Abstract
The lantibiotic mersacidin inhibits peptidoglycan biosynthesis by binding to the peptidoglycan precursor lipid II. Mersacidin contains an unsaturated thioether bridge, which is proposed to be synthesized by posttranslational modifications of threonine residue +15 and the COOH-terminal cysteine residue of the mersacidin precursor peptide MrsA. We show that the flavoprotein MrsD catalyzes the oxidative decarboxylation of the COOH-terminal cysteine residue of MrsA to an aminoenethiol residue. MrsD belongs to the recently described family of homo-oligomeric flavin-containing Cys decarboxylases (i.e., the HFCD protein family). Members of this protein family include the bacterial Dfp proteins (which are involved in coenzyme A biosynthesis), eukaryotic salt tolerance proteins, and further oxidative decarboxylases such as EpiD. In contrast to EpiD and Dfp, MrsD is a FAD and not an FMN-dependent flavoprotein. HFCD enzymes are characterized by a conserved His residue which is part of the active site. Exchange of this His residue for Asn led to inactivation of MrsD. The lantibiotic-synthesizing enzymes EpiD and MrsD have different substrate specificities.
Lantibiotics (for “lanthionine-containing antibiotics”) are a group of ribosomally synthesized and posttranslationally modified antibiotic peptides containing the thioether amino acid lanthionine as a characteristic building block (33). In the last few years, several lantibiotics with unsaturated thioether bridges at the COOH terminus have been characterized. S-[(Z)-2-aminovinyl]-d-cysteine is present in epidermin (2), gallidermin (12), cypemycin (27), and some of the mutacins (28-30, 34), whereas mersacidin contains a COOH-terminal S-[(Z)-2-aminovinyl]-3-methyl-d-cysteine residue (15, 32). Among these lantibiotics, mersacidin, which is produced by a Bacillus strain (9), is of particular interest because it is active against methicillin-resistant Staphylococcus aureus strains (8). Brötz et al. (6, 7) were able to show that mersacidin inhibits the transglycosylation reaction in peptidoglycan biosynthesis by binding to the cell wall precursor lipid II [undecaprenyl-diphosphoryl-N-acetylmuramic acid-(pentapeptide)-N-acetylglucosamine]. The complex formed by mersacidin and lipid II differed from the well-known vancomycin-lipid II complex, making mersacidin to a new lead structure for the development of antimicrobial compounds.
The biosynthesis of the S-[(Z)-2-aminovinyl]-d-cysteine residue of epidermin depends on the flavoenzyme EpiD. EpiD catalyzes the oxidative decarboxylation of the COOH-terminal cysteine residue of the epidermin precursor peptide EpiA to a (Z)-enethiol structure (see Fig. 7) (13, 17, 20, 21). The unsaturated thioether bridge is formed by addition of the enethiol group to a didehydroalanine residue, produced by dehydration of Ser at position +19 of EpiA (residues within the COOH-terminal propeptides of lantibiotic precursor peptides are labeled with “+”). Recently, molecular characterization of EpiD helped to identify homologous enzymes catalyzing the decarboxylation of 4"-phosphopantothenoylcysteine (PPC) to 4"-phosphopantetheine, a key step in coenzyme A biosynthesis. It was shown that in bacteria the NH2-terminal domain of the Dfp flavoproteins is responsible for this reaction (see Fig. 7) (16, 24), whereas the COOH-terminal domain catalyzes the biosynthesis of PPC (37). The generic name of the PPC decarboxylases is CoaC (37), since decarboxylation of PPC is the third step in coenzyme A biosynthesis from pantothenate. The PPC decarboxylase activity could also be attributed to a eukaryotic protein, namely, the plant flavoprotein AtHAL3a (19). Interestingly, there is another example of a eukaryotic counterpart of a lantibiotic-synthesizing enzyme, although in this case the enzymatic function of the enzyme pair is not exactly known. The rat protein LANCL1, which is highly expressed in testis and brain, is homologous to the so-called LanC proteins (25). LanC proteins are supposed to catalyze the thioether formation by addition of a cysteine residue to a dehydrated amino acid residue (26).
FIG. 7.
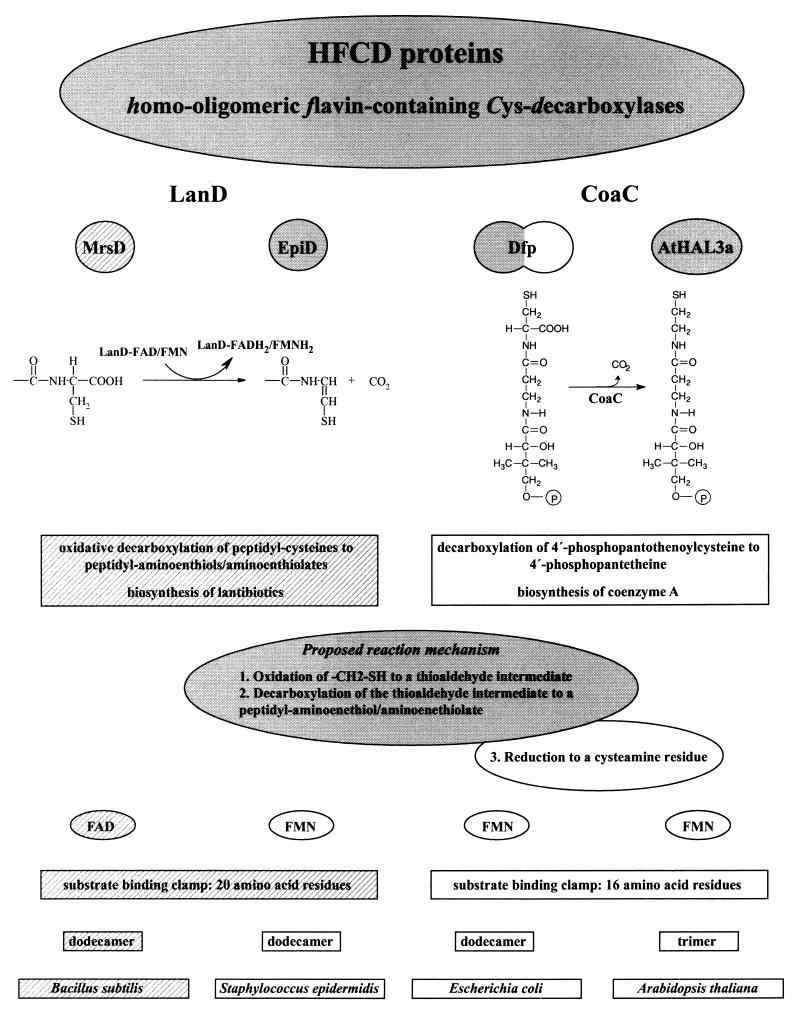
HFCD proteins with known enzymatic activity. The properties (from top to bottom: catalyzed reaction and proposed reaction mechanism, coenzyme requirement, structure of substrate binding clamp, oligomerization state, host organism) of the oxidative peptidyl-cysteine decarboxylases (LanD enzymes) MrsD (the present study), indicated by the hatched backgrounds, and EpiD (4, 20, 21, 23, 24) and the PPC decarboxylases (CoaC) Dfp (16, 24, 35, 37) and AtHAL3a (1, 11, 19) are compared in this figure. The proposed reaction mechanism for the LanD proteins is shaded in gray. The CoaC proteins reduce the oxidized intermediate to a cysteamine residue (reaction 3 of the proposed mechanism), so that in this case the overall reaction is a decarboxylation and not an oxidative decarboxylation. The length of the substrate binding clamp of MrsD is taken from the structure-based alignment of EpiD with MrsD (4). Dfp has PPC decarboxylase (NH2-terminal domain shaded in gray) and PPC synthetase activity (COOH-terminal domain).
