Abstract
Salmonella enterica forms polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. These organelles are thought to consist of a proteinaceous shell that encases coenzyme B12-dependent diol dehydratase and perhaps other enzymes involved in 1,2-propanediol degradation. The function of these organelles is unknown, and no detailed studies of their structure have been reported. Genes needed for organelle formation and for 1,2-propanediol degradation are located at the 1,2-propanediol utilization (pdu) locus, but the specific genes involved in organelle formation have not been identified. Here, we show that the pduA gene encodes a shell protein required for the formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. A His6-PduA fusion protein was purified from a recombinant Escherichia coli strain and used for the preparation of polyclonal antibodies. The anti-PduA antibodies obtained were partially purified by a subtraction procedure and used to demonstrate that the PduA protein localized to the shell of the polyhedral organelles. In addition, electron microscopy studies established that strains with nonpolar pduA mutations were unable to form organelles. These results show that the pduA gene is essential for organelle formation and indicate that the PduA protein is a structural component of the shell of these organelles. Physiological studies of nonpolar pduA mutants were also conducted. Such mutants grew similarly to the wild-type strain at low concentrations of 1,2-propanediol but exhibited a period of interrupted growth in the presence of higher concentrations of this growth substrate. Growth tests also showed that a nonpolar pduA deletion mutant grew faster than the wild-type strain at low vitamin B12 concentrations. These results suggest that the polyhedral organelles formed by S. enterica during growth on 1,2-propanediol are not involved in the concentration of 1,2-propanediol or coenzyme B12, but are consistent with the hypothesis that these organelles moderate aldehyde production to minimize toxicity.
Salmonella enterica degrades 1,2-propanediol in a coenzyme B12-dependent manner (13). The genes needed for this process were acquired by a horizontal gene transfer that is thought to be one of several related events important to the divergence of S. enterica and Escherichia coli (16, 26). In vivo expression technology has indicated that 1,2-propanediol utilization (pdu) genes may be important for growth in host tissues, and competitive index studies with mice have shown that pdu mutations confer a virulence defect (7, 11). Moreover, 1,2-propanediol degradation by S. enterica provides an important model system for understanding coenzyme B12-dependent processes, some of which are important in human physiology, industry, and the environment (29).
At first glance, the degradation of 1,2-propanediol appears to be a relatively simple process. The proposed pathway begins with the conversion of 1,2-propanediol to propionaldehyde, a process mediated by coenzyme B12-dependent diol dehydratase (21, 37). The aldehyde is then converted to propanol and propionic acid, presumably by alcohol dehydrogenase, coenzyme A-dependent aldehyde dehydrogenase, phosphotransacylase, and propionate kinase. This pathway generates one ATP, an electron sink, and a 3-carbon intermediate (propionyl-coenzyme A), which can feed into central metabolism via the methyl citrate pathway (12). In S. enterica, the degradation of 1,2-propanediol occurs aerobically, or anaerobically when tetrathionate is supplied as an electron acceptor (26).
The complexity of 1,2-propanediol degradation became apparent when the DNA sequence of the pdu locus was determined, and 23 pdu genes were identified: 6 pdu genes are thought to encode enzymes needed for the 1,2-propanediol degradative pathway; 2 are involved in transport and regulation; 2 are probably involved in diol dehydratase reactivation; 1 is needed for the conversion of vitamin B12 (CN-B12) to coenzyme B12; 5 are of unknown function; and 7 share similarity to genes involved in carboxysome formation (6). Carboxysomes are polyhedral organelles found in cyanobacteria and some chemoautotrophs (32, 34, 35). They are composed of a proteinaceous shell that houses most of the cell's ribulose bisphosphate carboxylase/oxygenase (RuBisCO). They are required for autotrophic growth at low CO2 concentrations and are thought to function as part of a CO2-concentrating mechanism (15, 23, 25).
Recently, S. enterica was shown to form polyhedral organelles that resemble carboxysomes during aerobic and anaerobic growth on 1,2-propanediol (6). These organelles are approximately 150 nm in cross-section and appear to consist of a proteinaceous shell and interior (6). However, there are significant differences between the S. enterica organelles and carboxysomes. S. enterica is not an autotroph and does not express RuBisCO. The S. enterica organelles are involved in coenzyme B12-dependent 1,2-propanediol degradation and are associated with coenzyme B12-dependent diol dehydratase and perhaps other enzymes (6). A role in CO2 concentration, similar to that of the carboxysome, is uncertain since there is no known association between CO2 and coenzyme B12-dependent 1,2-propanediol degradation in S. enterica. Thus, the function of the S. enterica organelles and the extent of their similarity to carboxysomes remain important questions.
