Abstract
ATP-binding cassette transporter LolCDE was previously identified, by using reconstituted proteoliposomes, as an apparatus catalyzing the release of outer membrane-specific lipoproteins from the inner membrane of Escherichia coli. Mutations resulting in defective LolD were previously shown to be lethal for E. coli. The amino acid sequences of LolC and LolE are similar to each other, but the necessity of both proteins for lipoprotein release has not been proved. Moreover, previous reconstitution experiments did not clarify whether or not LolCDE is the sole apparatus for lipoprotein release. To address these issues, a chromosomal lolC-lolD-lolE null mutant harboring a helper plasmid that carries the lolCDE genes and a temperature-sensitive replicon was constructed. The mutant failed to grow at a nonpermissive temperature because of the depletion of LolCDE. In addition to functional LolD, both LolC and LolE were required for growth. At a nonpermissive temperature, the outer membrane lipoproteins were mislocalized in the inner membrane since LolCDE depletion inhibited the release of lipoproteins from the inner membrane. Furthermore, both LolC and LolE were essential for the release of lipoproteins. On the other hand, LolCDE depletion did not affect the translocation of a lipoprotein precursor across the inner membrane and subsequent processing to the mature lipoprotein. From these results, we conclude that the LolCDE complex is an essential ABC transporter for E. coli and the sole apparatus mediating the release of outer membrane lipoproteins from the inner membrane.
Bacterial lipoproteins having a lipid-modified cysteine residue at the N terminus are anchored to membranes through the N-terminal fatty acyl chains. In gram-negative bacteria, lipoproteins are localized in either the inner or outer membrane. Lipoproteins are synthesized with signal peptides in the cytoplasm and then translocated across the inner membrane by Sec machinery (3, 15), followed by sequential modification reactions: formation of a thioether linkage between the N-terminal cysteine residue of the mature region and diacylglyceride, cleavage of the signal peptide by lipoprotein-specific signal peptidase, and aminoacylation of the N-terminal cysteine residue (5, 15). Since lipoproteins are more hydrophobic than nonlipidated outer membrane proteins, localization of lipoproteins in the outer membrane requires a distinct mechanism which enables the transport of lipoproteins through the hydrophilic periplasmic space (11). Furthermore, the mechanism should distinguish the lipoprotein-sorting signal since a subset of lipoproteins remains in the inner membrane (4, 16, 20).
We have identified five Lol proteins involved in the outer membrane localization of lipoproteins (10, 11, 21). Periplasmic chaperone LolA was found to form a hydrophilic complex with outer membrane-specific lipoproteins (11). The LolA-lipoprotein complex crosses the periplasm and then interacts with outer membrane receptor LolB, which mediates the anchoring of lipoproteins to the outer membrane (10). For formation of the LolA-lipoprotein complex, LolCDE in the inner membrane is required (21). LolCDE releases mature lipoproteins from the outer surface of the inner membrane in an ATP-dependent manner, leading to the formation of the LolA-lipoprotein complex (21). The lipoprotein-sorting signal is recognized at the release step, and the inner membrane-specific lipoproteins are not released. The functions of LolA and LolB have been clarified both in vivo and in vitro (10, 11, 18-22, 26). The LolCDE complex belonging to the ABC transporter superfamily comprises an ATPase subunit, LolD, and integral membrane proteins LolC and LolE. This complex was biochemically identified as a lipoprotein-releasing apparatus with proteoliposomes reconstituted from purified LolCDE and phospholipids (21). On the other hand, on the basis of our results, the functional complex could not be formed after its dissociation into subunits. Therefore, it is not clear whether or not both LolC and LolE are required for the apparatus. Moreover, it may be possible that other proteins can perform the function of LolCDE. In this study, we constructed a conditional lolC-lolD-lolE null strain and characterized the in vivo function of the LolCDE complex.
MATERIALS AND METHODS
Bacteria and plasmids.
The Escherichia coli K-12 strains and plasmids used in this study are listed in Table 1. DLP79-36 was a kind gift from Masaaki Wachi.
TABLE 1.
