Abstract
A direct sulfhydrylation pathway for methionine biosynthesis in Corynebacterium glutamicum was found. The pathway was catalyzed by metY encoding O-acetylhomoserine sulfhydrylase. The gene metY, located immediately upstream of metA, was found to encode a protein of 437 amino acids with a deduced molecular mass of 46,751 Da. In accordance with DNA and protein sequence data, the introduction of metY into C. glutamicum resulted in the accumulation of a 47-kDa protein in the cells and a 30-fold increase in O-acetylhomoserine sulfhydrylase activity, showing the efficient expression of the cloned gene. Although disruption of the metB gene, which encodes cystathionine γ-synthase catalyzing the transsulfuration pathway of methionine biosynthesis, or the metY gene was not enough to lead to methionine auxotrophy, an additional mutation in the metY or the metB gene resulted in methionine auxotrophy. The growth pattern of the metY mutant strain was identical to that of the metB mutant strain, suggesting that both methionine biosynthetic pathways function equally well. In addition, an Escherichia coli metB mutant could be complemented by transformation of the strain with a DNA fragment carrying corynebacterial metY and metA genes. These data clearly show that C. glutamicum utilizes both transsulfuration and direct sulfhydrylation pathways for methionine biosynthesis. Although metY and metA are in close proximity to one another, separated by 143 bp on the chromosome, deletion analysis suggests that they are expressed independently. As with metA, methionine could also repress the expression of metY. The repression was also observed with metB, but the degree of repression was more severe with metY, which shows almost complete repression at 0.5 mM methionine in minimal medium. The data suggest a physiologically distinctive role of the direct sulfhydrylation pathway in C. glutamicum.
Corynebacterium glutamicum is a gram-positive nonsporulating organism and has been widely used for the industrial production of amino acids. Due to the role of the organism in amino acid production, biosynthetic pathways leading to lysine and other industrially important amino acids have been studied in detail (16, 20, 23, 32).
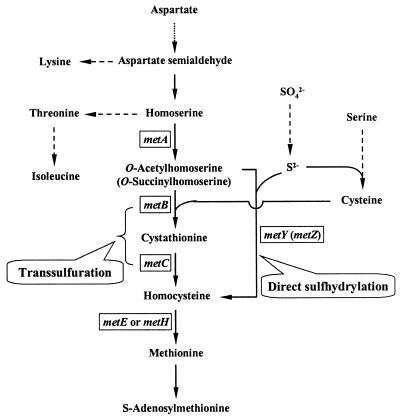
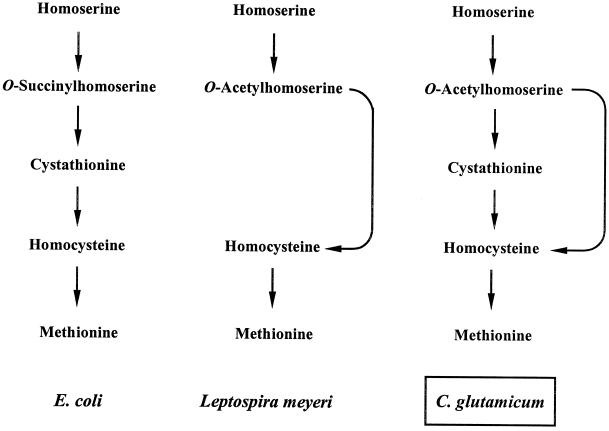
The biosynthetic pathways leading to methionine have been studied in diverse organisms and show similarities as well as differences (Fig. 1) (47). The first step, acylation of homoserine catalyzed by homoserine acetyl (or succinyl)-transferase (the product of metA), is common to all of the organisms, even though the source of the transferred acyl group is different. While the enzyme uses succinyl coenzyme A (succinyl-CoA) as the substrate in several prokaryotes such as enteric bacteria (3, 10, 38, 41), it uses acetyl-CoA in many other organisms, such as Saccharomyces cerevisiae (42), Neurospora crassa (18), Aspergillus nidulans (25), Leptospira meyeri (4), Brevibacterium flavum (26), and C. glutamicum (17, 29). As a result, the product of the step is either acetylhomoserine or succinylhomoserine, depending on the organism. Formation of homocysteine from acylhomoserine can occur in two different ways (Fig. 1). The transsulfuration pathway via cystathionine utilizes cysteine as the sulfur donor, while the direct sulfhydrylation pathway utilizes inorganic sulfur assimilated from sulfate. Escherichia coli uses the transsulfuration pathway which is catalyzed by cystathionine γ-synthase (the product of metB) and cystathionine β-lyase (the product of metC) (38). Organisms such as S. cerevisiae (6, 42), Rhizobium etli (41), Pseudomonas aeruginosa (10), and L. meyeri (2) utilize the direct sulfhydrylation pathway catalyzed by acylhomoserine sulfhydrylase (the product of metY or metZ). The last step, formation of methionine from homocysteine, is catalyzed by homocysteine methyltransferase encoded by metE (or metH). Although yeast, fungi, and green plants were reported to have functional transsulfuration and direct sulfhydrylation pathways (8, 13, 25, 42), no prokaryote has been clearly shown to have both pathways. Unlike the closely related Brevibacterium flavum, which may use only the direct sulfhydrylation pathway for methionine biosynthesis (28), enzyme activities of the transsulfuration pathway have been detected in the extracts of C. glutamicum cells, and the pathway has been shown to be functional for methionine biosynthesis in the organism (14, 17, 19).
FIG. 1.
Biosynthesis of methionine in eubacteria. In E. coli, the metA gene expresses homoserine succinyltransferase, which converts homoserine into O-succinylhomoserine. In Bacillus subtilis, Brevibacterium flavum, C. glutamicum, and L. meyeri, the metA gene expresses homoserine acetyltransferase which uses acetyl-CoA as the acyl donor to produce O-acetylhomoserine. Depending on the organism, formation of homocysteine from acylhomoserine can occur in two different routes. Transsulfuration is mediated by metB and metC, and direct sulfhydrylation is mediated by metY (or metZ). Unlike enteric bacteria, which preferentially use the transsulfuration pathway, Brevibacterium flavum (which is closely related to C. glutamicum) uses only the direct sulfhydrylation pathway to produce homocysteine.
Even though some genes involved in methionine biosynthesis in C. glutamicum were isolated in recent years, information on the biosynthesis of methionine in the organism is still very limited, and the biosynthetic pathways have not been clarified yet. We recently isolated the metA, metB, and metC genes of C. glutamicum and demonstrated the functionality of the transsulfuration pathway at genetic and biochemical levels. However, unlike the metA mutant of C. glutamicum, the metB and metC mutant strains showed methionine prototrophy, suggesting the presence of additional route(s) which may bypass the transsulfuration pathway (14, 19).
