Abstract
Cytochromes of the c type in the gram-positive bacterium Bacillus subtilis are all membrane anchored, with their heme domains exposed on the outer side of the cytoplasmic membrane. They are distinguished from other cytochromes by having heme covalently attached by two thioether bonds. The cysteinyls in the heme-binding site (CXXCH) in apocytochrome c must be reduced in order for the covalent attachment of the heme to occur. It has been proposed that CcdA, a membrane protein, transfers reducing equivalents from thioredoxin in the cytoplasm to proteins on the outer side of the cytoplasmic membrane. Strains deficient in the CcdA protein are defective in cytochrome c and spore synthesis. We have discovered that mutations in the bdbC and bdbD genes can suppress the defects caused by lack of CcdA. BdbC and BdbD are thiol-disulfide oxidoreductases. Our experimental findings indicate that these B. subtilis proteins functionally correspond to the well-characterized Escherichia coli DsbB and DsbA proteins, which catalyze the formation of disulfide bonds in proteins in the periplasmic space.
Gram-negative bacteria, such as Escherichia coli, have an inner and an outer membrane that confine the periplasmic space. Bacillus subtilis is a gram-positive bacterium with no outer membrane but a periplasm-like compartment confined by a thick cell wall and the cytoplasmic membrane (30, 33). Formation and disruption of disulfide bonds in proteins are important reactions in the periplasm of bacteria (12, 49). Disulfide bonds can form spontaneously at a slow rate. Formation of disulfide bonds in vivo must occur more efficiently than in vitro and is catalyzed by thiol-disulfide oxidoreductases. The available genome sequence data of bacteria belonging to, e.g., the genera Bacillus (26) and Mycobacterium (11) indicate that gram-positive bacteria contain several membrane-bound proteins involved in thiol redox chemistry in the cell envelope. Very little is known, however, about the physiological role and importance of these proteins.
Several proteins that function in formation and disruption of disulfide bonds in the periplasm of E. coli are known in considerable detail (for a review see reference 13). These proteins are located in the cytoplasmic membrane or the periplasmic space and contain thioredoxin sequence motifs (CXXC). DsbA and DsbB constitute an oxidative branch catalyzing formation of disulfide bonds (2, 3, 12). DsbA is a small, water-soluble periplasmic protein that directly oxidizes the substrate protein (23, 50). Reoxidation of DsbA is catalyzed by DsbB, which is an integral membrane protein (2, 20, 24). DsbD has a central function in a reductive branch. This protein transfers reducing equivalents from thioredoxin in the cytoplasm to various thiol-disulfide oxidoreductases in the periplasm (7, 14, 25, 31, 34, 42). The reducing equivalents are needed for isomerization of disulfide bonds (involves DsbC and DsbG) and for reduction of apocytochromes of the c type prior to ligation of heme (involves CcmG and CcmH). In cytochrome c the heme is covalently attached to the protein by two thioether bonds. Reduced cysteinyls at the heme-binding site (CXXCH) in the apocytochrome are necessary for the attachment of the heme cofactor (4).
Vegetative B. subtilis cells are known to contain three proteins with disulfide bonds. Two of them, ComGC and ComGG, are competence proteins. ComGC has one intramolecular disulfide bond, and ComGG has one intermolecular bond (9). The third protein is sublancin 168, which is a lantibiotic containing two intramolecular disulfide bonds and one proposed thioether lanthionine bond (32).
B. subtilis contains four c-type cytochromes, which are all membrane bound (6, 45, 46, 48). Proteins CcdA, ResB, and ResC are important for the synthesis of these cytochromes (28, 39). The exact functions of ResB and ResC have not been established. CcdA is an integral membrane protein functionally related to E. coli DsbD. CcdA is required for a late step in cytochrome c biogenesis, probably in keeping the two critical cysteinyls in apocytochrome reduced (38). Strains lacking CcdA are also defective in spore synthesis, and this is not a secondary effect due to the absence of cytochrome c (37).