EpiD, Dfp, and AtHAL3a belong to a new family of flavoproteins which are called HFCD proteins (for “homo-oligomeric flavin-containing Cys decarboxylases”) (4, 24). EpiD, AtHAL3a, and Dfp all bind the cofactor FMN (11, 23, 35), and molecular characterization of EpiD and crystal structure analysis of EpiD and AtHAL3a revealed a new flavin-binding motif (1, 4, 24). The crystal structure of the active-site mutant EpiD H67N with bound substrate peptide also revealed the active-site architecture of EpiD and gave first insights into the decarboxylation mechanism used by the HFCD proteins. Based on the geometry of the EpiD H67N-substrate complex it was suggested that Cα-Cβ dehydrogenation is not the initial reaction but rather oxidation of the thiol group by FMN. Thus, a thioaldehyde group-containing intermediate is formed, and its spontaneous decarboxylation (in analogy to the decarboxylation of β-keto acids) leads to the peptidyl-aminoenethiolate reaction product (4).
The enzymatic activity of EpiD was elucidated a couple of years ago, but until now no other lantibiotic-synthesizing enzyme activity has been demonstrated in vitro. Recently, the mersacidin gene cluster containing the mrsD gene has been cloned and sequenced (3). MrsD shows 25.9% sequence identity to EpiD and, despite this low sequence identity, it was assumed that MrsD is involved in formation of the COOH-terminal S-[(Z)-2-aminovinyl]-3-methyl-d-cysteine residue of mersacidin (3, 24). In the present study, we continued our work on the lantibiotic-synthesizing enzymes and HFCD proteins, and we characterized the MrsD protein as the second example of a so-called “LanD” enzyme (“Lan” is used for enzymes involved in lantibiotic biosynthesis). We proved that MrsD also is an oxidative peptidyl-cysteine decarboxylase, but it differs significantly from the prototype EpiD by its substrate specificity and coenzyme requirement. In total, the enzymatic function of four HFCD proteins (two oxidative peptidyl-cysteine decarboxylases and two PPC decarboxylases) is now known, giving insight into the relationship between sequence, structure and function of this new protein family.
MATERIALS AND METHODS
Growth of strains.
The E. coli strains used were grown to an A578 of 0.4 in 0.5 liter of B-broth (10 g of casein hydrolysate 140 [Gibco], 5 g of yeast extract [Difco], 5 g of NaCl, 1 g of glucose, and 1 g of K2HPO4/liter, pH 7.3) in 2-liter shaker flasks, induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and harvested 2 h after induction, if not otherwise stated. Escherichia coli TB1(pIH902) cells were grown in B-broth containing 100 μg of ampicillin/ml, and E. coli M15(pREP4, pQE) cells were grown in B-broth containing 100 μg of ampicillin and 25 μg of kanamycin/ml. The growth temperature was 37°C.
Amplification by PCR and DNA sequencing.
All PCR amplifications were performed with Vent DNA polymerase (New England Biolabs). For amplification of mrsD, the plasmid pKA3.1 was used as a template (3). For amplification of mrsA, plasmid pMER2 containing mrsA and mrsR1 (3) was used. The entire sequences of the mrsD and mrsA coding regions of the various constructed plasmids were verified. Used oligonucleotides were purchased from MWG Biotech.
Site-directed mutagenesis of mrsD.
mrsD mutants were constructed by sequential PCR steps (10) with appropriate mutagenesis primers. The 5"- and 3"-terminal primers used for amplification and subsequent cloning into pQE-12 are mentioned below.
Cloning of malE-mrsD and malE-mrsA fusions.
In order to purify MrsD and MrsA by affinity chromatography, the genes were expressed as malE fusions. malE encodes the maltose-binding protein (MBP) of E. coli, which can be purified in a single step by amylose chromatography. Construction of the malE-mrsD and malE-mrsA fusions with the malE expression vector pIH902 (New England Biolabs) was performed as described previously (23). mrsD and mrsA genes were amplified by PCR in such a way that the amplified DNA fragments started exactly with the ATG start codon and then were inserted into the StuI site of pIH902. To amplify mrsD, we used the forward primer 5"-ATGAGTATTTCAATATTAAAAGATAAAAAG-3" and the reverse primer 5"-CATTTATGTTAGTGAGGGGTGTTTTGTTC-3". To amplify the mrsA gene, the forward primer 5"-ATGAGTCAAGAAGCTATCATTCGT-3" and the reverse primer 5"-ATGCTGGATCCTTAACAAATACATTCAGAAGTTAGAG-3" were used (the introduced BamHI site was not used for cloning). The pIH902 derived plasmids were transformed into the expression strain E. coli TB1 (New England Biolabs) by electroporation.
Cloning of pQE12 mrsD and pQE8 mrsA.
To express mrsD and mutant mrsD, the corresponding genes were cloned into the single EcoRI and BglII sites of the pQE12 expression vector (Qiagen) in such a way that no fusion with the His tag codons of pQE12 occurred. The forward primer 5"-CCACAAAAAGAATTCTTGTGTAGTATGATTG-3" and the reverse primer 5"-GACTTTTGAGAGATCTCATTTATGTTAGTG-3" were used for PCR amplification of mrsD. For cloning of mrsA into the BamHI site of the pQE8 expression vector (Qiagen), the mrsA gene was amplified by PCR with the forward oligonucleotide 5"-TTGATTATGAGTGGATCCAATACTATACTTAATAAG-3" and the reverse oligonucleotide 5"-CCTTCTGAAGGGATCCAGCCTATAGAGCTCAAATTAACA-3" as primers. The introduced restriction sites BamHI and SstI (the latter of which was not used for cloning in this experiment) are underlined. The expression plasmid pQE8 mrsA encodes the N-terminal His tag fusion peptide of the MrsA precursor peptide MRGSHHHHHHGSNTILNKGVNT-MrsA. The pQE derived plasmids were transformed into the expression strain E. coli M15(pREP4) (Qiagen) by electroporation.
Purification of MBP-MrsD and MBP-EpiD fusion proteins.