E. coli K-12 strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or reference(s) |
|---|---|---|
| Strains | ||
| FS1576 | supE44 hsdR thi-1 thr-1 leuB6 lacY1 tonA21 recD1009 | 13, 17 |
| JC10240 | Hfr thi-300 recA56 srl::Tn10 relA1 ilv-318 spoT1 thi rpsE2300 | 1 |
| DLP79-36 | HfrC pps man lpp | This study |
| CDE4 | DLP79-36 ΔlolC-lolD-lolE::kan lpp recA56/pMAN1015 | This study |
| Plasmids | ||
| pJY310 | cat; lolC-lolD-lolE | 21 |
| pMAN997 | bla; temperature-sensitive replicon, multicloning sites | 19 |
| pMAN885 | cat; multicloning sites | 23 |
| pMAN885EH | cat; multicloning sites | 23 |
| pMAN1015 | bla; lolC-lolD-lolE temperature-sensitive replicon | This study |
| pNASCD | cat; arabinose promoter-controlled lolC-lolD | This study |
| pNASDE | cat; arabinose promoter-controlled lolD-lolE | This study |
| pNASCDE | cat; arabinose promoter-controlled lolC-lolD-lolE | This study |
| pKM601 | cat; arabinose promoter-controlled gene for L10PSR | This study |
| pKT100 | spc; tacPO lppSR | This study |
Media and chemicals.
Luria-Bertani (LB) medium was used as the standard medium. Labeling experiments were carried out in M63 (NaCl)-maltose minimal medium (23) supplemented with 20 μg of thiamine, 40 μg of thymine, 40 μg of uracil, and 40 μg of each of the amino acids except methionine and cysteine/ml. When required, ampicillin, kanamycin, tetracycline, chloramphenicol, and spectinomycin were added at concentrations of 50, 25, 10, 35, and 25 μg/ml, respectively. Restriction enzymes and DNA-modifying enzymes were obtained from Takara Shuzo Co. Proteinase K was from Merck, and l-arabinose was from Wako Pure Chemicals Industries Ltd. Tran35S-label (a mixture of 70% [35S]methionine and 20% [35S]cysteine; 1,000 Ci/mmol) was obtained from ICN. LolA was purified as described previously (11). Anti-LolC antibodies against a synthetic peptide corresponding to the K239-to-E255 region of LolC were raised in rabbits. Other antibodies against purified proteins were raised.
Growth measurement.
The growth of E. coli cells was monitored by measuring the optical density (OD) at 660 nm. Overnight cultures at 30°C were diluted with prewarmed fresh LB medium and then incubated at 42°C. When OD reached 1, the culture was diluted 10-fold with fresh LB medium and then incubated further for various times. To determine the number of viable cells, aliquots of the culture were taken at various times, plated onto LB agar, and then incubated overnight at 30°C.
Construction of plasmids.
For the construction of helper plasmid pMAN1015, the coding regions of LolC, LolD, and LolE genes were amplified by PCR using pJY310 as a template and oligonucleotide primers U52 (5"-GATGAATTCGGAGGTTTAAATTTATGTACCAACCTGTCGCTCTATTTA-3") and W32 (5"-GAATTCAAGCTTACTGGCCGCTAAGGACTCGCGCAG-3"). The amplified DNA was digested with EcoRI and HindIII and then cloned into the same sites of pMAN997. For pNASCD, the coding regions of LolC and LolD genes were amplified using primers U51 (5"-CAGAATTCGAAGGAGATATAAATATGTACCAACCTGTCGCTC-3") and V31 (5"-CACTCTGCAGTTACTCCGCCCCCATCAG-3") and then cloned into the EcoRI-PstI sites of pMAN885EH. For pNASDE, the coding regions of LolD and LolE genes were amplified using primers V51 (5"-ACGATGAGCTCGAAGGAGATATAAATATGAATAAGATCCTGTTGCAATGC-3") and W31 (5"-AAGCCTGCAGTTTTTGTTCCACCAATATCAAACCC-3") and then cloned into the SacI-PstI sites of pMAN885EH. For pNASCDE, the coding regions of LolC, LolD, and LolE genes were amplified using primers U51 and W31 and then cloned into the EcoRI-PstI sites of pMAN885EH.
To construct pKM601 carrying a gene for L10PSR under the control of PBAD, a 0.6-kb BamHI-Bpu1102I fragment of pJYL10P (22) and a 0.4-kb Bpu1102I-XbaI fragment of pJY851 (23) were ligated with a 3.2-kb BamHI-XbaI fragment of pMAN885. L10PSR, a derivative of major outer membrane lipoprotein Lpp, carried Pro instead of Leu at position +10 and lacked the C-terminal Lys. This derivative remained release competent for a long time after its expression (22) and did not inhibit growth when it accumulated in the inner membrane (23). To construct pKT100 carrying spc and lppSR under the control of tacPO, the spc gene was amplified by PCR with pHM45Ω (14) as a template and inserted into the ScaI site in the bla gene of pJY151 (23).