Mutations that suppress the defects caused by CcdA deficiency in B. subtilis have been isolated but not identified (37). Strains containing such suppressor mutations sporulate like the wild type and emerge as microcolonies within lysed colonies of a strain with ccdA deleted on plates incubated for several days at room temperature. In this work we have isolated CcdA suppressor mutations using transposon mutagenesis. The suppressor mutations in both the new and previous isolates were identified. They are all positioned in the yvgUV locus. The yvgU gene has recently been designated bdbC (Bacillus disulfide bond) (8). The yvgV gene is named bdbD. The physiological role of the BdbC and BdbD proteins in B. subtilis has been analyzed. Functions of different B. subtilis putative thiol-disulfide oxidoreductases in the cytoplasmic membrane are proposed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The B. subtilis strains and the plasmids used in this work are listed in Table 1. B. subtilis LUL10 was obtained by transformation of strain 1A1 with LUL7 chromosomal DNA. E. coli strain JM109 (recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ[lac-proAB] F"[traD36 proAB+ lacIq lacZΔM15]) was used for propagation of plasmids.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| B. subtilis strains | ||
| 1A1 | trpC2 | BGSCb |
| LU60A1 | trpC2 ΔccdA::ble Pmr | 37 |
| LU62A1 | trpC2 Δ(ccdA-yneI-yneJ)::ble Pmr | 37 |
| LUL3 | trpC2 bdbC::pLLE21 Emr | This work |
| LUL4 | trpC2 ΔccdA::ble bdbC::pLLE21 Pmr Emr | This work |
| LUL7 | trpC2 ΔccdA::ble, bdbDΩTn10(1) Pmr Spr | This work (Fig. 1) |
| LUL8 | trpC2 ΔccdA::ble bdbDΩTn10(2) Pmr Spr | This work (Fig. 1) |
| LUL10 | trpC2 bdbDΩTn10(1) Spr | This work |
| LU62A1R#1 | trpC2 Δ(ccdA-yneI-yneJ)::ble bdbC1 Pmr | 37 |
| LU62A1R#3 | trpC2 Δ(ccdA-yneI-yneJ)::ble bdbD3 Pmr | 37 |
| LU62A1R#10 | trpC2 Δ(ccdA-yneI-yneJ)::ble bdbD10 Pmr | 37 |
| Plasmids | ||
| pHP13 | Shuttle vector; Emr Cmr | 17 |
| pHPSK | Shuttle vector; Emr Cmr | 21 |
| pHPKS | Shuttle vector; Emr Cmr | 21 |
| pIC333 | Vector for transposon mutagenesis; Emr Spr | 41 |
| pMutin2 | Integration vector for B. subtilis; Emr Amr | 44 |
| pLLE21 | 101-bp internal fragment from bdbC in pMutin2; EmrAmr | This work (Fig. 1) |
| pLLE26 | dbdD on 0.8-kb fragment in pHPSK; Emr Cmr | This work (Fig. 1) |
| pLLE27 | bdbDC operon on 1.5-kb fragment in pHPKS; Emr Cmr | This work (Fig. 1) |
| pCPC23 | cccA-phoA fusion on 3.9-kb fragment in pHP13; Emr Cmr | 38 |
Pmr, Emr, Amr, Spr, and CmR, resistance to phleomycin, erythromycin, ampicillin, spectinomycin, and chloramphenicol, respectively.
Bacillus Genetic Stock Center, University of Ohio, Columbus.
Media and growth conditions.
E. coli cells were grown at 37°C in Luria-Bertani (LB) medium or on LB plates (35). B. subtilis strains were cultivated at 37°C in LB or nutrient sporulation medium with phosphate (NSMP) (15) or on tryptose blood agar base (TBAB) plates (Difco). Antibiotics were used at various concentrations when appropriate; for B. subtilis, spectinomycin (150 mg/liter), erythromycin (1 mg/liter), and chloramphenicol (4 to 5 mg/liter) were used; for E. coli, ampicillin (50 mg/liter) and chloramphenicol (12.5 mg/liter) were used.
Transposon insertional mutagenesis.
Plasmid pIC333 was used for transposon mutagenesis (41). The plasmid contains an erythromycin resistance gene, a temperature-sensitive B. subtilis origin of replication, a transposase gene, a Tn 10 transposon unit with an internal spectinomycin resistance gene, and a pUC origin of replication. B. subtilis strain LU60A1 was transformed with the plasmid and grown on TBAB plates with erythromycin at 28°C. Colonies were selected and individually grown at 28°C in LB medium supplemented with spectinomycin. The temperature was then elevated to 37°C, and the cultures were incubated for 4 h. Cultures were then grown for sporulation. Finally the cultures were incubated at 80°C for 15 min, and samples were plated on NSMP plates with spectinomycin. The plates were incubated at 37°C overnight, and colonies were screened for TMPD (N,N,N",N"-tetramethyl-p-phenylenediamine) oxidation activity
DNA techniques.
Standard DNA techniques were used (35). Plasmid DNA was isolated by using a Quantum prep plasmid miniprep kit (Bio-Rad) or by CsCl density gradient centrifugation. Chromosomal DNA from B. subtilis was isolated as described by Marmur (29). E. coli was transformed by electroporation, and B. subtilis was grown to natural competence essentially as described by Hoch (19). B. subtilis strains defective in competence were transformed using protoplast transformation (19).
Fluorescent DNA sequencing was carried out using the BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems) and analyzed on a ABI prism 3100 DNA sequencer.
Construction of plasmids.
Plasmid pLLE21 was constructed by amplifying an internal fragment of bdbC using primers 5"-AAAAGCTTCTGTGCTGGTACCAGCG-3" and 5"-AAGGATCCCGAGCACGGCACGCC-3" (the underlined sequences indicate the restriction sites added via the primers). The PCR product was cut with HindIII and BamHI and cloned into pMutin2 that had been cut with the same restriction enzymes. Plasmid pLLE26 was constructed using primers 5"-CCATCGATGTTACTTCCCTTTCAGC-3" and 5"-CGGGATCCAACGCCATGCGCGTCATG-3". The amplified bdbD gene was cut with BamHI and ClaI and cloned into vector pHPSK. Plasmid pLLE27 was obtained using primers 5"-CGGGATCCAACGCCATGCGCGTCATG-3" and 5"-CGGGATCCTTCCTCTTCCATCGCAAC-3". The amplified bdbDC region was cut with BamHI and XbaI and cloned into pHPKS.
Spore assay.
Cultures were grown in 25 ml NSMP at 30°C in baffled Erlenmeyer flasks for 2 days. The sporulation efficiency of strains was analyzed by heating 5-ml samples at 80°C for 10 min. Serial dilutions of heat-treated and unheated samples were spread on TBAB plates. After incubation of the plates at 37°C overnight, colonies were counted.
Alkaline phosphatase activity and Western blot analysis.