Seven genes found in the pdu operon have DNA sequence similarity to those required for carboxysome formation, and four of these (pduAJKT) encode proteins that have similarity to carboxysome shell proteins (6, 33). The roles of these pdu genes in organelle formation have not been investigated, and no genetic studies on organelle function have been reported. Here, we show that the pduA gene is required for the formation of polyhedral organelles by S. enterica. Results show that the PduA protein is a component of the organelle's shell. Physiological studies revealed that a pduA mutant grew similarly to the wild type on minimal medium containing lower concentrations of 1,2-propanediol but exhibited a period of interrupted growth at higher concentrations. This mutant also grew at a faster rate than the wild type at low CN-B12 concentrations, suggesting that the organelle's shell may present a barrier to B12 entrance. These results indicate that the organelles of S. enterica are not involved in concentrating 1,2-propanediol or CN-B12, but are consistent with a role in moderating aldehyde toxicity.
MATERIALS AND METHODS
Chemicals and reagents.
Formaldehyde (R,S), 1,2-propanediol, trichloroacetic acid, and antibiotics were from Sigma Chemical Company (St. Louis, Mo.). Isopropyl-β-d-thiogalactopyranoside (IPTG) was from Diagnostic Chemicals Limited (Charlottetown, Canada). Restriction enzymes were from New England Biolabs (Beverly, Mass.) or Promega (Madison, Wis.). T4 DNA ligase was from New England Biolabs. Electrophoresis supplies were from Bio-Rad (Hercules, Calif.). Bacterial Protein Extraction Reagent II and Micro BCA reagents were from Pierce (Rockford, Ill.). Other chemicals were from Fisher Scientific (Pittsburgh, Pa.).
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. The rich medium used was Luria-Bertani medium (Difco Laboratories, Detroit, Mich.) (20). The minimal medium used was the No-carbon-E (NCE) medium (4, 19, 38). Amino acids were provided at the following concentrations: valine, isoleucine, leucine, and threonine, 0.3 mM; and histidine, 0.1 mM. Antibiotics were provided in liquid or solid rich medium at the following concentrations unless otherwise stated: carbenicillin, 100 μg/ml; ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 20 μg/ml. Ampicillin was used at 15 μg/ml in minimal medium. Tetracycline was used at 2 μg/ml to induce expression out of TPOP insertions. IPTG was added to a final concentration of 1 mM for induction of genes cloned into pET vectors or placIqPO-BglII. MacConkey-1,2-propanediol-vitamin B12 indicator plates and aldehyde indicator plates were prepared as described previously (6, 13).
TABLE 1.
Bacterial strains
| Species and strains | Genotype |
|---|---|
| E. coli | |
| BE11 | (E. coli ER2267) F−e14− (MrcA−) endA1 supE44 thi-1 relA1? rfbD1? spoT1? Δ(mrcC-mrr) 114::IS10 Δ(argF-lac)U169 recA1/F−proA+B+laclq Δ(lacZ)M15 zzf::mini-Tn10 (Kanr)/pMGS2 |
| BE229 | (E. coli ER1992) F−λ− Δ(argF-lac)U169 supE44 e14−dind1::MudI1473 (KanrlacZ+) rtbD1? relA1? endA1 spoT1? thi-1 Δ(mrcL-mrr)114::IS10/pGH48 (His6-PduA Apr) |
| BE230 | BL21(DE3)/pGH48(His6-PduA Apr) and pSJS1240 (ileX argU Spr) |
| BE231 | BL21(DE3) RIL/pGH107(T7 expression vector with pduJ insert, Kanr) |
| BL21(DE3) | (E. coli B) F−ompT hsdS(rB− mB−) dcm+ Tetrgal λ (DE3) endA |
| BL21(DE3) RIL | (E. coli B) F−ompT hsdS(rB− mB−) dcm+ Tetrgal λ (DE3) endA Hte (argU ileY leuW Camr) |
| RT1679 | DH5α/pVJ70 (contains pduF"ABCDE ORF1" Apr) |
| S. enterica serovar Typhimurium LT2 | Wild-type |
| BE33 | pduA672:: TPOP1 |
| BE39 | pduA673 TPOP1::fragment (a short nonpolar insertion resulting from the spontaneous deletion of the majority of the pduA672::TPOP1 element) |
| BE103 | TR6579/pKD46a |
| BE182 | ΔpduA652 (precise nonpolar deletion of the pduA gene)b |
| BE228 | ΔpduA652/pTA749 (placIqPO-BglII with pduA insert, Apr) |
| BE232 | metE205 ara-9 Δ299/placIqPO-BglII (no insert, Apr) (in Δ299 the entire cob and pdu operons are deleted) |
| BE233 | metE205 ara-9 Δ299/pTA749 (pduA under plac control, Apr) (in Δ299 background the entire cob and pdu operons are deleted) |
| BE235 | pdu12::MudJ |
| TR6579 | metA22 metE551 trpD2 ivl-452 hsdLT6 hsdSA29 HsdB−strA120 GalE− Leu− Pro− |
See reference 8.