For purification of the MBP-LanD fusion proteins, 500 ml of IPTG-induced E. coli TB1(pIH902 mrsD) cells (this study) and E. coli TB1(pIH902 epiD) cells (23) were harvested, the pellet was resuspended in 10 ml of 20 mM Tris-HCl (pH 8.0) buffer, and the cells were disrupted by sonication for 2 min. Then, 20 ml of column buffer (20 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 0.25% Tween 20, 10 mM 2-mercaptoethanol) was added to the crude cell extract. A cleared lysate obtained by two centrifugation steps (each 20 min at 30,000 × g at 4°C) was then applied to a column containing 2 ml of amylose resin (New England Biolabs) equilibrated with column buffer. The resin was washed with 6 volumes of column buffer and then with 6 volumes of 20 mM Tris-HCl (pH 8.0)-500 mM NaCl buffer. Amylose-bound proteins were eluted with 10 mM maltose in 20 mM Tris-HCl (pH 8.0)-500 mM NaCl buffer. Residual malE expression was largely reduced by the addition of glucose to the medium. Cleavage of the purified MBP-MrsD fusion protein was carried out in 20 mM Tris-HCl-200 mM NaCl buffer containing 1 U of factor Xa/100 μl (Amersham Pharmacia Biotech). The mix was incubated for several hours at room temperature, and cleavage was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Flavin identification.
The coenzyme could be directly removed from purified MBP-MrsD by heat denaturation (4 min, 100°C). The denatured fusion protein was removed by a centrifugation step. The coenzyme-containing supernatant was analyzed by thin-layer chromatography on cellulose aluminium sheets (Merck) by using the solvent n-butyl alcohol-acetic acid-water (4:3:3) (14). FMN (Boehringer), FAD, and riboflavin (Sigma) were used as reference substances.
Purification of MrsD and mutant MrsD proteins.
E. coli M15(pREP4, pQE-12 mrsD) cells induced with 500 ml of IPTG (0.2 mM) were harvested and disrupted by sonication for 2 min in 10 ml of column buffer (20 mM Tris-HCl [pH 8.0]). Next, 3.5 ml of the cleared lysate obtained by two centrifugation steps (each 20 min at 30,000 × g at 4°C) were diluted with 1.5 ml of column buffer and loaded on a 1-ml HiTrap Q column equilibrated with column buffer. The column was then washed with 5 ml of column buffer. MrsD and mutant MrsD proteins were eluted with column buffer containing 0.1 M NaCl, and the yellow peak fractions (ca. 400 μl) were collected. For gel filtration, a 25-μl aliquot of the HiTrap Q eluate was subjected to a Superdex 200 PC 3.2/30 column equilibrated in running buffer (20 mM Tris-HCl [pH 8.0], 200 mM NaCl) at a flow rate of 40 μl/min. Eluted proteins were collected in 80-μl fractions starting with an elution volume of 0.7 ml. The MrsD-containing fraction 7 (elution volume, 1.18 to 1.26 ml) was used for activity assays. The HiTrap Q column, the Superdex 200 PC 3.2/30 column, and the standard proteins used for calibration were obtained from Amersham Pharmacia Biotech. Protein concentrations were measured by the Bradford assay with bovine serum albumin as a standard (5).
Purification of EpiD.
EpiD was purified from IPTG-induced E. coli M15(pREP4, pQE-12 epiD) cells (24) by a method similar to a published purification procedure (23, 24) by two anionic-exchange chromatographic steps (HiLoadQ Sepharose and MonoQ HR10/10 chromatography; columns were from Amersham Pharmacia Biotech) and additionally by gel filtration (Superdex 200 PC 3.2/30 column).
Purification of MrsA.
We harvested 4 liters of IPTG-induced E. coli TB1 (pIH902 mrsA) cells grown in rich medium (containing 2 g of glucose and tryptone per liter instead of casein hydrolysate) at an A600 of 1.5 and froze them in 50 ml of 20 mM Tris-HCl-1 mM EDTA (pH 8.0). After disruption by ultrasonic treatment on ice, cell debris was removed by centrifugation and the supernatant was concentrated to ca. 65 ml by ultrafiltration on an YM10 membrane (Millipore Corporation), diluted to 175 ml with 20 mM Tris-HCl-1 mM EDTA (pH 7.6; buffer A), and again reduced to ca. 60 ml. Since the MBP-MrsA fusion protein did not bind well to the amylose affinity resin, the sample was divided into two aliquots, cleared by centrifugation and filtration (25 mm Arodisc syringe filters, 0.2-μm pore size; Pall Corporation), diluted 1:5 with buffer A, and loaded onto a 5-ml HiTrap Q Sepharose HP anion-exchange column (Amersham Pharmacia Biotech) equilibrated in buffer A. After the column was washed with 16 bed volumes of buffer A, the fusion protein was eluted in 3-ml fractions with a gradient of 37.5 ml of buffer A and 37.5 ml of buffer A containing 0.5 M NaCl. The MBP-MrsA-containing fractions were adjusted to 1 mM CaCl2 and incubated with 3.3 μg of factor Xa (New England Biolabs)/ml for 8 h at room temperature. Afterward, 50 μl of 5 M NaCl/ml and 3 ml of 20 mM Tris-1 mM EDTA-200 mM NaCl (buffer B) were added to each fraction. Free MBP was then removed by affinity chromatography on amylose resin equilibrated in buffer B. The runthrough of the amylose column contained the MrsA peptide, and samples of 1.8 ml were mixed with 0.2 ml of 1% trifluoroacetic acid (TFA) in water, centrifuged and loaded onto a POROS R2 perfusion high-pressure liquid chromatography column (Applied Biosystems). The peptide was eluted with a gradient of acetonitrile-0.1% TFA (B) in water-0.1% TFA (A) (0 min, 0% B; 5 min, 20% B; 19 min, 41% B; 20 min, 100% B) and detected at 220 nm. The MrsA peptide which eluted at 35% B was lyophilized, preincubated for 10 min at room temperature in 20 mM Tris-HCl (pH 8.0)-5 mM dithiothreitol (DTT) and then used for the activity assays.
Purification of His-MrsA.
A total of 500 ml of IPTG (0.2 mM)-induced E. coli M15(pREP4, pQE8 mrsA) cells were harvested and disrupted by sonication for 2 min in 10 ml of 20 mM Tris-HCl (pH 8.0). The cleared lysate obtained by two centrifugation steps (each for 30 min at 30,000 × g at 4°C) was diluted with 5 ml of column buffer (50 mM Tris-HCl, pH 8.0; 300 mM NaCl; 25 mM imidazole) and applied to an equilibrated Ni-nitrilotriacetic acid (NTA) column containing 2 ml of Ni-NTA agarose (Qiagen). The column was then washed with 10 ml of column buffer. His-MrsA was eluted with column buffer containing 300 mM imidazole. The eluate was then successively separated by reversed-phase chromatography (RPC) with a μRPC C2/C18 SC 2.1/10 column on a SMART system (Amersham Pharmacia Biotech) to purify His-MrsA from proteolytic degradation products. Peptides were eluted with a linear gradient of 0 to 50% acetonitrile-0.1% TFA in 5.8 ml with a flow rate of 200 μl/min and detected at 214 nm. His-MrsA eluted at about 38.5% acetonitrile.
Purification of EpiA R30Q.
EpiA R30Q was obtained by factor Xa cleavage of purified fusion protein MBP-EpiA R30Q and then was purified by anion-exchange chromatography and RPC as described previously (22). The mutation R30Q prevents internal factor Xa cleavage of EpiA but not oxidative decarboxylation of the COOH-terminal cysteine residue of EpiA by EpiD (20).
SDS-PAGE and immunoblotting.