The protoplast supernatant subfraction was isolated from B. subtilis strains as described previously for strains 3G18 and LU6018 by Schiött et al. (38). Alkaline phosphatase activity was measured using p-nitrophenyl phosphate as the substrate as described before (47). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis protein separation was carried out using the Schägger and von Jagow system (36). Proteins were transferred from the gel to a polyvinylidene fluoride blotting membrane using a semidry electroblotter (KemEn Tec Semidry Blotter II). Rabbit antiserum against E. coli alkaline phosphatase was used as the primary antibody. A peroxidase-labeled antirabbit antibody from the ECL Western blotting analysis system (Amersham Pharmacia Biotech) was used for visualization of the bound primary antibody.
Cytochrome c oxidase activity assay.
Isolated membranes from B. subtilis strains were added at a final protein concentration of 40 μg/ml to a 40 μM reduced solution of Saccharomyces cerevisiae cytochrome c in 20 mM MOPS (morpholinepropanesulfonic acid), pH 7.4. Oxidation of the c-type cytochrome was measured by a dual-wavelength spectrometer using the wavelength pair 540 and 550 nm (45). An extinction coefficient (ɛ550-540) of 19.5 mM−1 cm−1 was used to calculate activities. The measurements were carried out at 30°C using a 3-ml cuvette with a magnetic stirrer
Other methods.
Membranes were isolated from strains grown in NSMP at 37°C essentially as described previously (18). Protein concentrations were determined using the bicinchoninic acid protein assay (Pierce Chemical Co.) with bovine serum albumin as the standard. TMPD oxidation by colonies on NSMP plates was assayed as described by Le Brun et al. (28) except when cells were grown on NSMP plates containing dithiothreitol (DTT) or l-cystine. In these cases an Immobilon-NC Triton X-100-free MCE filter (pore size, 0.45 μm) disk (Millipore) was laid on the surface of the agar in the plate. The bacterial strains were streaked directly on the filter, and the plate was then incubated at 37°C overnight. Next day, prior to the TMPD oxidation activity staining, the filter was removed from the plate and washed by floating it on distilled water. The same procedure and the TMPD staining solution used for plates were used for the filters except that the agar solution was replaced by water.
RESULTS
Isolation of CcdA suppressor mutations using Tn 10.
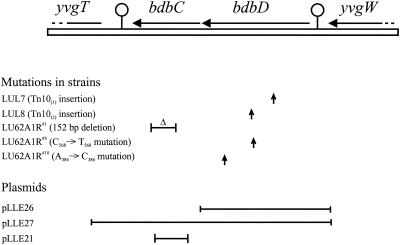
It was previously observed that strains containing CcdA suppressor mutations are proficient in sporulation (37). This property was exploited to isolate a new set of mutants. B. subtilis strain LU60A1, with the ccdA gene deleted, was subjected to random Tn10 insertion mutagenesis using pIC333. To select clones containing CcdA suppressor mutations, mutagenized cells were grown for sporulation. The cultures were heat treated, to kill vegetative cells, and samples were spread on NSMP plates. Colonies resulting after overnight growth at 37°C were screened in situ for TMPD oxidation activity. This activity is dependent on a functional cytochrome caa3 in the cytoplasmic membrane (45). CcdA-deficient strains lack cytochrome c and are therefore TMPD oxidation negative. Chromosomal DNA was isolated from independent TMPD oxidation-positive clones and used to transform LU60A1 to spectinomycin resistance to select for the transposon. TMPD oxidation-positive transformants LUL7 and LUL8 were analyzed. Both strains were found to have the bdbD gene disrupted by insertion of the transposon (Fig. 1), as determined by sequence analysis of the DNA flanking Tn 10.
FIG. 1.
Map of the bdbD-bdbC region in the B. subtilis chromosome. The positions of mutations identified in this work are indicated. Shown also are DNA fragments cloned in plasmids and used for complementation analysis (pLLE26 and pLLE27) or disruption of the bdbC gene (pLLE21).
Mutations in bdbC and bdbD can both suppress CcdA deficiency.
The bdbD gene is located in a dicistronic operon, bdbDC (Fig. 1), as deduced from the B. subtilis genome sequence (26). This gene organization and the fact that BdbD and BdbC are apparent orthologs of E. coli DsbA and DsbB, respectively, indicate that BdbC and BdbD function as a pair. The bdbC gene in strain LU60A1 was therefore inactivated by integration of the pMutin2 derivative pLLE21 (Fig. 1). Colonies of the resulting strain, LUL4, on NSMP plates oxidized TMPD. This showed that inactivation of bdbC also suppresses the cytochrome c synthesis defect of CcdA-deficient mutants.
To determine if previously isolated CcdA-suppressor strains are mutated in the bdbDC locus, LU62A1R#1 and LU62A1R#10 were transformed with plasmid pLLE27, carrying the B. subtilis bdbDC gene cluster, and pHP13, which is the corresponding vector. Colonies of the strains transformed with pHP13 were TMPD oxidation positive, as expected. However, colonies obtained after transformation with pLLE27 were TMPD oxidation negative. Thus, in the presence of the wild-type bdbDC locus on a plasmid the suppression of the CcdA deficiency in these strains was reversed. The suppressor phenotype of LU62A1R#10 was also reversed by pLLE26 carrying only bdbD. The results suggested that the previously isolated strains carry mutations in the bdbCD locus and that these mutant genes are recessive relative to the wild-type locus.