Construction of a conditional lolCDE null mutant.
To replace the chromosomal lolC-lolD-lolE genes with the kan gene, plasmid pJY310, carrying lolC-lolD-lolE and flanking regions, was digested with XhoI and PmaCI (Fig. 1A) and then ligated with a DNA fragment containing kan, which was obtained by digestion of pSY343 (25) with HindIII, followed by treatment with T4 DNA polymerase and digestion with XhoI. The resultant plasmid, pMAN1014, carrying the ΔlolCDE::kan gene, was linearized with EcoRI and HindIII and then transformed into recD strain FS1576 to allow homologous recombination. Kanamycin-resistant transformants could be obtained at 30°C only when the lolC-lolD-lolE genes were provided by pMAN1015, which also carries a temperature-sensitive replicon derived from pMAN997. The ΔlolCDE::kan gene was then introduced into lpp mutant strain DLP79-36, harboring pMAN1015, by P1 transduction, followed by introduction of mutant recA from JC10240 by conjugation to construct CDE4. The chromosomal deletion of the lolC-lolD-lolE genes was confirmed by PCR.
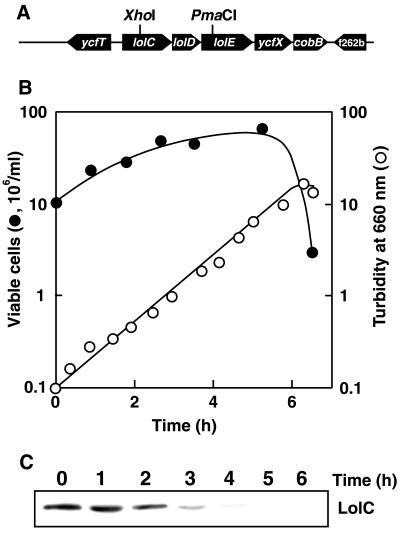
FIG. 1.
Construction of the ΔlolCDE::kan mutant and depletion of LolC at a nonpermissive temperature. (A) Restriction map for the lolC-lolD-LolE region. The XhoI-PmaCI region was replaced by the kan gene. (B) The CDE4 strain (ΔlolCDE::kan) harboring helper plasmid pMAN1015, which carries lolCDE and a temperature-sensitive replicon, was grown on LB medium at 30°C and then at 42°C for the indicated times by repeated inoculation of portions of the culture into fresh medium. Turbidity was monitored at 660 nm and plotted after correction for culture dilution. The numbers of viable cells at the indicated times were determined by plating aliquots of the culture onto LB plates, followed by overnight incubation at 30°C. The number of viable cells in 1 ml of culture was then calculated by correcting for the culture dilution. (C) CDE4 cells were grown at 42°C for the indicated times. Total membrane fractions derived from 2 × 107 cells were analyzed by SDS-PAGE and immunoblotting with anti-LolC antibodies.
Separation of the inner and outer membranes.
CDE4 cells harboring pKM601, which carries a gene for L10PSR under the control of PBAD, was grown at 30 or 42°C for 5 h on M63 (NaCl)-maltose minimal medium. L10PSR was induced for 5 min with 0.2% arabinose, followed by labeling for 1 min with Tran35S-label (10 μCi/ml). The labeled cells were converted into spheroplasts as described previously (11) and then disrupted by sonication. After removal of unbroken cells by centrifugation at 16,000 × g for 2 min, the envelope fraction was obtained by centrifugation at 100,000 × g for 30 min and then fractionated by 25 to 55% (wt/wt) sucrose density gradient centrifugation at 60,000 × g for 12 h to obtain the inner and outer membranes.
Release of LppSR from spheroplasts.
The release of LppSR from spheroplasts was examined as described previously (11). Briefly, CDE4 cells harboring the specified plasmids were grown at 42°C, induced with IPTG (isopropyl-β-d-thiogalactopyranoside), and then converted into spheroplasts. A suspension (300 μl) containing 5 × 108 spheroplasts was kept on ice for 3 min in the presence and absence of LolA (2 μg). M63 medium (750 μl) containing 0.25 M sucrose and 10 μCi of Tran35S-label was then added for 2-min labeling at 30°C. The labeling was chased by the addition of nonradioactive methionine and cysteine (each at 12 mM). The release of LppSR was terminated by chilling the reaction mixture in ice water and analyzed after fractionation into spheroplasts and medium by centrifugation at 16,000 × g for 2 min.