In this study, we provide evidence that the additional route is the direct sulfhydrylation pathway mediated by metY. The molecular characteristics of the metY gene, encoding O-acetylhomoserine sulfhydrylase, are also investigated. Finally, based on biochemical and genetic evidence, we propose a methionine biosynthetic pathway of C. glutamicum.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli and C. glutamicum cells were cultured at 37°C in Luria-Bertani (LB) medium (33) and at 30°C in MB (12), respectively. Minimal media for E. coli and C. glutamicum were M9 (33) and MCGC (45), respectively. Glucose was added to a final concentration of 1%. Sulfur-free minimal medium (pH 7.4) for C. glutamicum was composed of 1% glucose, 50 mM Na2HPO4, 25 mM KH2PO4, 20 mM NaCl, 30 mM NH4Cl, 2 mM MgCl2, 0.5 mM CaCl2, 0.1 mM FeCl2, 0.01 mM MnCl2, and 1 mg of biotin/liter. The sulfate concentration of the medium was adjusted with ammonium sulfate. Ampicillin and kanamycin were added to final concentrations of 50 and 25 mg/liter, respectively. All amino acids were added to a final concentration of 40 mg/liter.
TABLE 1.
Bacterial strains, plasmids, and primers
| Strain or plasmid | Relevant genotype, phenotype, or sequencea | Source or reference |
|---|---|---|
| E. coli | ||
| DH5αF" | φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 | Bethesda Research Laboratory |
| CGSC 2569 | Δ(gpt-proA)62 lacY1 tsx-29 glnV44 galK2 hisG4 xylA5 mtl-1 argE3 thi-1 metA28 | CGSCb |
| CGSC 4896 | argF58 relA1 spoT1 metB1 | CGSC |
| CGSC 6684 | ecfA4 metC186 spoT1 thi-1 | CGSC |
| C. lactofermentum ATCC 13869 | Glutamic acid producer | ATCCc |
| B. flavum ATCC 14067 | Glutamic acid producer | ATCC |
| C. glutamicum | ||
| ATCC 13032 | Wild type | ATCC |
| AS019 | Spontaneous rifampin-resistant mutant of ATCC 13059 | 49 |
| AS019E12 | Restriction-deficient variant of AS019 | 11 |
| HL921 | metY-disrupted AS019E12 | This study |
| HL938 | metB-disrupted AS019E12 | This study |
| HL939 | metY metB double-disrupted AS019E12 | This study |
| Plasmids | ||
| pUC19 | Cloning vector, Apr | 48 |
| pK19mobsacB | Integration vector, mob sacB Kmr | 34 |
| pMT1 | E. coli-C. glutamicum shuttle vector, Apr Kmr | 12 |
| pSL72 | pMT1 carrying metY and metA | 29 |
| pSL73 | pMT1 carrying metY and metA | 29 |
| pSL75 | pMT1 carrying 2.0-kb XhoI-SalI fragment of pSL73, metA | 29 |
| pSL79 | pMT1 carrying 3.0-kb SphI-SalI fragment of pSL73, metY and metA | 29 |
| pSL90 | pMT1 carrying 2.0-kb XhoI-ScaI fragment of pSL72, metY | 29 |
| pSL123 | pMT1 carrying metB | 14 |
| pSL191 | pMT1 carrying 2.2-kb SphI-ScaI fragment of pSL73, metY | This study |
| pSL312 | pK19mobsacB carrying ΔmetY | This study |
| pSL315 | pK19mobsacB carrying ΔmetB | This study |
| Oligonucleotidesd | ||
| ppYA | 5"-CGCGGATCCATGGGTGTTTCTGTATGC-3" | This study |
| ppYB | 5"-CCCATCCACTAAACTTAAACAGGTAGCGATGGTGTTGTC-3" | This study |
| ppYC | 5"-TGTTTAAGTTTAGTGGATGGGACCACCACCCATTCACAG-3" | This study |
| ppYD | 5"-CGCGGATCCTGCAGCCGGATTCGTAGT-3" | This study |
| ppBA | 5"-CGCGGATCCGACTGTTTCAGAAGTGAT-3" | This study |
| ppBB | 5"-CCCATCCACTAAACTTAAACAGATGTGATCGCCCGGCTT-3" | This study |
| ppBC | 5"-TGTTTAAGTTTAGTGGATGGGAATCACCCAGGCCACGAA-3" | This study |
| ppBD | 5"-CGCGGATCCGACTACACCTTTGACAAT-3" | This study |
r superscript indicates resistance. Ap, ampicillin.
E. coli Genetic Stock Center, Yale University, New Haven, Conn.
ATCC, American Type Culture Collection, Rockville, Md.
Sequences shown in italics indicate BamHI restriction sites for cloning into the integration vector pK19mobsacB. Underlined sequences are the annealing region for the secondary PCR (22).
DNA technology.
Standard molecular cloning, transformation, and electrophoresis procedures were used (33). ExTaq DNA polymerase, restriction endonucleases, and modifying enzymes were purchased from Takara (Takara Shuzo Co., Tokyo, Japan) and used as described in the manufacturer's instructions. E. coli DH5αF" and C. glutamicum AS019E12 were used as hosts for typical transformation. Transformation of C. glutamicum was performed by electroporation by using the methods of van der Rest et al. (44), and transformants were selected in MB containing kanamycin. Plasmid preparation for C. glutamicum was performed as previously described (49).
For nucleotide sequence analysis of metY, plasmids pSL72 and pSL73 were used as templates. The complete nucleotide sequence of metY was determined commercially at the Korea Research Center for Basic Sciences (Taejon, Korea) with universal and synthetic oligonucleotide primers. A sequence similarity search of nucleotide and amino acid sequences was performed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) by using the basic local alignment search tool (BLAST) (1). Pairwise sequence alignments were performed at the website of ExPASy Proteomics Tools (http://www.expasy.ch/tools/) by using the CLUSTAL W alignment method (43).
Construction of plasmids.