Amplification of the bdbDC region of strains LU62A1R#1, LU62A1R#3, and LU62A1R#10 by PCR and subsequent DNA sequence analysis demonstrated mutations in bdbD and bdbC (Fig. 1). Strain LU62A1R#1 was found to contain a 152-bp deletion in the bdbC gene (bases G138 to C290). LU62A1R#3 has a C568-to-T568 transition in bdbD, resulting in a nonsense mutation (Gln190→amber stop codon). LU62A1R#10 has an A386-to-C368 transversion, changing His129 in BdbD to a Pro residue.
Inactivation of bdbC and bdbD in a wild-type genetic background (strains LUL3 and LUL10) did not affect the TMPD oxidation activity of colonies on NSMP plates. Light-microscopic observations of LUL3 and LUL10 showed no apparent differences compared to the parental strain, 1A1, in terms of cell size, shape, or motility.
Cytochrome c oxidase activity of suppressor-containing strains.
The TMPD oxidation phenotype of strains with ccdA deleted and carrying bdbC or bdbD mutations was found to be dependent on the growth medium. Compared to colonies of the wild-type strain, colonies of suppressor-containing strains showed very low TMPD oxidation activity when grown on TBAB plates but high activity when grown on NSMP plates. This difference can be explained by variations in enzyme content and by the fact that TMPD oxidation activity is not apparent if the cytochrome caa3 content is below a certain threshold level. B. subtilis cells grown on NSMP contain about five times more cytochrome caa3 than cells grown on TBAB (J. Bengtsson and L. Hederstedt, unpublished data). To determine the extent of suppression of the CcdA deficiency, cytochrome c oxidase activity of isolated membranes from cells grown on NSMP was analyzed (Table 2). A strain with ccdA deleted completely lacked oxidase activity but showed 7 to 8% of wild-type activity when bdbC or bdbD was inactivated. This result showed that a lack of BdbC or BdbD only partially overcomes the defect in cytochrome c oxidase activity caused by CcdA deficiency.
TABLE 2.
TMPD oxidation phenotypes of colonies on NSMP plates and cytochrome c oxidase activities of isolated membranes
| Strain | Relevant genotype | TMPD phenotype | Cytochrome c oxidase activity (%)a |
|---|---|---|---|
| 1A1 | Wild type | Positive | 100 |
| LU62A1 | Δ(ccdA-yneI-yneJ)::ble | Negative | <0.1 |
| LU62A1R#1 | Δ(ccdA-yneI-yneJ)::ble bdbC1 | Positive | 8.3 |
| LU62A1R#10 | Δ(ccdA-yneI-yneJ)::ble bdbD10 | Positive | 6.9 |
Cytochrome c oxidase activities of isolated membranes are shown as percentages of activity compared to that for 1A1. The activity of 1A1 membranes was 0.21 μmol of cytochrome c oxidized per min per mg of protein.
Competence development and sporulation efficiency of BdbC- and BdbD-deficient strains.
It has been noted previously that strain LU62A1R#3 (37) and a B. subtilis BdbC-deficient strain (8) do not develop competence. We found that all our strains carrying mutated bdbD or bdbC (Table 1) were defective in competence irrespective of whether CcdA was present or absent in the strains.
The ability of strains to form heat-resistant spores during growth in NSMP for 2 days at 30°C was assessed (Table 3). The results showed that BdbC and BdbD are not important for sporulation. They also demonstrated that, in the absence of functional BdbC or BdbD, the CcdA protein is no longer required for efficient synthesis of spores.
TABLE 3.
Sporulation efficiencies of different B. subtilis strains
| Strain | Relevant genotype | Titer (CFU/ml)
|
Sporulation efficiencyb | |
|---|---|---|---|---|
| Total cell | Sporea | |||
| 1A1 | 4.8 × 108 | 4.4 × 108 | 92 | |
| LU60A1 | ΔccdA::ble | 1.4 × 108 | 4.2 × 106 | 3.0 |
| LU62A1 | Δ(ccdA-yneI-yneJ)::ble | 4.3 × 108 | 1.7 × 107 | 3.5 |
| LU62A1#1 | Δ(ccdA-yneI-yneJ)::ble bdbC1 | 3.7 × 108 | 3.2 × 108 | 87 |
| LU62A1#10 | Δ(ccdA-yneI-yneJ)::ble bdbD10 | 4.6 × 108 | 4.2 × 108 | 91 |
| LUL3 | bdbC::pLLE21 | 3.9 × 108 | 3.4 × 108 | 87 |
| LUL10 | bdbDΩTn 10 | 3.6 × 108 | 3.0 × 108 | 82 |
Titer after 10 min at 80°C.
Sporulation efficiency is calculated as 100 times the spore titer divided by the total cell titer.
BdbC- and BdbD-deficient strains are defective in the synthesis of a disulfide bond-containing protein.
To determine if bdbD and bdbC are involved in disulfide bond formation, we looked at the abilities of different strains to produce active E. coli alkaline phosphatase (PhoA). This enzyme requires formation of two intramolecular disulfide bonds to be active (40).
Alkaline phosphatase was expressed from pCPC23 as a CccA-PhoA fusion protein (38). The CccA part corresponds to the first 104 residues of the B. subtilis cytochrome c550. It replaces the native E. coli N-terminal signal sequence and directs the excretion of PhoA (47). With CccA-PhoA in B. subtilis, it has been demonstrated that the major portion of the alkaline phosphatase activity is found in the protoplast supernatant subfraction (38). It appears as if the PhoA part needs to be cleaved off from the CccA-PhoA polypeptide to become active. The activities of soluble alkaline phosphatase produced by different strains containing pCPC23 or plasmid vector pHP13 are shown in Table 4.