See Materials and Methods.
General protein and molecular methods.
Plasmid purification, bacterial transformation, electrophoresis, and other standard molecular and protein methods were performed as previously described (14, 30).
P22 transduction and transposon mutagenesis of the pdu region.
Transductional crosses were performed as described previously (9, 31). For TPOP1 mutagenesis of the pdu region, a pool of approximately 80,000 independent insertion mutants was prepared as described previously (5). The pool was then used as a donor in transductional crosses with strain BE235 (pdu12::MudJ). Tetracycline resistance was selected, and transductants were screened for loss of the pdu12::MudJ elements using MacConkey-lactose-1,2-propanediol indicator plates (5). Following transposon mutagenesis, 30,000 colonies were screened, and 120 TPOP1 insertions located near the pdu operon were isolated.
Localization of TPOP1 insertions.
PCR was used to amplify the region of DNA that included one join point between the TPOP1 element and the S. enterica chromosome. The primers used for PCR amplification were 5"-ACCTTTGGTCACCAACGCTTTTCC-3" and 5"-GTTCATATGCGAAACCACTTC-3". The DNA sequence of the PCR product was determined, and DNA sequence analysis showed that the downstream join point between the pduA672::TPOP1 and the S. enterica chromosome was bp 160 of the pduA coding sequence.
Growth curves.
Growth curves were determined as described previously (14). For physiological studies that employed various CN-B12 and 1,2-propanediol concentrations, cells were grown in 16- by 100-mm test tubes containing 5 ml of appropriate medium. Cultures were incubated as described for the aerobic growth curves, except the tubes were held in place at an angle of 45°. Cell growth was monitored by measuring the optical density at 600 nm (OD600) using a Spectronic 20D+ spectrophotometer. Inocula for the growth curves were prepared as described previously (14), and 0.125 ml was used to inoculate 5-ml cultures.
Cloning of pduA for high-level expression.
The following primers were used for PCR amplification of pduA DNA using pVJ70 (RT1679) as a template: forward, 5"-GGAATTCCATATGCAACAAGCACTAGGAATGG-3"; and reverse, 5"-CACCGATGGATCCTCATTGGCTAATTCCCTTCG-3". The PCR product was gel purified, restricted with NdeI and BamHI, and ligated to the similarly restricted plasmid pET15b. The ligation reaction mixture was heated to 70°C for 15 min and used to transform E. coli ER1992 by electroporation. The DNA sequence of the cloned DNA was shown to be in agreement with the previously reported pduA DNA sequence (6). For protein expression, E. coli BL21(DE3)/pSJS1240 was used as the host strain.
Cloning of pduJ.
The following primers were used for PCR amplification of pduJ using pMGS2 (BE11) as a template: forward, 5"-GGAATTCATATGAATAACGCACTGGGACTGG-3"; and reverse, 5"-AGGATCATGCTCGAGGGCTGATTTCGGTAAAATGG-3". The PCR product was gel purified, restricted with NdeI and XhoI, and ligated to the similarly restricted plasmid pET41a. The ligation reaction was heated to 70°C for 15 min and used to transform E. coli DH5α by electroporation. The sequence of the cloned DNA was shown to be in agreement with the previously reported pduJ DNA sequence (6). For protein expression, E. coli BL21(DE3) RIL was used as the host strain.
Purification of the PduA protein.