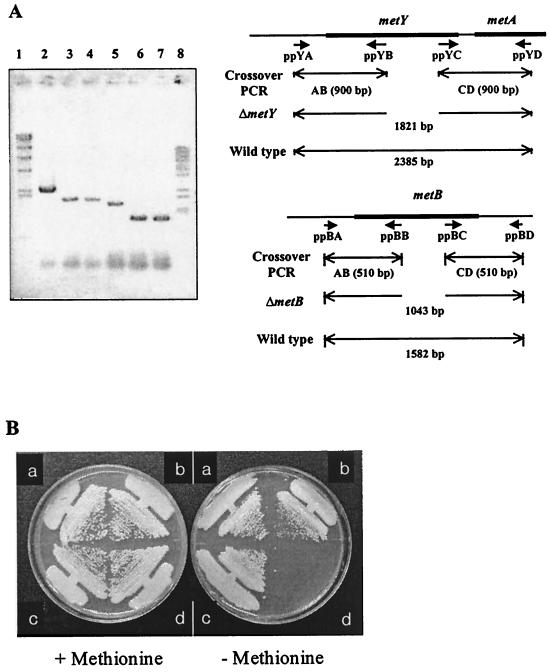
Plasmid pSL191 was constructed by inserting the blunt-ended 2.2-kb SphI-ScaI fragment of pSL73 into the SmaI-digested pMT1. Plasmids pSL312 and pSL315 were constructed by the crossover PCR method as described previously (22). The primers used are listed in Table 1, and their annealing regions are shown in Fig. 6A. The primary PCR products, AB and CD, were amplified with 600 nM concentrations of the outer primers and 60 nM concentrations of the inner primers. These products were directly used as templates for the secondary PCR with 600 nM concentrations of the outer primers. Secondary PCR products were digested with BamHI and ligated into the BamHI-digested pK19mobsacB. The ligation mixture was used to transform E. coli DH5αF", and the transformants were selected on LB plates containing 25 mg of kanamycin and 40 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/liter.
FIG. 6.
Construction of C. glutamicum AS019E12 mutant in metY and metB genes (A) and growth properties of the mutants (B). (A) Chromosomal deletion of the gene was identified in agarose gels by PCR amplification. To test for the deletion, primers of ppYA and ppYD were used for metY (lanes 2, 3, and 4) and primers of ppBA and ppBD were used for metB (lanes 5, 6, and 7). The predicted lengths of the amplified fragment are shown. Lanes: 1, λ-HindIII size marker; 2 to 4, C. glutamicum AS019E12, HL921, and HL939, respectively; 5 to 7, C. glutamicum AS019E12, HL938, and HL939, respectively; 8, λ-BstEII size marker. (B) Abilities of the constructed mutant strains to grow on MCGC minimal medium. The left plate contains supplemental methionine, whereas the right plate does not. a to d, C. glutamicum AS019E12, HL938, HL921, and HL939, respectively.
Site-specific gene disruption.
Site-specific gene disruption was performed by using the nonreplicable integration vector pK19mobsacB, which enables marker-free deletion of the target gene (34). Plasmids pSL312 and pSL315 were introduced into C. glutamicum AS019E12 by electroporation (44). Integration of the introduced plasmid into the chromosome by a single crossover was monitored on LB plates containing 25 mg of kanamycin/liter. The single crossover was confirmed by the inability of the cells to grow on LB plates containing 10% sucrose. For the deletion of the target gene and the vector by another round of a crossover, the kanamycin-resistant (Kmr) and sucrose-sensitive cells were grown for 8 h in the LB medium and spread onto LB plates containing 10% sucrose. Cells growing on the plates were picked and tested for the deletion of the target gene by PCR.
Biochemical analysis.
Cell extracts were prepared as described previously (15). O-Acetylhomoserine was synthesized by the method of Nagai and Flavin (27). The activities of cystathionine γ-synthase were determined by the method of Ravanel et al. (30), which measures the disappearance of cysteine by the ninhydrin reaction. O-Acetylhomoserine sulfhydrylase was assayed as homocysteine formation by using the nitroprusside reaction (10). Protein was measured by the method of Bradford (5), with bovine serum albumin as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as previously described (21).
GenBank accession number.
The nucleotide sequence of metY was deposited in GenBank under accession number AF220150.
RESULTS
Characteristics of the C. glutamicum metY gene.
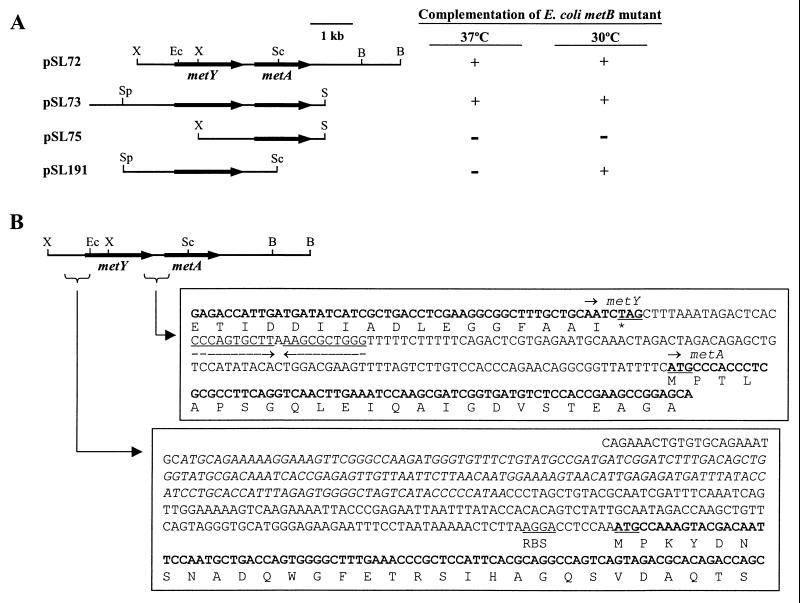
We previously isolated and characterized the metA gene required for methionine biosynthesis in C. glutamicum (29). In order to find additional met genes, we extended our sequence analysis to the upstream and downstream region of the metA gene by using plasmids pSL72 and pSL73 (Fig. 2A) as the templates. In the immediate upstream of metA, we found an open reading frame (ORF) that consisted of 1,314 bp and was separated by 143 bp from the metA gene (Fig. 2B). The ATG start site was chosen based on the similarities of the amino acid sequences with other O-acetylhomoserine sulfhydrylases. Judging from the length of the ORF, no other upstream ATGs were expected to serve as the start site. A potential ribosome-binding site (36) of AGGA was located 8 bp upstream from the ATG (Fig. 2B). A sequence of TAG was identified as the stop codon for the ORF. The GC content of the ORF was 57%, which is typical of C. glutamicum genes. The codon preference was also very similar to that of previously reported corynebacterial genes and, interestingly, it also indicated that the ORF could encode a protein that is expressed at a high level (24).
FIG. 2.
Clones and subclones of metY (A) and nucleotide sequences of the metY-flanking regions (B). (A) E. coli metB mutant cells were transformed with the clones and tested for growth on the M9 minimal media. (B) Nucleotide sequences of the upstream and downstream region of metY gene. RBS, a putative ribosome-binding site. Sequences underlined with facing arrows indicate a putative rho-independent transcription stop signal. Sequences shown in italics indicate a putative leader sequence. Abbreviations: B, BamHI; Ec, EcoO109I; S, SalI; Sc, ScaI; Sp, SphI; X, XhoI.