TABLE 4.
Alkaline phosphatase activities of the protoplast supernatant fractions of different B. subtilis strains containing pHP13 or pCPC23
| Strain | Alkaline phosphatase activity (U/ml of culture)a for plasmid:
|
|
|---|---|---|
| pHP13 | pCPC23 | |
| 1A1 | <0.01 | 4.4 |
| LU62A1 | <0.01 | 3.9 |
| LU62A1R#1 | <0.01 | 1.3 |
| LU62A1R#10 | <0.01 | 1.2 |
| LUL3 | <0.01 | 1.5 |
| LUL10 | <0.01 | 1.4 |
The values are the averages obtained from three independent experiments. The variation between experiments was less than 15%. One unit corresponds to 1 nmol of phosphoester bonds hydrolyzed per min per amount of cell fraction corresponding to 1 ml of culture.
Strains 1A1 (wild type) and LU62A1 (ΔccdA) containing pCPC23 showed similar alkaline phosphatase activities. BdbC- and BdbD-deficient strains containing pCPC23 produced less alkaline phosphatase activity; the difference in activity was 3- to 3.5-fold. The effects of BdbC and BdbD deficiency were about the same. Western blot analysis was consistent with these results, showing a reduced level of alkaline phosphatase protein in extracts from BdbC- and BdbD-defective strains (blot not shown). The results indicated that disulfide bond formation in proteins is impaired in the absence of BdbC or BdbD. We consider it unlikely that the observed decrease in alkaline phosphatase activity of BdbC- and BdbD-deficient strains occurs because these strains have defects in translation of PhoA or export of the polypeptide compared to the parental strain.
Effects of low-molecular-weight redox compounds.
CcdA is thought to transfer reducing equivalents from the cytoplasm to thioredoxin-like protein domains on the outer side of the cytoplasmic membrane (37). Consistent with such a function, we found that the defect in TMPD oxidation activity of colonies of CcdA-deficient strains can be suppressed if the reducing thiol reagent DTT is added to the growth medium (Table 5). Strains deficient in CcdA and BdbC or BdbD, in contrast, lost the ability to oxidize TMPD if the oxidizing thiol compound l-cystine was added to the growth medium. Full effect was obtained at 5 mM l-cystine in plates, but a 1 mM concentration of the reagent also had an effect. The presence of l-cystine (5 mM) or DTT (15 mM) did not affect the TMPD oxidation phenotypes of strains defective in only BdbC or BdbD. These results suggest that B. subtilis BdbC and BdbD, similar to the E. coli orthologs, DsbB and DsbA, catalyze formation of disulfide bonds in proteins on the outer side of the cytoplasmic membrane. Control experiments showed that DTT (15 mM) or l-cystine (5 mM) in the growth medium had no apparent effect on the growth of strains and did not complement the defect in TMPD oxidation activity of colonies of cytochrome caa3- (ΔctaCD), ResB-, or ResC-deficient strains.
TABLE 5.
TMPD oxidation activity of colonies on supplemented NSMP plates
| Strain | Activity with:
|
||
|---|---|---|---|
| No addition | 15 mM DTT added | 5 mM l-cystine added | |
| 1A1 | Positive | Positive | Positive |
| LU62A1 | Negative | Positive | Negative |
| LU62A1R#1 | Positive | Positive | Negative |
| LU62A1R#10 | Positive | Positive | Negative |
DISCUSSION
In this work we show that the defects in spore and cytochrome c synthesis caused by a lack of CcdA protein in B. subtilis can be suppressed by mutations in the bdbC or the bdbD gene. The two genes are organized in a dicistronic operon. Our results from the analysis of BdbC- and BdbD-deficient strains combined with results recently reported by Bolhuis et al. (8) and a comparison with the well-characterized E. coli DsbA and DsbB proteins indicate that BdbC and BdbD function as a pair in catalyzing disulfide bond formation in proteins on the outer side of the cytoplasmic membrane in B. subtilis.
It is suggested that CcdA in the B. subtilis cytoplasmic membrane transfers reducing equivalents from thioredoxin in the cytoplasm to an as yet unidentified thiol-disulfide protein(s) in the periplasm. One proposed role of this transmembrane electron transfer pathway involving CcdA is to keep the cysteine residues in the heme-binding site in apocytochrome c reduced so that heme can be covalently bound. During transport across the cytoplasmic membrane the cysteine residues in the apocytochrome are most likely in the reduced state. Our results suggest that BdbD and BdbC oxidize the two thiol groups in the heme-binding site of apocytochrome c to form a disulfide bond. CcdA, together with other proteins (as discussed below), seemingly counteracts the effect of BdbC and BdbD by breaking (reducing) the disulfide bond in apocytochrome c (Fig. 2). This proposed function of CcdA is consistent with our finding that addition of the reducing agent DTT to a ΔccdA strain restores the ability to synthesize cytochrome c (Table 5) and also increases the efficiency of sporulation (our unpublished data). Furthermore, the disulfide bond-containing compound l-cystine was found to compensate for the lack of BdbC and BdbD, i.e., in the presence of l-cystine, inactivation of bdbC and bdbD no longer suppressed the cytochrome c defect of a CcdA-deficient strain. l-Cystine and also oxidized glutathione can compensate for the effects seen with E. coli DsbA- and DsbB-deficient strains. A concentration of 0.6 mM l-cystine is required for the production of an oxidized λ102MalF-βgal fusion protein in a strain lacking DsbB, but 8.6 mM is required in a strain lacking DsbA (2). We found no clear difference in the levels of l-cystine required to reverse the TMPD oxidation-positive phenotype of different CcdA-deficient B. subtilis strains defective in BdbC and BdbD.