Proteins were separated by Tricine-SDS-10% PAGE (31) under reducing conditions. After SDS-PAGE, proteins were electrophoretically transferred to a polyvinylidene difluoride membrane (38). Proteins were stained with Coomassie brilliant blue R250 (Sigma); prestained molecular weight markers were obtained from Life Technologies. MrsD and MrsD H75N were detected by using polyclonal anti-MrsD antiserum developed in rabbits, and immunoreactive proteins were visualized by enhanced chemiluminescence as described previously (23).
Oxidative decarboxylation assay.
The oxidative decarboxylase activity of MrsD was analyzed with the three different peptide substrates MrsA, His-MrsA, and EpiA R30Q. A total of 20 μl of the MrsD-containing gel filtration eluate (ca. 8 μg) was incubated with ca. 15 to 20 μg of MrsA and His-MrsA, respectively, for 15 min at 37°C in a total volume of 600 μl of 20 mM Tris-HCl (pH 8.0) buffer containing 5 mM DTT. The mutant protein MrsD H75N, which was supposed to be inactive, was used at an approximately double concentration. The reaction mixtures were kept at −80°C, and then 500-μl aliquots were successively separated by RPC with a μRPC C2/C18 SC 2.1/10 column on a SMART system. Compounds were eluted with a linear gradient of 0 to 50% acetonitrile-0.1% TFA in 5.8 ml with a flow rate of 200 μl/min. The absorbance was measured simultaneously at 214, 260, and 280 nm to allow identification of the reaction products (17, 20, 21).
High-resolution MS.
Electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry (ESI-FTICR-MS) measurements were carried out with a passively shielded 4.7-T APEXII-ESI/MALDI-FTICR mass spectrometer (Bruker Daltonik). On a Silicon Graphics O2 workstation, the MS software XMASS v.5.0.10 (Bruker Daltonik) was used for mass calculation, data acquisition, and processing. Dried peptide samples were dissolved in 50 μl of acetonitrile-water (50:50 [vol/vol]), and then 1 μl of acetic acid was added to provide protons for positive mode detection. Samples containing the MrsD reaction products separated by RPC as described above were stored at −80°C and then analyzed by direct infusion with a syringe pump. In general, 524,288 data points were acquired, and a mass range of 200 to 3,500 Da was covered. A maximum of 1,000 scans were averaged to increase the signal-to-noise ratio. ESI was performed in the positive mode with a grounded capillary sprayer needle mounted 60° off axis (Analytica of Branford). No nebulizer gas was necessary to support the spray process. For the external four-point-calibration, a known compound mixture was used.
RESULTS AND DISCUSSION
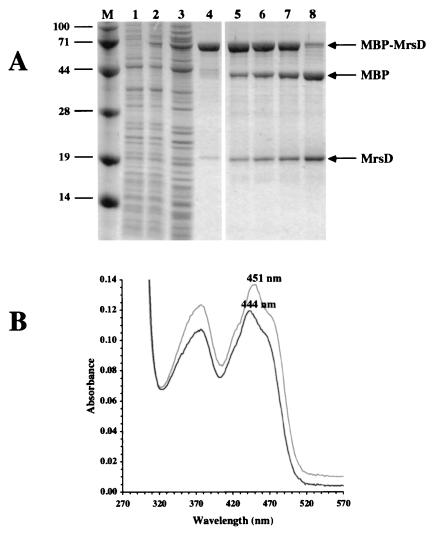
Purification and characterization of MBP-MrsD. mrsD encodes a 194-amino-acid protein that is similar to the FMN-dependent flavoproteins EpiD from Staphylococcus epidermidis, to Dfp from bacteria, and to AtHAL3a from Arabidopsis thaliana (24). EpiD catalyzes the oxidative decarboxylation of peptidyl-cysteines (20, 21), whereas AtHAL3a and Dfp decarboxylate PPC to 4"-phosphopantetheine (16, 19, 24) (see Fig. 7). To obtain more information on its biochemical properties, we first expressed mrsD in E. coli by using the MBP fusion system. The MBP-MrsD fusion protein was purified to homogeneity in one step by amylose affinity chromatography (Fig. 1A). The MBP-MrsD fusion protein was identified as a flavoprotein by its absorption spectrum with maxima at 278, 377, and 444 nm (Fig. 1B). In comparison to MBP-EpiD, the third maximum is shifted significantly to lower wavelengths (from 451 to 444 nm). The coenzyme could be removed by heat denaturation of the fusion protein and was identified by thin-layer chromatography as FAD. In contrast to MBP-EpiD (23), MBP-MrsD could be cleaved by factor Xa (Fig. 1A). Interestingly, the affinity chromatography-purified MBP-MrsD fraction already contained a protein with a size similar to that of MrsD (Fig. 1A, lane 4). NH2-terminal sequencing showed that this protein is MrsD lacking the NH2-terminal methionine residue (data not shown). This indicates that not only malE-mrsD but also mrsD is expressed from the pIH902 mrsD plasmid, that the NH2-terminal Met residue is cleaved off by methionine aminopeptidase activity in E. coli, and that, finally, MrsD forms oligomers with MBP-MrsD enabling copurification by affinity chromatography.
FIG. 1.
Purification and factor Xa cleavage of MBP-MrsD fusion protein. (A) Purification and factor Xa cleavage of MBP-MrsD was followed by SDS-PAGE. Lanes: M, molecular mass marker (sizes are in kilodaltons in the left margin); 1 and 2, whole-cell lysate of E. coli TB1(pIH902 mrsD) before and after induction with IPTG; 3, cleared cell lysate; 4, MBP-MrsD purified by amylose affinity chromatography; 5 to 8, cleavage of MBP-MrsD with factor Xa after 1, 2, 3, and 17.5 h of incubation at room temperature, respectively. (B) Absorption spectra of purified MBP-MrsD (black line) and purified MBP-EpiD (gray line).
Purification and characterization of MrsD and MrsD H75N.