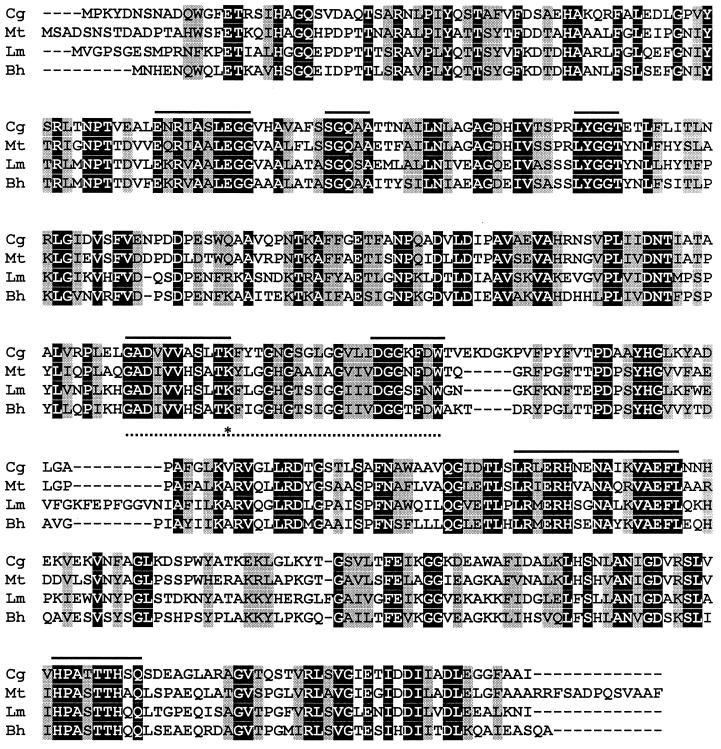
The putative gene product consisted of 437 amino acids encoding a 46,751-Da protein with a predicted isoelectric point of 5.0. The translated amino acid sequence of the ORF was compared with the sequences in the protein database. Among the known proteins, the putative O-succinylhomoserine (thiol)-lyase of Mycobacterium tuberculosis (7) gave the highest score, with an amino acid identity of 58% (Fig. 3). O-Acetylhomoserine sulfhydrylases of L. meyeri and Bacillus halodurans (2, 40) showed 47% identity. Although the identical amino acids were fairly well distributed throughout the sequences, close analysis of the amino acid sequences revealed seven conserved motifs that may be involved in the catalytic activity of the enzyme. In addition, a binding motif for pyridoxal 5"-phosphate was identified (Fig. 3). We named the corynebacterial gene metY, based on its amino acid sequence similarities.
FIG. 3.
Multiple sequence alignment of O-acetylhomoserine sulfhydrylases. Conserved and functionally similar amino acids are indicated by black and shaded boxes, respectively. Conserved motifs are indicated by solid bars. The binding motif for pyridoxal 5"-phosphate is shown with a dotted bar. An asterisk indicates the lysine residue which binds pyridoxal 5"-phosphate. Abbreviations: Cg, O-acetylhomoserine sulfhydrylase of C. glutamicum (AF220150); Mt, O-succinylhomoserine (thiol)-lyase of M. tuberculosis (AL021841); Lm, O-acetylhomoserine sulfhydrylase of L. meyeri (T44655); Bh, O-acetylhomoserine sulfhydrylase of Bacillus halodurans (AP001516).
Although located in close proximity, the metA gene appeared to be expressed independently of the metY gene, since a putative rho-independent transcriptional stop signal was found at the downstream region of metY gene , as shown in Fig. 2. In addition, deleting the metY gene did not affect the metA expression and vice versa (Fig. 2 and Table 2).
TABLE 2.
Enzymatic activities of O-acetylhomoserine sulfhydrylase and cystathionine γ-synthasea
| Strain | Plasmid | Phenotype | Sp act (nmol min−1 mg−1) of:
|
Growth on minimal mediumb | |
|---|---|---|---|---|---|
| OAHSH | CGS | ||||
| AS019E12 | |||||
| pMT1 | Empty vector | 14 | 17 | + | |
| pSL73 | metY-metA clone | 426 | ND | + | |
| pSL75 | metA clone | 17 | ND | + | |
| pSL191 | metY clone | 415 | ND | + | |
| pSL123 | metB clone | NDd | 113 | + | |
| HL921 | ΔmetYc | 1.4 | 13 | + | |
| HL938 | ΔmetBc | 10.1 | 4.0 | + | |
| HL939 | ΔmetY ΔmetBc | 1.4 | 3.8 | − | |
The enzymes were induced by growth of C. glutamicum cells to the stationary phase on MCGC minimal medium containing 1% glucose. Cells were harvested, disrupted, and assayed for activity as described in Materials and Methods. Enzyme reactions were carried out for 10 min at 30°C. The enzyme activities represent the mean of three independent experiments. OAHSH, O-acetylhomoserine sulfhydrolase; CGS, cystathione γ-synthase.
MCGC minimal medium was used. Growth was monitored on agar plates.
The mutants were generated in this study.
ND, not determined.
Complementation of E. coli metB and metA mutants.