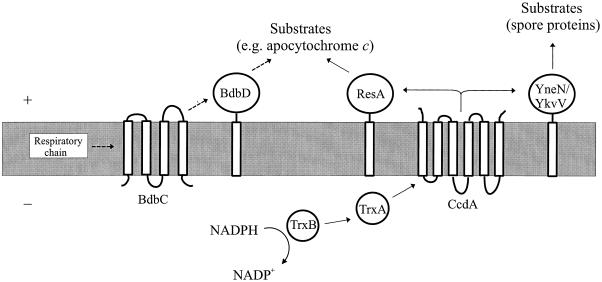
FIG. 2.
Suggested function of B. subtilis thiol-disulfide oxidoreductases in the cytoplasmic membrane. + and −, positive and negative sides of the membrane. Dashed and solid arrows indicate roles in oxidation and reduction, respectively. The indicated functions of TrxA, ResA, YneN, and YkvV are speculative.
The low sporulation efficiency of strains lacking CcdA was increased to the wild-type level by inactivation of bdbC or bdbD. Synthesis of the spore coat during sporulation involves proteins that are rich in cysteine residues, and many spore coat proteins are heavily cross-linked by disulfide bonds in the final spore (1). BdbD and BdbC do not seem important for spore synthesis, indicating that disulfide bonds may not be essential in spores or that in B. subtilis there exists thiol-disulfide oxidoreductases other than BdbD and BdbC. The reason why CcdA is required for efficient sporulation is not known, but it might be needed to break or reorganize disulfide bonds during spore maturation and germination.
Strains deficient in BdbC or BdbD but sufficient in CcdA are not able to develop competence. Two proteins in B. subtilis known to have disulfide bonds are the essential competence proteins, ComGC and ComGG. They are located on the cell surface and have a role in binding DNA to the cell surface (9, 10). ComGC is stabilized by BdbDC (D. Dubnau, personal communication). Thus is seems as if BdbC and BdbD catalyze disulfide bond formation in ComGC and ComGG.
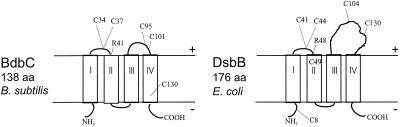
Searches for homologues of E. coli Dsb proteins in B. subtilis using the genome sequence have revealed several candidates. DsbA and B. subtilis BdbD show 41% sequence similarity and 21% identity. Each has a thioredoxin motif (CXXC), and the two overlap in the alignment. The crystal structures of both oxidized and reduced DsbA are known (16). DsbA contains two domains: domain A, which is very similar to that of E. coli thioredoxin, and domain B. Based on the structural data and sequence comparisons B. subtilis BdbD most likely also has two domains. E. coli DsbA is a soluble protein located in the periplasm and is synthesized with a N-terminal cleaved export signal sequence. BdbD has a putative signal peptide and a type I signal peptidase (SPase) cleavage site at residue 27 (43). However, since predictions for type I SPase cleavable signal peptides often turn out to be wrong, BdbD might well be membrane bound by an N-terminal single transmembrane segment (Fig. 2). E. coli DsbB and B. subtilis BdbC show 52% sequence similarity and 21% identity, and the thioredoxin motifs overlap. BdbC is a 138-amino-acid protein that has four predicted transmembrane regions (prediction of transmembrane regions was obtained using the TMHMM server [http://www.cbs.dtu.dk/services/TMHMM-1.0/]). BdbC contains five cysteine residues, and four of them, Cys34, Cys37, Cys95, and Cys101, correspond in position to the four essential cysteines (Cys41, Cys44, Cys104, and Cys130) in DsbB, which also has four transmembrane regions (Fig. 3) (2). An arginine residue in DsbB with a role in the interaction with quinone in the membrane is also conserved in B. subtilis BdbC (22).
FIG. 3.
Comparison of the B. subtilis BdbC and E. coli DsbB proteins in the cytoplasmic membrane. The predicted topology of DsbB is confirmed by experimental data (20). There is a notable difference in the size of the loop between transmembrane segments III and IV in the two proteins. DsbB has four essential cysteine residues (C41, C44, C104, and C130) facing the periplasmic space (20). The position of a conserved arginine residue important for reoxidation of E. coli DsbB (22) is indicated.
B. subtilis BdbA and BdbB, are also apparent homologues of E. coli DsbA and DsbB. The bdbA and bdbB genes are located in an operon, sunA-sunT-bdbA-yolJ-bdbB, which is part of the prophage SPβ genome (27). This prophage resides in the chromosomes of all the strains used in this work. The YolJ protein is of unknown function, SunA is a sublancin 168 lantibiotic antimicrobial peptide that is secreted into the medium, and SunT is a potential ABC-type transporter with a proteolytic domain and an ATP-binding domain. Sublancin 168 has two disulfide bonds and one thioether lanthionine bond (32). BdbA and BdbB possibly function as thiol-disulfide oxidoreductases specific for this lantibiotic. Deletion of the bdbA-yolJ-bdbB genes in B. subtilis strain LU60A1 did not suppress the TMPD oxidation-negative phenotype or affect the ability of the cell to develop natural competence (our unpublished data). It has been demonstrated that inactivation of bdbB in B. subtilis has only a small effect on the yield of active recombinant E. coli alkaline phosphatase compared to disruption of the bdbC gene (8). Thus, it seems as if BdbC and BdbD constitute a major enzyme system for disulfide bond formation in B. subtilis.