Immunoprecipitation, SDS-PAGE, and Western blot analyses.
Immunoprecipitation was carried out as described previously (12). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described previously (6). LolC was analyzed on a 10% polyacrylamide gel as described by Laemmli (8). Proteins labeled with Tran35S-label were analyzed by SDS-PAGE, followed by fluorography with Enlightning (NEN Life Science Products, Inc.). Western blot analyses were carried out as described previously (24).
RESULTS AND DISCUSSION
Construction of a conditional lolCDE null mutant.
We previously identified LolCDE as a lipoprotein-releasing apparatus of the inner membrane using proteoliposomes reconstituted from purified LolCDE and phospholipids (21). Moreover, a defective LolD mutant having a single amino acid substitution in the Walker A motif exhibited a dominant-negative effect and inhibited LolA-dependent lipoprotein release from spheroplasts (21). These biochemical data clarified the function of LolCDE. However, whether all three proteins are required and whether any other proteins can perform the function of LolCDE are still open to question. We addressed these issues by disrupting the lolCDE genes. We recently constructed a temperature-sensitive LolB null mutant and found that LolB depletion at a nonpermissive temperature was very efficient (19). Furthermore, a basal level of LolB was found to be sufficient for the outer membrane localization of lipoproteins except Lpp, suggesting that efficient depletion of LolCDE is also critical for characterization of the in vivo function of LolCDE. Based on these observations, a conditional lolCDE null mutant was constructed, as described in Materials and Methods.
The lolCDE genes are essential for E. coli.
CDE4 cells grown overnight on LB medium at 30°C were harvested and then incubated at 42°C, a nonpermissive temperature for the replication of pMAN1015. The turbidity of the culture stopped increasing after about eight generations at 42°C (Fig. 1B), whereas the number of viable cells in the culture ceased to increase earlier than the culture turbidity. SDS-PAGE and immunoblotting analysis with anti-LolC antibodies of the membrane fraction revealed the depletion of LolC with increasing incubation time at 42°C (Fig. 1C). These results indicate that disruption of the lolC-lolD-lolE genes is lethal for E. coli.
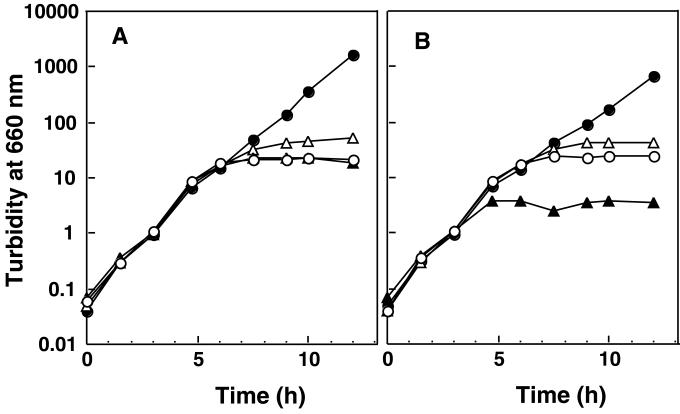
Both LolC and LolE are integral membrane proteins that are considered to constitute the membrane-spanning domains of the LolCDE complex (21). Both proteins are predicted to span the membrane four times and exhibit 26% identity in amino acid sequence. Moreover, the N-terminal 60 residues of the two proteins exhibit 55% identity (21). It seemed possible, therefore, that LolC and LolE could substitute for each other and that a functional complex could be formed from one of the two proteins and LolD. To determine whether or not both LolC and LolE are essential for growth, the lolC-lolD and lolD-lolE regions were cloned into plasmid vector pMAN885EH under the control of the PBAD promoter to construct pNASCD and pNASDE, respectively. Both pNASCD and pNASDE, as well as pMAN885EH, failed to support the growth of CDE4 at 42°C, irrespective of the presence or absence of arabinose (Fig. 2). When LolC and LolD were induced in the absence of LolE, the growth of CDE4 was more severely inhibited (Fig. 2B). On the other hand, pNASCDE carrying the lolC-lolD-lolE genes on pMAN885EH complemented the ΔlolCDE::kan mutation of CDE4 even in the absence of arabinose (Fig. 2A), suggesting that a basal level of LolCDE can support growth. These results indicate that both LolC and LolE are essential.