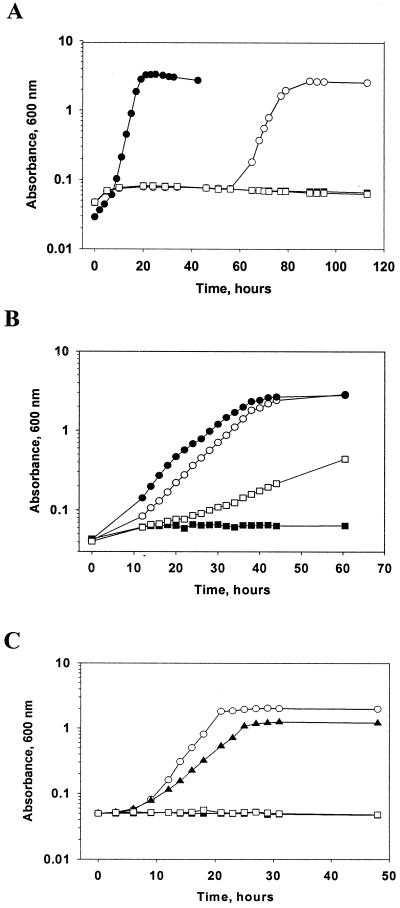
To test for the possibility of complementation, we introduced the C. glutamicum metY gene into E. coli CGSC 4896, an E. coli metB mutant strain, and tested for the growth of the strain on a minimal medium. When the complementation tests were performed at 37°C, a clone carrying both metY and metA (metY-metA) could complement the E. coli metB mutant strain, although a clone carrying only the metY or metA gene could not (Fig. 2). However, as shown in Fig. 4A, the complemented strain showed a long lag period of 60 h. The observed growth rate of the strain complemented with the clone carrying metY-metA was 0.184 h−1, which is 50% slower than that of the strain complemented with the clone carrying C. glutamicum metB gene. However, as shown in Fig. 4B, the long lag disappeared soon when the complementation test was performed at 30°C, suggesting that the metA product of C. glutamicum may show temperature sensitivity, as found with those of E. coli (31) and Bacillus polymyxa (46). Unlike the complementation of the E. coli metB mutation with corynebacterial metY at 37°C, some degree of growth was observed at 30°C (Fig. 4B), which may also suggest temperature sensitivity of the metY product. For E. coli CGSC 2569, a metA mutant, a fragment carrying the C. glutamicum metY-metA complemented better than the one carrying only the metA gene, and it was the same at 37°C (Fig. 4C) or 30°C (data not shown). The observed growth rate at 37°C after complementation by the metA gene was 0.074 h−1, only a 51% level compared to that achieved by the metY-metA genes. The enzyme activities of homoserine acetyltransferase, the product of the metA gene, were similar in these two strains. E. coli CGSC 2569 cells carrying metY-metA or metA showed specific activities of 694 and 768 nmol mg−1 min−1, respectively, showing that the different growth pattern was not caused by the enzyme activity. Even though these data suggest that the methionine biosynthesis of C. glutamicum shows similarities in the overall pathways to that of E. coli, it also shows a clear difference presumably mediated by the corynebacterial metY. Based on these data, we concluded that (i) O-acetylhomoserine produced by the corynebacterial homoserine acetyltransferase (metA) is inefficiently utilized in E. coli; (ii) while the protein product of the metY gene utilize O-succinylhomoserine very poorly as the substrate, it can efficiently use O-acetylhomoserine produced from the corynebacterial metA; (iii) the metY gene of C. glutamicum encodes an enzyme that can replace the E. coli metB or metC gene product, but with different substrate specificity; and (iv) the metY gene may encode O-acetylhomoserine sulfhydrylase catalyzing the direct sulfhydrylation pathway which is not present in E. coli. Our effort to complement E. coli metC mutant with either metY-metA or metY was unsuccessful due to the leakiness of the metC mutation (37).
FIG. 4.
Complementation of E. coli metB and metA mutants. Clones of metY, metA, metY-metA, or metB were transformed into E. coli metB (A and B) or metA (C) mutant strains, and the transformed strains were grown in M9 minimal medium lacking supplemental methionine at 37°C (A and C) or 30°C (B). Growth measurements were performed with two independent cultures. Symbols: ▪, pMT1 (empty vector); □, pSL191 (metY); •, pSL123 (metB); ○, pSL73 (metY metA); ▴, pSL75 (metA).
Expression of O-acetylhomoserine sulfhydrylase from metY.
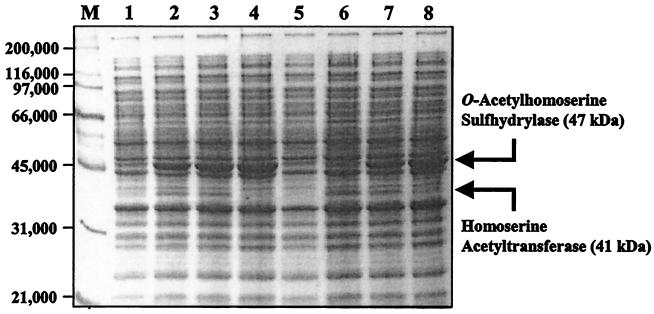
The ability of the metY clones to express O-acetylhomoserine sulfhydrylase was tested by enzymatic assays. Crude extracts were prepared from the various C. glutamicum AS019E12 cells harboring metY and/or metA clone(s) and assayed (Table 2). Introduction of the metY clone pSL73 or pSL191 into C. glutamicum AS019E12 increased O-acetylhomoserine sulfhydrylase activity ca. 30-fold. As evidenced by the complementation analysis, no O-acetylhomoserine sulfhydrylase activity was detected with O-succinylhomoserine as the substrate (data not shown). SDS-PAGE analysis of the crude extracts obtained from the C. glutamicum ASO19E12 cells harboring plasmid pSL73 or pSL191 revealed a putative O-acetylhomoserine sulfhydrylase band with an approximate Mr of 47,000 (Fig. 5). The data are in good agreement with the predicted molecular mass of 46,751 Da. The intensity of the protein band was roughly proportional to the activities observed in Table 2. The presence or absence of the protein band agreed with the enzymatic assay data. It is unique to observe an approximate 30-fold increase in the O-acetylhomoserine sulfhydrylase activity by the introduction of cloned metY gene into C. glutamicum, since it is common to observe only a 7- to 9-fold amplification for other genes cloned into the vector pMT1 (12). For example, introduction of the metB cloned pSL123 into C. glutamicum achieved a sevenfold increase in the cystathionine γ-synthase activity (Table 2). Introduction of the clones carrying metA and metY, such as pSL72 and pSL73, into the C. glutamicum AS019E12 cells resulted in the expression of an additional polypeptide with approximate Mr of 41,000 (Fig. 5). The protein band was not detected in the extracts of the cells carrying plasmid pSL191, a clone that does not carry metA (Fig. 2). The result indicates that the additional band may be homoserine acetyltransferases expressed from the metA gene. The level of the protein O-acetylhomoserine sulfhydrylase expression achieved by the metY gene was higher than that of the protein homoserine acetyltransferase by the metA gene, as evidenced by the strong intensity of the protein band (Fig. 5) and higher enzymatic activities of O-acetylhomoserine sulfhydrylase (46.1 nmol min−1 mg−1) than those of homoserine acetyltransferase (32.7 nmol min−1 mg−1). The data agree with the codon preference data (24), which indicated an efficient expression of the metY gene, and they also suggest a greater efficiency in the expression of the metY gene compared to that of the metA gene.
FIG. 5.