There are several proteins in B. subtilis that might accept reducing equivalents from CcdA and play a role in keeping the cysteine residues in apocytochrome reduced or function in sporulation. These proteins, ResA, YneN, and YkvV, all have high sequence similarity to CcsX of the gram-negative bacterium Bordetella pertussis. CcsX is known to be involved in cytochrome c synthesis (5). It has one transmembrane region in the N-terminal part and contains a thioredoxin motif. The resA gene in the B. subtilis genome sequence in the SubtiList database has an error affecting the reading frame (N. E. Le Brun, personal communication). Based on the corrected sequence, ResA has 179 amino acid residues. The N-terminal part of ResA probably functions both as a signal sequence for export of the rest of the protein and as a membrane anchor. YneN and YkvV each also have one transmembrane region and contain one thioredoxin motif. Deletion of the yneN gene in strain 1A1 does not affect the TMPD oxidation phenotype or sporulation efficiency, indicating that YneN does not play an important role in these processes (our unpublished data). ResA is encoded by the same operon as the ResB and ResC proteins, which are required for cytochrome c synthesis (28). We predict that ResA functions in cytochrome c biosynthesis. The paralogous YneN and YkvV might be involved in sporulation but have overlapping functions, as illustrated in Fig. 2.
Acknowledgments
This work was supported by a grant from NorFa to L.S.E. and by a grant from the Swedish Natural Science Research Council (grant B 650-1998 1074/2001) to L.H.
We thank Lars Rutberg and Nick E. Le Brun for comments on the manuscript and David Dubnau for communicating results prior to publication.
REFERENCES
- 1.Aronson, A. I., and P. Fitz-James. 1976. Structure and morphogenesis of the bacterial spore coat. Bacteriol. Rev. 40:360-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell, J. C., J. O. Lee, G. Jander, N. Martin, D. Belin, and J. Beckwith. 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. USA 90:1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 4.Barker, P. D., and S. J. Ferguson. 1999. Still a puzzle: why is haem covalently attached in c-type cytochromes? Structure Fold. Des. 7:R281-R290. [DOI] [PubMed]
- 5.Beckett, C. S., J. A. Loughman, K. A. Karberg, G. M. Donato, W. E. Goldman, and R. G. Kranz. 2000. Four genes are required for the system II cytochrome c biogenesis pathway in Bordetella pertussis, a unique bacterial model. Mol. Microbiol. 38:465-481. [DOI] [PubMed] [Google Scholar]
- 6.Bengtsson, J., C. Rivolta, L. Hederstedt, and D. Karamata. 1999. Bacillus subtilis contains two small c-type cytochromes with homologous heme domains but different types of membrane anchors. J. Biol. Chem. 274:26179-26184. [DOI] [PubMed] [Google Scholar]
- 7.Bessette, P. H., J. J. Cotto, H. F. Gilbert, and G. Georgiou. 1999. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J. Biol. Chem. 274:7784-7792. [DOI] [PubMed] [Google Scholar]
- 8.Bolhuis, A., G. Venema, W. J. Quax, S. Bron, and J. M. van Dijl. 1999. Functional analysis of paralogous thiol-disulfide oxidoreductases in Bacillus subtilis. J. Biol. Chem. 274:24531-24538. [DOI] [PubMed] [Google Scholar]
- 9.Chung, Y. S., F. Breidt, and D. Dubnau. 1998. Cell surface localization and processing of the ComG proteins, required for DNA binding during transformation of Bacillus subtilis. Mol. Microbiol. 29:905-913. [DOI] [PubMed] [Google Scholar]
- 10.Chung, Y. S., and D. Dubnau. 1998. All seven comG open reading frames are required for DNA binding during transformation of competent Bacillus subtilis. J. Bacteriol. 180:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 12.Dailey, F. E., and H. C. Berg. 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:1043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabianek, R. A., H. Hennecke, and L. Thöny-Meyer. 2000. Periplasmic protein thiol:disulfide oxidoreductases of Escherichia coli. FEMS Microbiol. Rev. 24:303-316. [DOI] [PubMed] [Google Scholar]
- 14.Fabianek, R. A., T. Hofer, and L. Thöny-Meyer. 1999. Characterization of the Escherichia coli CcmH protein reveals new insights into the redox pathway required for cytochrome c maturation. Arch. Microbiol. 171:92-100. [DOI] [PubMed] [Google Scholar]
- 15.Fortnagel, P., and E. Freese. 1968. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J. Bacteriol. 95:1431-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guddat, L. W., J. C. Bardwell, and J. L. Martin. 1998. Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization. Structure 6:757-767. [DOI] [PubMed] [Google Scholar]
- 17.Haima, P., S. Bron, and G. Venema. 1987. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol. Gen. Genet. 209:335-342. [DOI] [PubMed] [Google Scholar]
- 18.Hederstedt, L. 1986. Molecular properties, genetics, and biosynthesis of Bacillus subtilis succinate dehydrogenase complex. Methods Enzymol. 126:399-414. [DOI] [PubMed] [Google Scholar]
- 19.Hoch, J. A. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol. 204:305-320. [DOI] [PubMed] [Google Scholar]
