe, C. Tardif, C. Gaudin, and J.-P. Belaich. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localisation of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, J. T., and M. J. Johnson. 1949. A submicrodetermination of glucose. J. Biol. Chem. 181:149-151. [PubMed] [Google Scholar]
- 26.Parsiegla, G., M. Juy, C. Reverbel-Leroy, C. Tarfif, J. P. Belaich, H. Driguez, and R. Haser. 1998. The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 A resolution. EMBO J. 17:5551-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reverbel-Leroy, C., A. Bélaich, A. Bernadac, C. Gaudin, J.-P. Bélaich, and C. Tardif. 1996. Molecular study and overexpression of the Clostridium cellulolyticum celF cellulase gene in Escherichia coli. Microbiology 142:1013-1023. [DOI] [PubMed] [Google Scholar]
- 28.Reverbel-Leroy, C., S. Pagès, A. Bélaich, J.-P. Bélaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome—purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakon, J., D. Irwin, D. B. Wilson, and P. A. Karplus. 1997. Structure and mechanism of endo/exocellulase E4 from Thermonospora fusca. Nat. Struct. Biol. 4:810-818. [DOI] [PubMed] [Google Scholar]
- 30.Shima, S., Y. Igarashi, and T. Kodama. 1991. Nucleotide sequence analysis of the endoglucanase-encoding gene, celCCD, of Clostridium cellulolyticum. Gene 104:33-38. [DOI] [PubMed] [Google Scholar]
- 31.Shima, S., Y. Igarashi, and T. Kodama. 1993. Purification and properties of two truncated endoglucanases produced in Escherichia coli harbouring Clostridium cellulolyticum endoglucanase gene celCCD. Appl. Microbiol. Biotechnol. 38:750-754. [DOI] [PubMed] [Google Scholar]
- 32.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 33.Tamaru, Y., C. C. Liu, A. Ichi-ishi, L. Malburg, and R. H. Doi. 1999. The Clostridium cellulovorans cellulosome and noncellulosomal cellulases, p. 488-494. In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimuna (ed.), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers Co., Ltd., Tokyo, Japan.
- 34.Walseth, C. S. 1952. Occurrence of cellulase in enzyme preparations from microorganisms. TAPPI 35:228-233.