A 200-ml culture of BE230 was prepared using LB-ampicillin-spectinomycin-1% glucose medium incubated at 37°C in a 1-liter baffled flask with shaking at 275 rpm. When the cells reached an OD600 of 0.8, expression of recombinant His6-PduA protein was induced by addition of 1 mM IPTG. Cells were lysed, and inclusion bodies were isolated by treatment with Bacterial Protein Extraction Reagent II according to the manufacturer's protocol. The inclusion bodies were then solubilized in binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9], 6 M guanidine hydrochloride) and filtered though a 0.45-μm-pore size cellulose acetate filter. His6-PduA was purified from the solubilized inclusion bodies using a 1-ml Amersham Pharmacia HiTrap Chelating (Ni2+) column. The manufacturer's directions were followed with the following modifications: a 20-ml 20 to 200 mM linear imidazole gradient was used to elute the recombinant His6-PduA, and 6 M guanidine hydrochloride was added to the binding, wash, and elution buffers.
Antibody preparation.
His6-PduA protein obtained from Ni2+ affinity chromatography (see above) was resolved on a 12% polyacrylamide gel. The portion of the gel containing the PduA protein was excised and used as a source of antigen. Polyclonal antibodies were prepared in a New Zealand White rabbit by Cocalico Biologicals (Reamstown, Pa.).
For subtraction of antibodies reacting with PduJ and E. coli proteins, two 200-ml cultures of BE231 were prepared using Luria broth-kanamycin-1% glucose medium incubated at 37°C in 1-liter baffled flasks with shaking at 275 rpm. When the cells reached an OD600 of 0.8, expression of recombinant PduJ-His8 protein was induced by addition of 1 mM IPTG. An acetone powder made out of whole cells was used to subtract antibodies reacting with the PduJ protein as described previously (10).
Western blots.
Cultures were grown in NCE minimal medium supplemented with 0.4% propanediol, 1% succinate, and the amino acids valine, leucine, isoleucine, and threonine. The pellet from 1 ml of cells was mixed with enough Tris-Tricine loading buffer to obtain an OD600 of 1. Samples were boiled for 8 min at 100°C, and 20 μl (equivalent to protein from cells with an OD600 of 0.02) was separated on a 16.5% Tris-Tricine gel. Electroblotting was performed in 10 mM morpholineethanesulfonic acid-20% MeOH, pH 6.0, using a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad) run at a constant 20 V for 14.5 h at 4°C using a PowerPac 1000 power supply (Bio-Rad). The membrane used was Hybond-P (Amersham Pharmacia Biotech, Buckinghamshire, England). Membranes were probed as described previously (10) using anti-PduA polyclonal antibodies diluted 1:7,000 in blocking buffer as the primary antibody and goat anti-rabbit immunoglobulin G-alkaline phosphatase conjugate (Bio-Rad) diluted 1:3,000 in blocking buffer as the secondary antibody. Color developing reagents were used as specified in the manufacturer's directions (Bio-Rad).
Electron microscopy.
For electron microscopy, cells were grown on minimal medium supplemented with 1% succinate and 0.4% propanediol. Cultures (10 ml) were incubated in 125-ml shake flasks at 37°C, with shaking at 275 rpm in a New Brunswick C24 Incubator Shaker. For the observation of ultrastructure, cells were sectioned, fixed, observed, and photographed as described previously (6).
For immunogold localization of the PduA protein and diol dehydratase, cells were sectioned, fixed, observed, and photographed as described previously (6). The primary antibodies used were either rabbit polyclonal antibody against the PduA protein (anti-PduA) diluted 1:100 in phosphate-buffered saline (PBS), rabbit polyclonal antibody against diol dehydratase diluted 1:1,000 in PBS, or preimmune serum diluted 1:100 in PBS. The secondary antibody used was goat anti-rabbit immunoglobulin G conjugated to 12-nm colloidal gold (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) diluted 1:30 in PBS.
Construction of a nonpolar pduA deletion.