- 20.Jander, G., N. L. Martin, and J. Beckwith. 1994. Two cysteines in each periplasmic domain of the membrane protein DsbB are required for its function in protein disulfide bond formation. EMBO J. 13:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson, P., and L. Hederstedt. 1999. Organization of genes for tetrapyrrole biosynthesis in gram-positive bacteria. Microbiology 145:529-538. [DOI] [PubMed] [Google Scholar]
- 22.Kadokura, H., M. Bader, H. Tian, J. C. Bardwell, and J. Beckwith. 2000. Roles of a conserved arginine residue of DsbB in linking protein disulfide-bond-formation pathway to the respiratory chain of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:10884-10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishigami, S., Y. Akiyama, and K. Ito. 1995. Redox states of DsbA in the periplasm of Escherichia coli. FEBS Lett. 364:55-58. [DOI] [PubMed] [Google Scholar]
- 24.Kishigami, S., E. Kanaya, M. Kikuchi, and K. Ito. 1995. DsbA-DsbB interaction through their active site cysteines. Evidence from an odd cysteine mutant of DsbA. J. Biol. Chem. 270:17072-17074. [DOI] [PubMed] [Google Scholar]
- 25.Krupp, R., C. Chan, and D. Missiakas. 2001. DsbD-catalyzed transport of electrons across the membrane of Escherichia coli. J. Biol. Chem. 276:3696-3701. [DOI] [PubMed] [Google Scholar]
- 26.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 27.Lazarevic, V., A. Dusterhoft, B. Soldo, H. Hilbert, C. Mauel, and D. Karamata. 1999. Nucleotide sequence of the Bacillus subtilis temperate bacteriophage SPβc2. Microbiology 145:1055-1067. [DOI] [PubMed] [Google Scholar]
- 28.Le Brun, N. E., J. Bengtsson, and L. Hederstedt. 2000. Genes required for cytochrome c synthesis in Bacillus subtilis. Mol. Microbiol. 36:638-650. [DOI] [PubMed] [Google Scholar]
- 29.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 30.Merchante, R., H. M. Pooley, and D. Karamata. 1995. A periplasm in Bacillus subtilis. J. Bacteriol. 177:6176-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Missiakas, D., F. Schwager, and S. Raina. 1995. Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. EMBO J. 14:3415-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paik, S. H., A. Chakicherla, and J. N. Hansen. 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 273:23134-23142. [DOI] [PubMed] [Google Scholar]
- 33.Pooley, H. M., R. Merchante, and D. Karamata. 1996. Overall protein content and induced enzyme components of the periplasm of Bacillus subtilis. Microb. Drug Resist. 2:9-15. [DOI] [PubMed] [Google Scholar]
- 34.Rietsch, A., P. Bessette, G. Georgiou, and J. Beckwith. 1997. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol. 179:6602-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 37.Schiött, T., and L. Hederstedt. 2000. Efficient spore synthesis in Bacillus subtilis depends on the CcdA protein. J. Bacteriol. 182:2845-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiött, T., M. Throne-Holst, and L. Hederstedt. 1997. Bacillus subtilis CcdA-defective mutants are blocked in a late step of cytochrome c biogenesis. J. Bacteriol. 179:4523-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiött, T., C. von Wachenfeldt, and L. Hederstedt. 1997. Identification and characterization of the ccdA gene, required for cytochrome c synthesis in Bacillus subtilis. J. Bacteriol. 179:1962-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sone, M., S. Kishigami, T. Yoshihisa, and K. Ito. 1997. Roles of disulfide bonds in bacterial alkaline phosphatase. J. Biol. Chem. 272:6174-6178. [DOI] [PubMed] [Google Scholar]
- 41.Steinmetz, M., and R. Richter. 1994. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart, E. J., F. Katzen, and J. Beckwith. 1999. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. EMBO J. 18:5963-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 45.van der Oost, J., C. von Wachenfeld, L. Hederstedt, and M. Saraste. 1991. Bacillus subtilis cytochrome oxidase mutants: biochemical analysis and genetic evidence for two aa3-type oxidases. Mol. Microbiol. 5:2063-2072. [DOI] [PubMed] [Google Scholar]
- 46.von Wachenfeldt, C., and L. Hederstedt. 1990. Bacillus subtilis 13-kilodalton cytochrome c-550 encoded by cccA consists of a membrane-anchor and a heme domain. J. Biol. Chem. 265:13939-13948. [PubMed] [Google Scholar]
- 47.von Wachenfeldt, C., and L. Hederstedt. 1990. Bacillus subtilis holo-cytochrome c-550 can be synthesised in aerobic Escherichia coli. FEBS Lett. 270:147-151. [DOI] [PubMed] [Google Scholar]
- 48.Yu, J., and N. E. Le Brun. 1998. Studies of the cytochrome subunits of menaquinone:cytochrome c reductase (bc complex) of Bacillus subtilis. Evidence for the covalent attachment of heme to the cytochrome b subunit. J. Biol. Chem. 273:8860-8866. [DOI] [PubMed] [Google Scholar]
- 49.Yu, J., H. Webb, and T. R. Hirst. 1992. A homologue of the Escherichia coli DsbA protein involved in disulphide bond formation is required for enterotoxin biogenesis in Vibrio cholerae. Mol. Microbiol. 6:1949-1958. [DOI] [PubMed] [Google Scholar]
- 50.Zapun, A., J. C. Bardwell, and T. E. Creighton. 1993. The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry 32:5083-5092. [DOI] [PubMed] [Google Scholar]