FIG. 2.
Complementation of the ΔlolCDE::kan mutation of CDE4. CDE4 was transformed with pMAN885EH, a cloning vector (open circles), pNASCD carrying lolC-lolD (solid triangles), pNASDE carrying lolDE (open triangles), or pNASCDE carrying lolCDE (solid circles) and then grown on LB medium at 42°C for the indicated times by repeated inoculation of portions of the cultures into fresh LB medium. Turbidity at 660 nm was measured to monitor the growth and plotted after correction for the culture dilution. (A) Growth without arabinose; (B) growth with arabinose.
Membrane localization of outer membrane lipoproteins in CDE4.
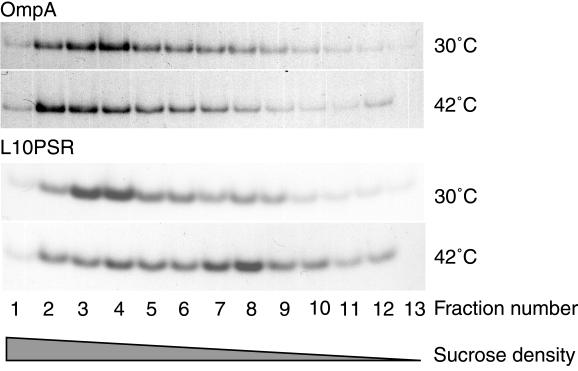
The LolCDE complex has been shown to catalyze the release of outer membrane lipoproteins from reconstituted proteoliposomes (21). Depletion of LolCDE is therefore expected to perturb the localization of outer membrane lipoproteins in vivo. To test this, we examined the membrane localization of L10PSR, a derivative of Lpp, under LolCDE-depleted conditions. CDE4 cells were transformed with pKM601, which carries a gene for L10PSR under the control of PBAD, and grown at 30 or 42°C on M63 (NaCl)-maltose minimal medium, and then 0.2% arabinose was added to induce L10PSR. The cells were labeled with Tran35S-label and then converted into spheroplasts. Cell envelope fractions prepared after sonic disruption of spheroplasts were separated by sucrose density gradient centrifugation. When cells were grown at 30°C, 35S-labeled L10PSR was recovered in higher-density fractions together with OmpA (Fig. 3). In contrast, when cells were grown at 42°C, the majority of 35S-labeled L10PSR was recovered in lower-density fractions, whereas the distribution of 35S-labeled OmpA at 42°C was essentially the same as that at 30°C. These results indicate that LolCDE is essential for the correct localization of outer membrane lipoproteins in vivo.
FIG. 3.
Effect of temperature on the in vivo localization of L10PSR in the lolCDE::kan mutant. CDE4 cells harboring pKM601 were grown at 30 or 42°C for 5 h and then induced for 5 min with 0.2% arabinose. Envelope fractions were prepared from 35S-labeled cells as described in Materials and Methods and separated by 25 to 55% sucrose density gradient centrifugation into fractions 1 to 13 from the bottom to the top. Each fraction was immunoprecipitated with anti-OmpA or anti-Lpp antibodies and then analyzed by SDS-PAGE and fluorography.
Both LolC and LolE are required for the release of outer membrane lipoproteins from the inner membrane.
To determine whether or not both LolC and LolE are essential for the release of lipoproteins, we constructed plasmid pKM100, which carries spectinomycin resistance gene spc and lppSR under the control of tacPO. This plasmid was transformed into CDE4 cells harboring pMAN885EH, pNASCD, pNASDE, or pNASCDE. The transformants were grown at 42°C, and then the expression of LppSR was induced. The induced cells were converted into spheroplasts and then labeled with Tran35S-label at 30°C in the presence and absence of LolA, followed by a chase with cold methionine and cysteine. LppSR remaining in spheroplasts or released into the spheroplast supernatant was examined by SDS-PAGE and fluorography (Fig. 4A). Spheroplasts prepared from cells harboring pNASCDE exhibited efficient release of LppSR in a LolA-dependent manner. In contrast, when LolC or LolE was depleted, the release of LppSR was severely inhibited (Fig. 4A), indicating that both LolC and LolE are essential for lipoprotein release.