A nonpolar deletion of pduA was constructed as described by Datsenko and Wanner (8) with some modifications. The primers used for PCR amplification of the kanamycin resistance cassette from plasmid pKD4 were 5"-GTGTCCCAACTATCGGAACACTCCATGCGAGGTCTTTTGTAGGCTGGAGCTGCTTCG-3" and 5"-CTGCGCCATGATCTGTTCCACCAGCTCATTGCTGCATGAATATCCTCCTTAGTTC-3". Strain BE103 was used as the host strain for linear transformation. The primers used to verify the chromosomal location of the kanamycin cassette insertion were 5"-GTCCTGGCCAGCGCAAGTTTCGGC-3", 5"-CAGTCATAGCCGAATAGCCT-3", 5"-CGGTGCCCTGAATGAACTGC-3", and 5"-GCTTTTTCCAGCGCATAGCTGGCGCGAGC-3". After the insertion site was verified by PCR, the kanamycin cassette was moved into LT2 via transduction. This cassette was then removed using the FlP recombinase as described previously (8). DNA sequencing showed that the expected deletion was formed; the entire pduA coding sequence was deleted except for the last 29 bp, which included the native ribosome binding site of the pduB gene.
DNA sequencing and analysis.
DNA sequencing was carried out by the University of Florida Interdisciplinary Center for Biotechnology Research, DNA Sequencing Core Facility, as described previously (6) and the University of Florida Department of Microbiology and Cell Science, DNA Sequencing Facility, using a LI-COR model 4000L DNA sequencer, automated sequencing equipment, and Base ImagIR analysis software version 04.1 h (LI-COR, Lincoln, Nebr.). BLAST software was used for sequence similarity searching (1).
RESULTS
Purification of recombinant His6-PduA protein.
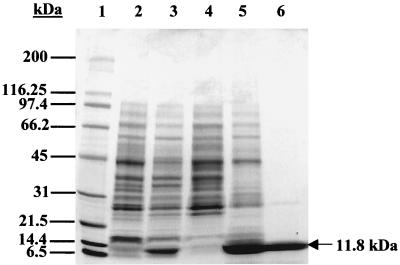
E. coli strain BE230 was constructed to produce high levels of recombinant His6-PduA protein. Samples of induced and uninduced boiled cells of this strain were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to determine if the His6-PduA protein was being expressed. When induced by IPTG, strain BE230 produced high levels of an 11.8-kDa protein (Fig. 1, lane 3), which was not seen in the uninduced sample (Fig. 1, lane 2). This observed mass correlates well with the predicted mass of 11.4 kDa for the recombinant His6-PduA protein. Electron microscopy revealed that strain BE230 produced rod-like structures in the cytosol when induced with IPTG (data not shown), and SDS-PAGE showed that these inclusion bodies were composed mainly of the His6-PduA protein (Fig. 1, lane 5). Significant amounts of the His6-PduA protein were not found in the soluble fraction (Fig. 1, lane 4).
FIG. 1.
Overexpression and purification of the His6-PduA protein. Lane 1, molecular mass markers; lane 2, uninduced boiled cell lysate; lane 3, induced boiled cell lysate; lane 4, soluble fraction; lane 5, inclusion body preparation; lane 6, combined fractions 12 to 15 from Ni2+ column elution. Molecular masses in kilodaltons are shown at the left.
The His6-PduA protein was purified from inclusion bodies by Ni2+ chromatography under denaturing conditions. The His6-PduA protein eluted from the column at approximately 160 to 200 mM imidazole. The fractions containing the His6-PduA protein were analyzed by SDS-PAGE. A single band at the predicted mass for the His6-PduA protein was observed, as well as three faint bands at higher molecular masses (Fig. 1, lane 6). About 600 μg of the partially purified His6-PduA protein was isolated on a preparatory SDS-PAGE gel, and the band of interest was excised and used as a source of antigen for polyclonal antibody production.
Preparation of PduA-specific antibody.
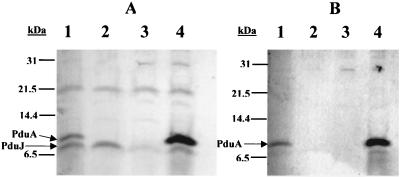
To determine the specificity of the polyclonal antibodies generated against the His6-PduA protein, Western blots were performed on boiled cell lysates. Anti-PduA polyclonal antibodies recognized a major protein band at 9.5 kDa in the wild-type strain but not in strain BE182 (which contains a nonpolar pduA deletion mutation) (Fig. 2A, lanes 1 and 2). This indicated that the band at 9.5 kDa corresponded to the native PduA protein. Anti-PduA antibodies also recognized a protein band at 9.5 kDa in strain BE233 (Fig. 2A, lane 4), which carries a plasmid with the pduA gene under control of an IPTG-inducible promoter but lacks the entire pdu operon due to a deletion mutation. No band at 9.5 kDa was observed in an isogenic strain carrying the same plasmid without an insert (BE232) (Fig. 2A, lane 3). These results confirmed that the band at 9.5 kDa was the native PduA protein.