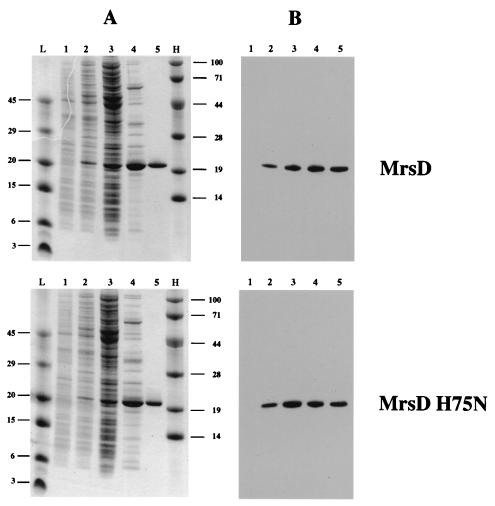
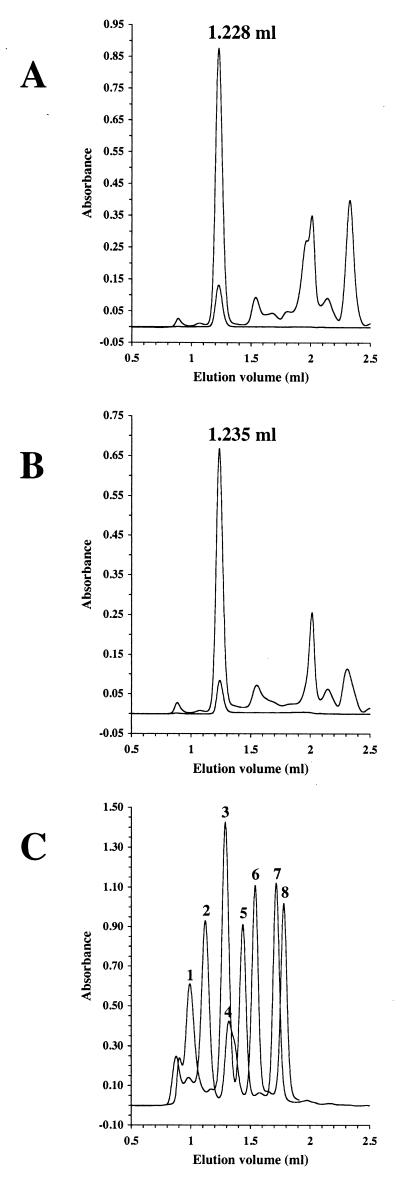
For further biochemical studies, mrsD was expressed without any affinity tag. Previous work on EpiD, Dfp, and AtHAL3a had shown that these HFCD proteins all have an essential histidine residue within the active site (19, 24). Therefore, mrsD H75N was expressed, purified, and characterized. His75 of MrsD aligns with His67 of EpiD, His90 of AtHAL3a and His75 of Dfp. MrsD and MrsD H75N were purified from induced E. coli M15(pREP4, pQE12 mrsD/mrsD H75N) cells by using anion-exchange chromatography and gel filtration (Fig. 2 and 3). MrsD eluted at an apparent molecular mass of ca. 237 kDa from the gel filtration column. The calculated molecular mass of MrsD Ser2-Thr194-FAD is 22.3 kDa. Although the observed molecular mass deviates by 30 kDa from the theoretical value for a homododecamer, the similarity of MrsD with EpiD indicates that MrsD also builds up homododecamers with trimers disposed at the vertices of a tetrahedron (4). MrsD H75N has the same oligomeric state in solution as MrsD and still binds the coenzyme, indicating that the overall three-dimensional structure of MrsD is not significantly altered by the exchange of His75.
FIG. 2.
Purification of MrsD and MrsD H75N. Purification of MrsD (upper part of the figure) and MrsD H75N (lower part of the figure) was followed by SDS-PAGE (A) and immunoblotting (B). Lanes: L and H, low- and high-molecular-mass markers (sizes are in kilodaltons in the left and right margins); 1 and 2, whole-cell lysate of E. coli M15(pREP4)(pQE12 mrsD or pQE12 mrsD H75N) before and after induction with IPTG; 3, cleared cell lysate; 4, HiTrap Q-enriched proteins; 5, MrsD or MrsD H75N purified by gel filtration by using a Superdex 200 PC 3.2/30 column.
FIG. 3.
Gel filtration of MrsD and MrsD H75N. HiTrap Q-enriched MrsD (A) and MrsD H75N (B) proteins were separated by gel filtration, and the elution was followed by measuring the absorbance at 280 nm (upper line), 378 nm (not shown), and 450 nm (lower line). The proteins eluted at 1.228 and 1.235 ml, respectively, corresponding to apparent molecular masses of ca. 236.9 and 228.8 kDa. The elution volume was not changed by preincubation of MrsD with 10 mM DTT for 15 min at room temperature. (C) Calibration of Superdex 200 PC3.2/30 column. To correlate the elution volume with molecular mass information, the elution volumes of standard proteins (thyroglobulin [peak 1, 669 kDa, 0.992 ml], ferritin [peak 2, 440 kDa, 1.121 ml], catalase [peak 3, 232 kDa, 1.287 ml], aldolase [peak 4, 158 kDa, 1.320 ml], albumin [peak 5, 67 kDa, 1.436 ml], ovalbumin [peak 6, 43 kDa, 1.539 ml], chymotrypsinogen A [peak 7, 25 kDa, 1.714 ml], and RNase [peak 8, 13.7 kDa, 1.779 ml]) were determined. The void volume of the column was determined to be 0.933 ml by using dextran blue 2000.
Purification and characterization of the substrate peptides MrsA and His-MrsA.
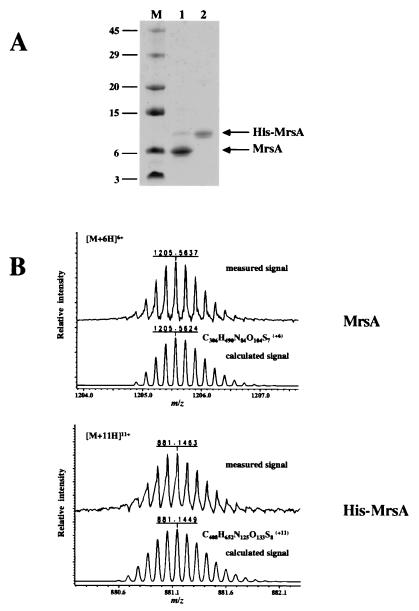
One of the problems in characterizing lantibiotic-synthesizing enzymes is the purification or synthesis of the substrate peptides. We have chosen two different approaches to obtain the MrsA precursor peptide. On the one hand, mrsA was expressed as malE-mrsA fusion, and the 68-amino-acid MrsA precursor peptide could be purified by factor Xa cleavage of MBP-MrsA, as has been described recently for EpiA (22) (Fig. 4). On the other hand, mrsA was expressed as His tag fusion peptide (see, for example, reference 18) and the 90-amino-acid His-MrsA was purified by Ni-NTA chromatography and RPC (Fig. 4). The identity of both peptides was verified by ESI-FTICR-MS analysis (Fig. 4B). Both peptides were used to determine the activity of MrsD. His-MrsA bound to Ni-NTA resin may also be used to analyze the interaction with the MrsM enzyme, which is proposed to catalyze the thioether formation from hydroxyamino acid residues and cysteine residues (3).
FIG. 4.
Purification of MrsA and His-MrsA. The RPC-purified substrate peptides MrsA and His-MrsA were analyzed by SDS-PAGE (A) and ESI-FTICR-MS (B). Lanes (panel A): M, molecular weight standard; 1, purified MrsA; 2, purified His-MrsA. It was also proven that His-MrsA reacted with monoclonal anti-RGS-His4 antibody (Qiagen) (data not shown). In panel B the experimentally determined isotope pattern was compared with the calculated one. Only the signals for the 6-fold (MrsA) and the 11-fold (His-MrsA) charged ions, which had the highest intensity, are shown here (the relative errors were 1.1 ppm for MrsA and 1.6 ppm for His-MrsA, respectively).
MrsD but not MrsD H75N catalyzes the oxidative decarboxylation of mersacidin precursor peptides.