FIG. 4.
Both LolC and LolE are essential for the release of lipoproteins from spheroplasts. (A) CDE4 cells harboring pMAN885EH (vector), pNASCD carrying lolCD, pNASDE carrying lolDE, or pNASCDE carrying lolCDE were transformed with pKM100 carrying lppSR and then grown at 42°C for 9 h. LppSR was induced with 1 mM IPTG for 5 min immediately prior to conversion of the cells into spheroplasts. Spheroplasts were labeled with Tran35S-label in the presence and absence of LolA for 2 min at 30°C, followed by a 2-min chase with cold methionine and cysteine. The reaction mixture, containing 5 × 108 spheroplasts, was fractionated into spheroplasts (p) and a supernatant (s). LppSR in each fraction was immunoprecipitated and then analyzed by SDS-PAGE and fluorography. (B) CDE4 cells harboring pKM601 were grown at 30 or 42°C for 5 h and then converted into spheroplasts, followed by labeling with Tran35S-label at 30°C as described above in the absence of LolA. The suspension, containing 5 × 108 spheroplasts, was treated with or without proteinase K (PK; 0.2 mg/ml) for 30 min at 4°C. After precipitation with trichloroacetic acid and resolubilization, the mixtures were immunoprecipitated with anti-elongation factor TU (EF-Tu) or anti-Pal antibodies and then analyzed by SDS-PAGE and fluorography.
We previously observed that both the LolD mutation (21) and LolB depletion (19) inhibited the release of lipoproteins from the inner membrane without perturbation of the translocation of lipoprotein precursors across the inner membrane and subsequent processing to mature lipoproteins. The mobility of lipoproteins remaining in the inner membrane (Fig. 3 and 4) upon LolCDE depletion was indistinguishable from that of lipoproteins localized in the outer membrane (Fig. 3) or released into the spheroplast supernatant (Fig. 4). These results, taken together, strongly indicate that the Sec-dependent translocation and modification of the lipoprotein precursors are completely independent of and not affected by the Lol-mediated reactions. Since the LolCDE complex mediates the first step of lipoprotein localization reactions, we examined whether or not lipoproteins remaining in spheroplasts upon LolCDE depletion are exposed on the outer surface of the inner membrane and therefore sensitive to external proteinase K. Since Lpp forms a proteinase K-resistant structure (11), we examined the proteinase K sensitivity of outer membrane lipoprotein Pal, expressed from the chromosome. CDE4 cells harboring pKM601 were grown at 30 or 42°C and then converted into spheroplasts, followed by pulse labeling. Like L10PSR expressed from pKM601, Pal remained in LolCDE-depleted spheroplasts even in the presence of LolA (data not shown). When the spheroplasts were treated with proteinase K, Pal was completely digested (Fig. 4B). Pal remaining in LolCDE-containing spheroplasts due to the omission of LolA was also proteolyzed. On the other hand, cytoplasmic elongation factor Tu was resistant to proteinase K. These results indicate that the accumulation of mature lipoproteins in the inner membrane due to LolCDE depletion does not affect upstream reactions, which lead to the formation of mature lipoproteins on the outer surface of the inner membrane.
Depletion of the LolCDE complex or its components nearly completely inhibited the release of lipoproteins from spheroplasts (Fig. 4A), indicating that LolCDE is the sole lipoprotein-releasing apparatus functioning in spheroplasts. On the other hand, a portion of L10PSR was localized in the outer membranes of LolCDE-depleted cells (Fig. 3), suggesting that other proteins, i.e., in addition to LolCDE, might be involved in the outer membrane localization of lipoproteins in cells. However, the presence of a second system seems to be unlikely for the following reasons. The outer membrane localization in cells takes place very rapidly even in the presence of a basal level of Lol proteins (19). Since the depletion of any one of the Lol factors causes severe inhibition of not only growth but also protein synthesis, radiolabeling of lipoproteins had to start immediately before growth inhibition occurred. It is therefore likely that a portion of L10PSR was localized in the outer membrane due to incomplete depletion of LolCDE.
E. coli has been predicted to possess 57 ABC transporters (9). As far as is known, LolCDE and MsbA (7) are the only essential ABC transporters of E. coli. MsbA was recently found to be involved in the transport of lipids and lipopolysaccharides from the inner membrane to the outer membrane (2). Whether or not lipid transport through the periplasm involves a periplasmic carrier protein such as LolA is of great interest.