Expression of O-acetylhomoserine sulfhydrylase and homoserine acetyltransferase from various metY and/or metA clones. Plasmids were introduced into the C. glutamicum AS019E12 cells, and crude extracts were prepared from the cells grown in the MCGC minimal media. Proteins were separated by SDS-12% PAGE. The protein bands corresponding to the O-acetylhomoserine sulfhydrylase (metY) and homoserine acetyltransferase (metA) are indicated. Lane M, molecular size markers; lane 1, AS019E12/pMT1 (empty vector); lane 2, AS019E12/pSL72 (metY metA); lane 3, AS019E12/pSL73 (metY metA); lane 4, AS019E12/pSL191 (metY); lane 5, HL921 (ΔmetY); lane 6, AS019E12/pSL75 (metA); lane 7, AS019E12/pSL79 (metY metA); lane 8, AS019E12/pSL90 (metY).
Evidence for the direct sulfhydrylation pathway in C. glutamicum.
To address the role of metY, we constructed a metY mutant strain by using the cloned gene. The constructed mutant strain (HL921) carried an internally deleted metY gene (Fig. 6). As it was observed in metB (14) and metC (19) mutant strains of C. glutamicum HL921 also showed prototrophy for methionine (Fig. 6B), but the O-acetylhomoserine sulfhydrylase activity of the strain was negligible (Table 2). Because C. glutamicum HL938, a metB mutant strain, also showed methionine prototrophy (Fig. 6B), we constructed a metB metY double mutant strain by using the technique developed by Schäfer et al. (34) and tested for methionine requirement. Unlike the single mutant strains, as shown in Fig. 6B, the metB metY double mutant strain (HL939) was unable to grow on a minimal medium lacking supplemental methionine. Both the O-acetylhomoserine sulfhydrylase and the cystathionine γ-synthase activities of the double mutant strain were negligible (Table 2). Supplementation with homocysteine or methionine was enough to support the growth of the mutant strain (data not shown). Additionally, the growth characteristics of both metY and metB single mutants in MB and minimal medium were identical (data not shown). These data clearly show the functionality of the metY gene in C. glutamicum and also demonstrate, in addition to the transsulfuration mediated by metB gene, the presence of direct sulfhydrylation pathway, mediated by metY, for methionine biosynthesis in C. glutamicum (Fig. 7).
FIG. 7.
Model for the methionine biosynthetic pathway in C. glutamicum. Unlike other pathways, both the transsulfuration and direct sulfhydrylation pathways appear to be functional in C. glutamicum, as shown in the present study.
We also examined the presence of direct sulfhydrylation and transsulfuration pathways in bacteria related to C. glutamicum. Like C. glutamicum AS019E12, enzyme activities of O-acetylhomoserine sulfhydrylase (metY) and cystathionine γ-synthase (metB) were detected in several coryneform bacteria, such as C. glutamicum ATCC 13032, C. lactofermentum ATCC 13869 , and Brevibacterium flavum ATCC 14067, and the level of activity was comparable to that of C. glutamicum AS019E12 (data not shown).
Regulation of metY expression.
As the first step to investigate the role of direct sulfhydrylation pathway, we examined the effect of various amino acids on the expression of the metY gene. Among the aspartate family of amino acids tested, such as methionine, S-adenosylmethionine, threonine, lysine, isoleucine, and leucine, only methionine was effective to repress the expression of metY. The addition of 0.5 mM methionine to the MCGC minimal medium resulted in a 99% reduction in the O-acetylhomoserine sulfhydrylase activity (Table 3). The gene metB also appeared to be repressed by methionine, but the degree of repression was less than that by caused metY (Table 3). In agreement with the data, only the marginal activity of O-acetylhomoserine sulfhydrylase was also detected from the cells grown in a complex medium. The activity of cystathionine γ-synthase observed in the same cells was not changed significantly compared to that observed in the cells grown in a synthetic medium (Table 3). Assuming that sulfur is incorporated through the direct sulfhydrylation pathway, we tested the effect of sulfate concentration on the expression of metY. Although sulfate was added to the growth media up to 40 mM, no differences in growth pattern and O-acetylhomoserine sulfhydrylase activities were detected (data not shown). Similar results were obtained with cysteine.
TABLE 3.
Activities of O-acetylhomoserine sulfhydrylase and cystathionine γ-synthase in complex and minimal media containing methioninea
| Culture condition | Concn (mM) of methionine added to the medium | Sp act (nmol min−1 mg−1) of:
|
|
|---|---|---|---|
| OAHSH | CGS | ||
| MCGC minimal medium | None | 42 | 44 |
| 0.1 | 7.8 | 23 | |
| 0.5 | 0.4 | 14 | |
| 5 | 0.4 | 10 | |
| 10 | 0.3 | 9 | |
| MB complex medium | None | 1.1 | 19 |
| 5 | 0.2 | 14 | |
| 10 | 0.2 | 14 | |
| 20 | 0.1 | 10 | |
The enzymes were induced by growing C. glutamicum AS019E12 cells to the stationary phase on the described medium containing the appropriate amount of methionine. When MCGC minimal medium was used, glucose was added to a final concentration of 1%. Cells were harvested, disrupted, and assayed for activity as described in Materials and Methods. Enzyme reactions were carried out for 10 min at 30°C. The enzyme activities represent the mean of three independent experiments. OAHSH, O-acetylhomoserine sulfhydrolase; CGS, cystathione r-synthase.
DISCUSSION
In this report, we describe the presence of two parallel pathways, called transsulfuration and direct sulfhydrylation, for methionine biosynthesis in C. glutamicum (Fig. 7). The presence of the direct sulfhydrylation pathway in C. glutamicum was shown by (i) the complementation of the E. coli metB strain with metY-metA, (ii) the expression of O-acetylhomoserine sulfhydrylase activity from the metY gene, (iii) the requirement of chromosomal metY metB double disruption for methionine auxotrophy, and (iv) the amino acid sequence similarity of the metY gene product with other O-acetylhomoserine sulfhydrylases. In addition, previous reports on the metB and metC mutants of C. glutamicum which showed methionine prototrophy (14, 19) also support our findings. The presence of O-acetylhomoserine in the direct sulfhydrylation pathway was shown by the complementation of E. coli metA and metB mutants and by enzymatic assays. C. glutamicum shows uniqueness in methionine biosynthesis because it possesses functional transsulfuration and direct sulfhydrylation pathways and catalyzes each pathway with independent enzymes (Fig. 7). As evidenced in this work, direct sulfhydrylation and transsulfuration pathways also appear to be functional in related coryneform bacteria, such as C. lactofermentum and Brevibacterium flavum. M. tuberculosis, which is related to C. glutamicum, may also possess both pathways, since its metB mutant shows methionine prototrophy (39). Although S. cerevisiae, Neurospora spp., and green plants were reported to have enzyme activities of both pathways, unlike C. glutamicum, only one of the pathways might be physiologically meaningful and responsible for the methionine biosynthesis in the organism (8, 13, 18, 30, 42).