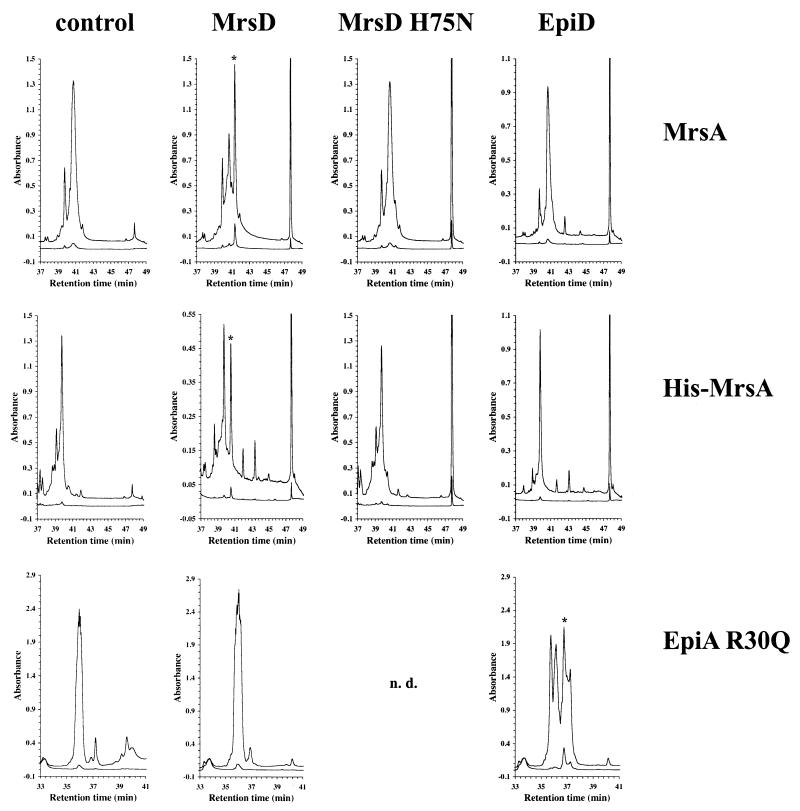
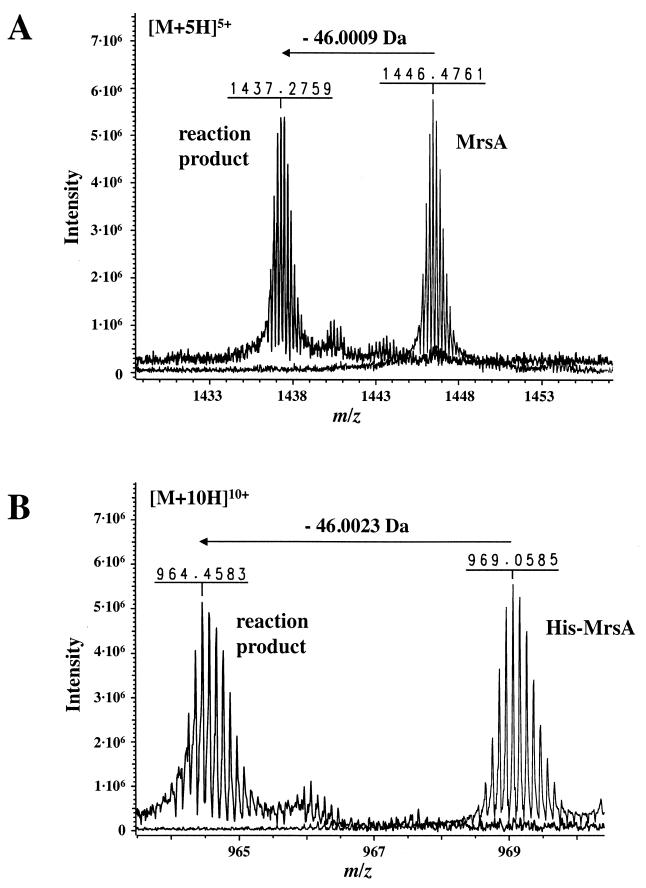
Sequence similarity between EpiD and MrsD and the presence of an unsaturated thioether ring in mature mersacidin suggested that MrsD is able to oxidize and decarboxylate COOH-terminal cysteine residues (3, 24). In order to verify this theory, MrsD was incubated with the substrate peptides MrsA and His-MrsA, respectively. RPC separation of the reaction mixture revealed that the substrate peptides were converted to peptides characterized by higher retention times and increased absorbance at 260 nm (Fig. 5). These same properties were observed for the EpiA-EpiD reaction products (16-18) (Fig. 5). Since peptidyl-aminoenethiols are converted nonenzymatically to other compounds 20; unpublished data), it is important to use short reaction times and to store the reaction products immediately at −80°C. Oxidative decarboxylation of MrsA and His-MrsA to peptidyl-aminoenethiols was confirmed by high-resolution ESI-FTICR-MS analysis (Fig. 6). The observed mass losses of 46.0009 and 46.0023 Da, respectively, are in excellent agreement with the removal of two hydrogen atoms and one molecule of CO2 (“H2CO2”), leading to a calculated monoisotopic mass difference of 46.0055 Da. In 1993, when the EpiD reaction was analyzed, MS methods enabling very high-resolution measurements were not available, and additional experiments were necessary to verify the oxidative decarboxylation reaction.
FIG. 5.
MrsD catalyzes the oxidative decarboxylation of MrsA and His-MrsA. The RPC-purified peptides MrsA (top), His-MrsA (middle), and EpiA R30Q (bottom) were incubated without any enzyme (control), with MrsD, with MrsD H75N, and with EpiD as indicated. The reaction mixtures were separated by RPC, and the elution was followed by measuring the absorbance at 214 nm (upper line), 260 nm (lower line), and 280 nm (data not shown). The LanD reaction products show an increased absorbance at 260 nm (and at 280 nm) due to the formation of the aminoenethiol chromophore and are labeled with an asterisk. The LanD reaction products are not very stable and are converted nonenzymatically to other compounds (20), as can be seen in the case of the reaction between EpiD and EpiA R30Q (positive control for the oxidative decarboxylase assay). The reaction of MrsD H75N with EpiA R30Q was not determined (n.d.) because this peptide was already not a substrate of wild-type MrsD enzyme.
FIG. 6.
ESI-FTICR-MS analysis of the reaction products formed by MrsD. The substrate peptides MrsA and His-MrsA, respectively, were incubated with MrsD, and the reaction mixtures were then analyzed by RPC. Reaction product containing fractions characterized by an increased absorbance at 260 nm (compare Fig. 5) were analyzed by ESI-FTICR-MS, and the obtained spectra were compared with the spectra of the substrate peptides. (A) The mass difference between MrsA and the reaction product is 46.0009 Da. (B) The mass difference between His-MrsA and the reaction product is 46.0023 Da. For calculation of the mass differences, only m/z values of the 5-fold charged ions (MrsA) and the 10-fold charged ions (His-MrsA; peaks with the highest intensity), respectively, were used. The observed mass differences are explained by oxidative decarboxylation of the COOH-terminal cysteine residue of the substrate peptides. The monoisotopic mass of “H2CO2” is 46.0055 Da (used atomic masses [in atomic mass units]: C, 12.00000000; H, 1.00782504; and O, 15.9949146).
In a control experiment, MrsD H75N was used. However, nearly no activity was observed in this case, although higher enzyme concentrations had been used (Fig. 5). This indicates that as in the case of EpiD, Dfp, and AtHAL3a, the conserved His residue of the HFCD proteins is an active-site residue.