Acknowledgments
We thank Masaaki Wachi for E. coli DLP79-36 and Rika Ishihara for technical assistance and secretarial support.
This work was supported by grants to H.T. from CREST of the Japan Science and Technology Corporation and from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Csonka, L. N., and A. J. Clark. 1980. Construction of an Hfr strain useful for transferring recA mutations between Escherichia coli strains. J. Bacteriol. 143:529-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doerrler, W. T., M. C. Reedy, and C. R. Raetz. 2001. An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 276:11461-11464. [DOI] [PubMed] [Google Scholar]
- 3.Duong, F., J. Eichler, A. Price, M. R. Leonard, and W. Wickner. 1997. Biogenesis of the Gram-negative bacterial envelope. Cell 91:567-573. [DOI] [PubMed] [Google Scholar]
- 4.Gennity, J. M., and M. Inouye. 1991. The protein sequence responsible for lipoprotein membrane localization in Escherichia coli exhibits remarkable specificity. J. Biol. Chem. 266:16458-16464. [PubMed] [Google Scholar]
- 5.Hayashi, S., and H. C. Wu. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 164:451-471. [DOI] [PubMed] [Google Scholar]
- 6.Hussain, M., S. Ichihara, and S. Mizushima. 1980. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J. Biol. Chem. 255:3707-3712. [PubMed] [Google Scholar]
- 7.Karow, M., and C. Georgopoulos. 1993. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutants in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 7:69-79. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 9.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 10.Matsuyama, S., Y. Yokota, and H. Tokuda. 1997. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 16:6947-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama, S., T. Tajima, and H. Tokuda. 1995. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 14:3365-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuyama, S., Y. Fujita, and S. Mizushima. 1993. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 12:265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogura, T., P. Bouloc, H. Niki, R. D'Ari, S. Hiraga, and A. Jaffe. 1989. Penicillin-binding protein 2 is essential in wild-type Escherichia coli but not in lov or cya mutants. J. Bacteriol. 171:3025-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prentki, P., and H. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 15.Pugsley, A. 1993. The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seydel, A., P. Gounon, and A. P. Pugsley. 1999. Testing the “+2 rule” for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34:810-821. [DOI] [PubMed] [Google Scholar]
- 17.Stahl, F. W., I. Kobayashi, D. Thaler, and M. M. Stahl. 1986. Direction of travel of RecBC recombinase through bacteriophage lambda DNA. Genetics 113:215-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tajima, T., N. Yokota, S. Matsuyama, and H. Tokuda. 1998. Genetic analysis of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins. FEBS Lett. 439:51-54. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka, K., S. Matsuyama, and H. Tokuda. 2001. Deletion of lolB, encoding an outer membrane lipoprotein, is lethal for Escherichia coli and causes the accumulation of lipoprotein localization intermediates in the periplasm. J. Bacteriol. 183:6538-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terada, M., K. Kuroda, S. Matsuyama, and H. Tokuda. 2001. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 276:47690-47694. [DOI] [PubMed] [Google Scholar]
- 21.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212-218. [DOI] [PubMed] [Google Scholar]
- 22.Yakushi, T., N. Yokota, S. Matsuyama, and H. Tokuda. 1998. LolA-dependent release of a lipid-modified protein from the inner membrane of Escherichia coli requires nucleoside triphosphate. J. Biol. Chem. 273:32576-32581. [DOI] [PubMed] [Google Scholar]
- 23.Yakushi, T., T. Tajima, S. Matsuyama, and H. Tokuda. 1997. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J. Bacteriol. 179:2857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada, H., S. Matsuyama, H. Tokuda, and S. Mizushima. 1989. A high concentration of SecA allows proton motive force-independent translocation of a model secretory protein into Escherichia coli membrane vesicles. J. Biol. Chem. 264:18577-18581. [PubMed] [Google Scholar]
- 25.Yasuda, S., and T. Takagi. 1983. Overproduction of Escherichia coli replication proteins by the use of runaway-replication plasmids. J. Bacteriol. 154:1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokota, N., T. Kuroda, S. Matsuyama, and H. Tokuda. 1999. Characterization of the LolA-LolB system as the general lipoprotein localization mechanism of Escherichia coli. J. Biol. Chem. 274:30995-30999. [DOI] [PubMed] [Google Scholar]