The methionine biosynthetic pathway of C. glutamicum also shows a clear distinction from that of E. coli, which is known to possess only the transsulfuration pathway (Fig. 7). In addition to the presence of the direct sulfhydrylation pathway in C. glutamicum, the enzymatic substrates for the metA and metB gene products of C. glutamicum are also different from those of E. coli (Fig. 7). Unlike E. coli, which favors succinylated substrates (8), C. glutamicum utilizes only acetylated substrates. Complementation of a metB mutation with a corynebacterial metY metA mutation, but very slow complementation with metY alone, shows that corynebacterial O-acetylhomoserine sulfhydrylase (metY) prefers O-acetylhomoserine to O-succinylhomoserine as the substrate. In addition, improved complementation of an E. coli metA mutation by a corynebacterial metY metA mutation shows that O-acetylhomoserine produced by corynebacterial metA can be more efficiently utilized by corynebacterial O-acetylhomoserine sulfhydrylase than E. coli cystathionine γ-synthase. Complementation of E. coli metA mutation by corynebacterial metA also confirms the previous findings that the E. coli metB gene product (cystathionine γ-synthase) also utilizes O-acetylhomoserine in place of its natural substrate, O-succinylhomoserine (8).
Interestingly, in many organisms, the metY and metA genes (or their homologs) appear to be located next to each other. This was true of L. meyeri (2, 4), M. leprae, Thermotoga maritima, M. tuberculosis, Deinococcus radiodurans, and Thermus thermophilus. Except for L. meyeri, the data were obtained by genome sequencing and therefore the physiological importance has not been addressed yet. Although the metY and metA genes of C. glutamicum were also located in close proximity to each other, they appeared to be expressed independently. This hypothesis is supported by the deletion analysis performed in this work and the presence of a putative transcriptional terminator in the downstream of metY. If cells decide to utilize only the transsulfuration pathway, it will be unnecessary for them to express the metY gene. Therefore, the simultaneous expression of metY and metA genes will be energetically inefficient, since metA should always be expressed when methionine is absent but the expression of metB or metY is optional thereafter. Thus, the independent expression of metY from metA is logical. This will be logical only if one of the metB or metY genes is expressed in a given physiological condition. In addition, as with the O-succinylhomoserine sulfhydrylase of P. aeruginosa (10), no methionine and cysteine residues were found in the coding region of metY. This suggests that amino acids or compounds containing sulfur may regulate metY expression. Although a putative leader sequence encoding a polypeptide composed of 60 amino acids was found in the upstream region of metY, the regulation mechanism of the gene expression by attenuation requires a further research.
Although we are uncertain about the importance of each pathway, our results suggest that the direct sulfhydrylation pathway is highly regulated. The evidence is for this is as follows: (i) the repressibility of metY gene by methionine, (ii) the unusually high level of metY expression in C. glutamicum, and (iii) the dramatic decrease of metY activity in complex media. These findings suggest that, compared to the transsulfuration pathway, the direct sulfhydrylation pathway is a low-affinity and energy-wasting route for methionine biosynthesis. Demonstration of our hypothesis will require further analysis with purified enzymes.
In conclusion, C. glutamicum possesses both transsulfuration and direct sulfhydrylation pathways for methionine biosynthesis (Fig. 7) and utilizes the pathways with almost equal efficiency, as evidenced by genetic and biochemical data. This situation is unique among prokaryotes and may provide evolutionary clues because methionine biosynthesis is known to show variations among organisms. Although we do not understand the role of each route, each pathway, analogous to the biosynthesis leading to lysine in C. glutamicum (35), which utilizes the succinylase or dehydrogenase pathway depending on the availability of ammonium ion, may play a critical role under a given physiological condition.
Acknowledgments
We thank Lothar Eggeling (Biotechnologie 1, Forschungszentrum Jülich, Jülich, Germany) for kindly providing us with pK19mobsacB.
This work was supported by grants from BASF Korea (to H.-S.L.) and Korea University (to H.-S.L.).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Belfaiza, J., A. Martel, D. Margarita, and I. Saint Girons. 1998. Direct sulfhydrylation for methionine biosynthesis in Leptospira meyeri. J. Bacteriol. 180:250-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Born, T. L., and J. S. Blanchard. 1999. Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded homoserine transsuccinylase. Biochemistry 38:14416-14423. [DOI] [PubMed] [Google Scholar]
- 4.Bourhy, P., A. Martel, D. Margarita, I. Saint Girons, and J. Belfaiza. 1997. Homoserine O-acetyltransferase, involved in the Leptospira meyeri methionine biosynthetic pathway, is not feedback inhibited. J. Bacteriol. 179:4396-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brzywczy, J., and A. Paszewski. 1993. Role of O-acetylhomoserine sulfhydrylase in sulfur amino acid synthesis in various yeasts. Yeast 9:1335-1342. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, S. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, S. Skelton, S. Squares, R. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Datko, A. H., J. Giovanelli, and S. H. Mudd. 1974. Homocysteine biosynthesis in green plants. O-Phosphorylhomoserine as the physiological substrate for cystathionine γ-synthase. J. Biol. Chem. 249:1139-1155. [PubMed] [Google Scholar]
- 9.Eggeling, L., S. Morbach, and H. Sahm. 1997. The fruits of molecular physiology: engineering the l-isoleucine biosynthesis pathway in Corynebacterium glutamicum. J. Biotechnol. 56:167-182. [Google Scholar]
- 10.Foglino, M., F. Borne, M. Bally, G. Ball, and J. C. Patte. 1995. A direct sulfhydrylation pathway is used for methionine biosynthesis in Pseudomonas aeruginosa. Microbiology 141:431-439. [DOI] [PubMed] [Google Scholar]
- 11.Follettie, M. T., and A. J. Sinskey. 1986. Recombinant DNA technology for Corynebacterium glutamicum. Food Technol. 40:88-94. [Google Scholar]
- 12.Follettie, M. T., O. P. Peoples, C. Agoropoulou, and A. J. Sinskey. 1993. Gene structure and expression of the Corynebacterium flavum N13 ask-asd operon. J. Bacteriol. 175:4096-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giovanelli, J., S. H. Mudd, and A. H. Datko. 1978. Homocysteine biosynthesis in green plants. Physiological importance of the transsulfuration pathway in Chlorella sorokiniana growing under steady-state conditions with limiting sulfate. J. Biol. Chem. 253:5665-5677. [PubMed] [Google Scholar]
- 14.Hwang, B.-J., Y. Kim, H.-B. Kim, H.-J. Hwang, J.-H. Kim, and H.-S. Lee. 1999. Analysis of Corynebacterium glutamicum methionine biosynthetic pathway: isolation and analysis of metB encoding cystathionine γ-synthase. Mol. Cell 9:300-308. [PubMed] [Google Scholar]
- 15.Jetten, M. S. M., and A. J. Sinskey. 1993. Characterization of phosphoenolpyruvate carboxykinase from Corynebacterium glutamicum. FEMS Microbiol. Lett. 111:183-188. [Google Scholar]
- 16.Jetten, M. S. M., and A. J. Sinskey. 1995. Recent advances in the physiology and genetics of amino acid-producing bacteria. Crit. Rev. Biotechnol. 15:73-103. [DOI] [PubMed] [Google Scholar]
- 17.Kase, H., and K. Nakayama. 1974. Production of O-acetyl-l-homoserine by methionine analog-resistant mutants and regulation of homoserine-O-transacetylase in Corynebacterium glutamicum. Agric. Biol. Chem. 38:2021-2030. [Google Scholar]
- 18.Kerr, D. S., and M. Flavin. 1970. The regulation of methionine synthesis and the nature of cystathionine γ-synthase in Neurospora. J. Biol. Chem. 245:1842-1855. [PubMed] [Google Scholar]
- 19.Kim, J.-W., H.-J. Kim, Y. Kim, M.-S. Lee, and H.-S. Lee. 2001. Properties of the Corynebacterium glutamicum metC gene encoding cystathionine β-lyase. Mol. Cell 11:220-225. [PubMed] [Google Scholar]
- 20.Kinoshita, S. 1985. Glutamic acid bacteria, p. 115-142. In A. L. Demain and N. A. Solomon (ed.), Biology of industrial microorganisms. The Benjamin/Cummings Publishing Company, London, England.