Formation of the COOH-terminal S-[(Z)-2-aminovinyl]-3-methyl-d-cysteine residue of mersacidin can be explained by addition of the enethiol group (formed by the MrsD reaction) to the didehydrobutyrine residue at position +15 of MrsA which is introduced by dehydration of Thr.
The LanD enzymes EpiD and MrsD have different substrate specificities.
EpiD and MrsD both catalyze the oxidative decarboxylation of peptidyl-cysteines. It has been shown by analysis of the reaction of EpiD with single peptides and peptide libraries that EpiD is not very specific and that most of the peptides with the COOH-terminal sequence [V/I/L/(M)/F/Y/W]-[A/S/V/T/C/(I/L)]-C are substrates of EpiD (21). However, the COOH-terminal cysteine residue is essential and peptidyl-serines are not decarboxylated by EpiD. These results could then be explained by analysis of the crystal structure of EpiD H67N with bound substrate peptide (4). The consensus sequence of the EpiD substrates is not present at the COOH terminus of MrsA and, therefore, it is not surprising that neither MrsA (Fig. 5) nor the COOH-terminal peptide NH2-TLTSECIC-COOH (21) was a substrate of EpiD. Nothing is known about the substrate specificity of MrsD, so we sought to determine whether MrsD is able to oxidize and decarboxylate the epidermin precursor peptide. (EpiA R30Q and not native EpiA was used for this experiment. The mutation R30Q simplifies the purification of the precursor peptide from MBP fusion proteins and has no influence on the oxidative decarboxylation of the COOH-terminal cysteine residue by EpiD.) Also in this case, no reaction was observed, indicating a different and not overlapping substrate specificity of both enzymes. At present, it is not clear whether MrsD only recognizes substrates with the MrsA leader sequence or if shorter peptides are also substrates, as has been observed for EpiD (the synthetic carboxy-terminal peptide NH2-TLTSECIC-COOH of MrsA is not a substrate of MrsD; data not shown).
Comparison of characterized HFCD proteins and conclusions.
The enzymatic function of four HFCD proteins is now known. The LanD enzymes EpiD and MrsD have different substrate specificities but both catalyze the oxidative decarboxylation of peptidyl-cysteines, whereas Dfp and AtHAL3a decarboxylate 4"-phosphopantothenoylcysteine to 4"-phosphopantetheine (Fig. 7). The binding of different substrates by CoaC and LanD enzymes is related to different lengths of the substrate binding clamps (4, 16, 19). At first sight, the HFCD family is quite heterogeneous, although several residues are conserved in all enzymes. There are homododecameric and trimeric proteins, FAD- and FMN-dependent enzymes, enzymes involved in lantibiotic and coenzyme A biosynthesis, and prokaryotic and eukaryotic enzymes. However, it seems that all four protein types catalyze the decarboxylation of cysteine residues by using the same biochemical principle. The -CH2-SH side chain of the COOH-terminal cysteine residue is first oxidized to a thioaldehyde or to the tautomeric enethiol. Crystal structure data of EpiD H67N with bound substrate peptide (4) and later biochemical studies on the mechanism of Dfp (36) indicated direct oxidation of the thiol group to the thioaldehyde group by the flavin cofactor. Independently of the mechanism used for oxidation, decarboxylation occurs via the thioaldehyde tautomer in analogy to the decarboxylation of β-keto acids (24) and leads to peptidyl-aminoenethiols and/or enthiolates. In the case of the PPC decarboxylases, the “peptidyl”-aminoenethiol intermediate is reduced to 4"-phosphopantetheine. The existence of oxidatively decarboxylated reaction products in the case of EpiD and MrsD is a direct hint that the homologous enzymes Dfp and AtHAL3a also use an oxidation step in order to decarboxylate the PPC substrate. Why the oxidized intermediates are reduced by the CoaC enzymes but not by the LanD proteins remains open for investigation. Crystal structural studies on Dfp, AtHAL3a, and MrsD will help to answer this question.
The work presented here and in earlier studies shows how cysteine residues are decarboxylated. Decarboxylation by initial oxidation is a beautiful example of the great variety of flavoprotein-catalyzed reactions.
Acknowledgments
We thank Günther Jung for providing the high-resolution MS equipment for our experiments, Regine Stemmler for excellent technical assistance, and Stefan Stevanovic for NH2-terminal sequencing of MrsD. The polyclonal antibodies directed against MrsD were from BioGenes (Berlin, Germany).
This work was supported by Deutsche Forschungsgemeinschaft grants KU 869/4-1 and KU 869/4-2 to T.K. and Deutsche Forschungsgemeinschaft grants BI 504/1-2 and BI 504/1-3 to G.B.
REFERENCES
- 1.Albert, A., M. Martínez-Ripoll, A. Espinosa-Ruiz, L. Yenush, F. A. Culiáñez-Macià, and R. Serrano. 2000. The X-ray structure of the FMN-binding protein AtHal3 provides the structural basis for the activity of a regulatory subunit involved in signal transduction. Struct. Fold. Des. 8:961-969. [DOI] [PubMed] [Google Scholar]
- 2.Allgaier, H., G. Jung, R. G. Werner, U. Schneider, and H. Zähner. 1985. Elucidation of the structure of epidermin, a ribosomally synthesized, tetracyclic heterodetic polypeptide antibiotic. Angew. Chem. Int. Ed. Engl. 24:1051-1053. [Google Scholar]
- 3.Altena, K., A. Guder, C. Cramer, and G. Bierbaum. 2000. Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl. Environ. Microbiol. 66:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaesse, M., T. Kupke, R. Huber, and S. Steinbacher. 2000. Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate. EMBO J. 19:6299-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brötz, H., G. Bierbaum, K. Leopold, P. E. Reynolds, and H.-G. Sahl. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brötz, H., G. Bierbaum, P. E. Reynolds, and H.-G. Sahl. 1997. The lantibiotic mersacidin inhibits peptidoglycan biosynthesis at the level of transglycosylation. Eur. J. Biochem. 246:193-199. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, S., D. K. Chatterjee, R. H. Jani, J. Blumbach, B. N. Ganguli, N. Klesel, M. Limbert, and G. Seibert. 1992. Mersacidin, a new antibiotic from Bacillus: in vitro and in vivo antibacterial activity. J. Antibiot. 45:839-845. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee, S., S. J. Lad, M. S. Phansalkar, R. H. Rupp, B. N. Ganguli, H. W. Fehlhaber, and H. Kogler. 1992. Mersacidin, a new antibiotic from Bacillus: fermentation, isolation, purification and chemical characterization. J. Antibiot. 45:832-838. [DOI] [PubMed] [Google Scholar]
- 10.Cormack, B. 1991. Directed mutagenesis using the polymerase chain reaction, p. 8.5.1-8.5.9. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y. [Google Scholar]
- 11.Espinosa-Ruiz, A., J. M. Bellés, R. Serrano, and F. A. Culiáñez-Macià. 1999. Arabidopsis thaliana AtHal3: a flavoprotein related to salt and osmotic tolerance and plant growth. Plant J. 20:529-539. [DOI] [PubMed] [Google Scholar]
- 12.Kellner, R., G. Jung, T. Hörner, H. Zähner, N. Schnell, K.-D. Entian, and F. Götz. 1988. Gallidermin: a new lanthionine-containing polypeptide antibiotic. Eur. J. Biochem. 177:53-59. [DOI] [PubMed] [Google Scholar]
- 13.Kempter, C., T. Kupke, D. Kaiser, J. W. Metzger, and G. Jung. 1996. Thioenols from peptidyl-cysteines: oxidative decarboxylation of a 13C-labeled substrate. Angew. Chem. Int. Ed. Engl. 35:2104-2107. [Google Scholar]
- 14.Kilgour, G. L., S. P. Felton, and F. M. Huennekens. 1957. Paper chromatography of flavins and flavin nucleotides. J. Am. Chem. Soc. 79:2254-2256. [Google Scholar]
- 15.Kogler, H., M. Bauch, H.-W. Fehlhaber, C. Griesinger, W. Schubert, and V. Teetz. 1991. NMR-spectroscopic investigation on mersacidin, p. 159-170. In G. Jung and H.-G. Sahl (ed.), Nisin and novel lantibiotics. Escom, Leiden, The Netherlands.
- 16.Kupke, T. 2001. Molecular characterization of the 4"-phosphopantothenoylcysteine decarboxylase domain of bacterial Dfp flavoproteins. J. Biol. Chem. 276:27597-27604. [DOI] [PubMed] [Google Scholar]
- 17.Kupke, T., and F. Götz. 1997. The enethiolate anion reaction products of EpiD: pKa value of the enethiol side chain is lower than that of the thiol side chain of peptides. J. Biol. Chem. 272:4759-4762. [DOI] [PubMed] [Google Scholar]
- 18.Kupke, T., and F. Götz. 1997. In vivo reaction of affinity-tag-labelled epidermin precursor peptide with flavoenzyme EpiD. FEMS Microbiol. Lett. 153:25-32. [DOI] [PubMed] [Google Scholar]
- 19.Kupke, T., P. Hernández-Acosta, S. Steinbacher, and F. A. Culiáñez-Macià. 2001. Arabidopsis thaliana flavoprotein AtHAL3a catalyzes the decarboxylation of 4"-phosphopantothenoylcysteine to 4"-phosphopantetheine, a key step in coenzyme A biosynthesis. J. Biol. Chem. 276:19190-19196. [DOI] [PubMed] [Google Scholar]
- 20.Kupke, T., C. Kempter, V. Gnau, G. Jung, and F. Götz. 1994. Mass spectroscopic analysis of a novel enzymatic reaction: oxidative decarboxylation of the lantibiotic precursor peptide EpiA catalyzed by the flavoprotein EpiD. J. Biol. Chem. 269:5653-5659. [PubMed] [Google Scholar]
- 21.Kupke, T., C. Kempter, G. Jung, and F. Götz. 1995. Oxidative decarboxylation of peptides catalyzed by flavoprotein EpiD: determination of substrate specificity using peptide libraries and neutral loss mass spectrometry. J. Biol. Chem. 270:11282-11289. [DOI] [PubMed] [Google Scholar]
- 22.Kupke, T., S. Stevanovic, B. Ottenwälder, J. W. Metzger, G. Jung, and F. Götz. 1993. Purification and characterization of EpiA, the peptide substrate for posttranslational modifications involved in epidermin biosynthesis. FEMS Microbiol. Lett. 112:43-48. [DOI] [PubMed] [Google Scholar]
- 23.Kupke, T., S. Stevanovic, H.-G. Sahl, and F. Götz. 1992. Purification and characterization of EpiD, a flavoprotein involved in the biosynthesis of the lantibiotic epidermin. J. Bacteriol. 174:5354-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kupke, T., M. Uebele, D. Schmid, G. Jung, M. Blaesse, and S. Steinbacher. 2000. Molecular characterization of lantibiotic-synthesizing enzyme EpiD reveals a function for bacterial Dfp proteins in coenzyme A biosynthesis. J. Biol. Chem. 275:31838-31846. [DOI] [PubMed] [Google Scholar]
- 25.Mayer, H., H. Bauer, J. Breuss, S. Ziegler, and R. Prohaska. 2001. Characterization of rat LANCL1, a novel member of the lanthionine synthetase C-like protein family, highly expressed in testis and brain. Gene 269:73-80. [DOI] [PubMed] [Google Scholar]
- 26.Meyer, C., G. Bierbaum, C. Heidrich, M. Reis, J. Süling, M. I. Iglesias-Wind, C. Kempter, E. Molitor, and H.-G. Sahl. 1995. Nucleotide sequence of the lantibiotic Pep5 biosynthetic gene cluster and functional analysis of PepP and PepC: evidence for a role of PepC in thioether formation. Eur. J. Biochem. 232:478-489. [DOI] [PubMed] [Google Scholar]
- 27.Minami, Y., K.-I. Yoshida, R. Azuma, A. Urakawa, T. Kawauchi, T. Otani, K. Komiyama, and S. Omura. 1994. Structure of cypemycin, a new peptide antibiotic. Tetrahedron Lett. 35:8001-8004. [Google Scholar]
- 28.Mota-Meira, M., C. Lacroix, G. LaPointe, and M. C. Lavoie. 1997. Purification and structure of mutacin B-Ny266: a new lantibiotic produced by Streptococcus mutans. FEBS Lett. 410:275-279. [DOI] [PubMed] [Google Scholar]
- 29.Qi, F., P. Chen, and P. W. Caufield. 2000. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Appl. Environ. Microbiol. 66:3221-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi, F., P. Chen, and P. W. Caufield. 1999. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl. Environ. Microbiol. 65:3880-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schägger, H., and G. Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 32.Schneider, T. R., J. Karcher, E. Pohl, P. Lubini, and G. M. Sheldrick. 2000. Ab initio structure determination of the lantibiotic mersacidin. Acta Crystallogr. D Biol. Crystallogr. 56:705-713. [DOI] [PubMed] [Google Scholar]
- 33.Schnell, N., K.-D. Entian, U. Schneider, F. Götz, H. Zähner, R. Kellner, and G. Jung. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333:276-278. [DOI] [PubMed] [Google Scholar]
- 34.Smith, L., J. Novak, J. Rocca, S. McClung, J. D. Hillman, and A. S. Edison. 2000. Covalent structure of mutacin 1140 and a novel method for the rapid identification of lantibiotics. Eur. J. Biochem. 267:6810-6816. [DOI] [PubMed] [Google Scholar]
- 35.Spitzer, E. D., and B. Weiss. 1985. dfp gene of Escherichia coli K-12, a locus affecting DNA synthesis, codes for a flavoprotein. J. Bacteriol. 164:994-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strauss, E., and T. P. Begley. 2001. Mechanistic studies on phosphopantothenoylcysteine decarboxylase. J. Am. Chem. Soc. 123:6449-6450. [DOI] [PubMed] [Google Scholar]
- 37.Strauss, E., C. Kinsland, Y. Ge, F. W. McLafferty, and T. P. Begley. 2001. Phosphopantothenoylcysteine synthetase from Escherichia coli: identification and characterization of the last unidentified coenzyme A biosynthetic enzyme in bacteria. J. Biol. Chem. 276:13513-13516. [DOI] [PubMed] [Google Scholar]
- 38.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]