- 21.Laemmli, U. K. 1970. Cleavage of structure proteins during the assembly of head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malumbres, M., and J. F. Mart|$$|Aa|fin. 1996. Molecular control mechanisms of lysine and threonine biosynthesis in amino acid-producing corynebacteria: redirecting carbon flow. FEMS Microbiol. Lett. 143:103-114. [DOI] [PubMed] [Google Scholar]
- 24.Malumbres, M., J. A. Gil, and J. F. Mart|$$|Aa|fin. 1993. Codon preference in corynebacteria. Gene 134:15-24. [DOI] [PubMed] [Google Scholar]
- 25.Marzluf, G. A. 1997. Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu. Rev. Microbiol. 51:73-96. [DOI] [PubMed] [Google Scholar]
- 26.Miyajima, R., and I. Shiio. 1973. Regulation of aspartate family amino acid biosynthesis in Brevibacterium flavum. VII. Properties of homoserine O-transacetylase. J. Biochem. 73:1061-1068. [DOI] [PubMed] [Google Scholar]
- 27.Nagai, S., and M. Flavin. 1971. Synthesis of O-acetylhomoserine. Methods Enzymol. 17B:423-424.
- 28.Ozaki, H., and I. Shiio. 1982. Methionine biosynthesis in Brevibacterium flavum: properties and essential role of O-acetylhomoserine sulfhydrylase. J. Biochem. 91:1163-1171. [DOI] [PubMed] [Google Scholar]
- 29.Park, S.-D., J.-Y. Lee, Y. Kim, J.-H. Kim, and H.-S. Lee. 1998. Isolation and analysis of metA, a methionine biosynthetic gene encoding homoserine acetyltransferase in Corynebacterium glutamicum. Mol. Cell 8:286-294. [PubMed] [Google Scholar]
- 30.Ravanel, S., M. Droux, and R. Douce. 1995. Methionine biosynthesis in higher plants. I. Purification and characterization of cystathionine γ-synthase from spinach chloroplasts. Arch. Biochem. Biophys. 316:572-584. [DOI] [PubMed] [Google Scholar]
- 31.Ron, E. Z., and M. Shani. 1971. Growth rate of Escherichia coli at elevated temperatures: reversible inhibition of homoserine trans-succinylase. J. Bacteriol. 107:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahm, H., L. Eggeling, B. Eikmanns, and R. Krämer. 1995. Metabolic design in amino acid producing bacterium Corynebacterium glutamicum. FEMS Microbiol. Rev. 16:243-252. [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 35.Schrumpf, B., A. Schwarzer, J. Kalinowski, A. Pühler, L. Eggeling, and H. Sahm. 1991. A functionally split pathway for lysine synthesis in Corynebacterium glutamicum. J. Bacteriol. 173:4510-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shine, J., and L. Dalgarno. 1975. Determinant of cistron specificity in bacterial ribosomes. Naure 254:34-38. [DOI] [PubMed] [Google Scholar]
- 37.Simon, M., and J.-S. Hong. 1983. Direct homocysteine biosynthesis from O-succinylhomoserine in Escherichia coli: an alternate pathway that bypasses cystathionine. J. Bacteriol. 153:558-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, D. A. 1971. S-Amino acid metabolism and its regulation in Escherichia coli and Salmonella typhimurium. Adv. Genet. 16:141-165. [DOI] [PubMed] [Google Scholar]
- 39.Smith, D. A., T. Parish, N. G. Stoker, and G. J. Bancroft. 2001. Characterization of auxotrophic mutants of Mycobacterium tuberculosis and their potential as vaccine candidates. Infect. Immun. 69:1142-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taté, R., A. Riccio, E. Caputo, M. Iaccarino, and E. J. Patriarca. 1999. The Rhizobium etli metZ gene is essential for methionine biosynthesis and nodulation of Phaseolus vulgaris. Mol. Plant-Microbe Interact. 12:24-34. [DOI] [PubMed] [Google Scholar]
- 42.Thomas, D., and Y. Surdin-Kerjan. 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61:503-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 45.von der Osten, C. H., C. Gioannetti, and A. J. Sinskey. 1989. Design of a defined medium for growth of Corynebacterium glutamicum in which citrate facilitates iron uptake. Biotechnol. Lett. 11:11-16. [Google Scholar]
- 46.Wyman, A., E. Shelton, and H. Paulus. 1975. Regulation of homoserine transacetylase in whole cells of Bacillus polymyxa. J. Biol. Chem. 250:3904-3908. [PubMed] [Google Scholar]
- 47.Yamagata, S. 1989. Roles of O-acetyl-l-homoserine sulfhydrylases in micro-organisms. Biochimie 71:1125-1143. [DOI] [PubMed] [Google Scholar]
- 48.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 49.Yoshihama, M., K. Higashiro, E. A. Rao, M. Akedo, W. G. Shanabruch, M. T. Follettie, G. C. Walker, and A. J. Sinskey. 1985. Cloning vector system for Corynebacterium glutamicum. J. Bacteriol. 162:591-597. [DOI] [PMC free article] [PubMed] [Google Scholar]