FIG. 2.
Western analysis with unsubtracted (A) and subtracted (B) anti-PduA polyclonal antibody preparations. For both panels: lane 1, S. enterica serovar Typhimurium LT2; lane 2, BE182 (ΔpduA mutant); lane 3, BE232 (isogenic to BE233, except that the expression plasmid lacks an insert); lane 4, BE233 (PduA expression strain). Molecular masses in kilodaltons are shown at the left of each blot. Total protein loaded in each lane was equivalent to that from cells at an OD600 of 0.02.
The anti-PduA antibody preparation also reacted with an 8-kDa protein expressed by the wild-type strain and BE182 (Fig. 2A, lanes 1 and 2) and a 7.5-kDa protein band expressed by BE233 and BE232 (Fig. 2A, lanes 3 and 4). Additional Western blots of strains overexpressing the PduJ protein indicated that this 8-kDa protein was the PduJ protein (data not shown). The observed 7.5-kDa protein band was apparently plasmid encoded, as this band was detected in Western blots of cells carrying plasmid pTA749 but not in blots of isogenic strains that lacked this plasmid (data not shown). In order to improve the specificity of the polyclonal antibody preparation, a subtraction procedure was performed.
An acetone powder was made from whole cells of strain BE231, a derivative of BL21(DE3) RIL that expressed PduJ at high levels, and then used to subtract antibodies recognizing the PduJ protein and E. coli proteins from the anti-PduA polyclonal antibody preparation. After subtraction, the antibody preparation recognized a single band at 9.5 kDa in the wild-type strain (Fig. 2B, lane 1), and no labeling was observed in the pduA deletion mutant, BE182 (Fig. 2B, lane 2). The subtracted polyclonal antibody preparation also recognized a 9.5-kDa protein band in strain BE233, which overexpresses the PduA protein (Fig. 2B, lane 4). No labeling at 9.5 kDa was observed in control strain BE232, which carried the plasmid without insert (Fig. 2B, lane 3). These results showed that the subtracted anti-PduA antibody preparation was highly specific for the PduA protein, and this antibody preparation was used for subsequent immunolabeling experiments.
Localization of PduA by immunoelectron microscopy.
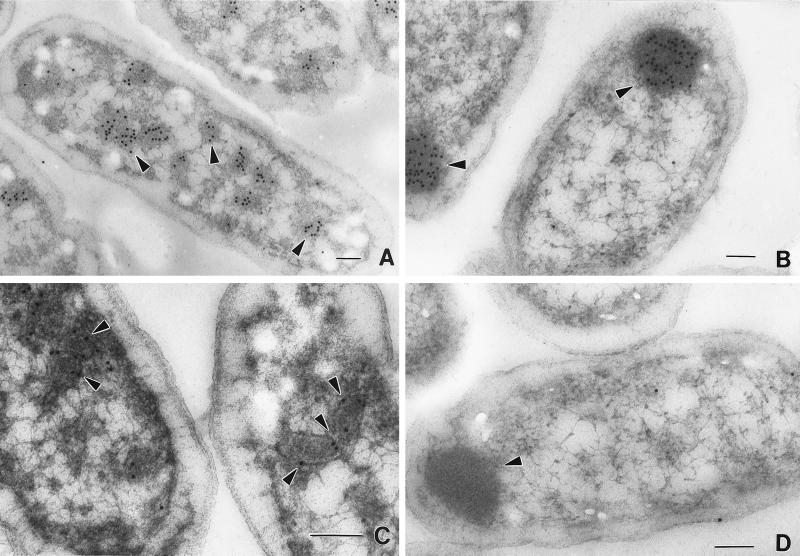
Immunogold labeling of S. enterica cells with highly specific subtracted anti-PduA antibodies (see above) indicated that the PduA protein is a component of the shell of the polyhedral organelles. In the micrograph shown in Fig. 3C, the antibody-conjugated gold particles (solid black circles) indicate the location of the PduA protein. The majority of the gold particles localized to the periphery of the polyhedral organelles. Preimmune serum failed to label the polyhedra (data not shown), and only a small amount of spurious labeling was observed when strain BE182 (ΔpduA) was labeled with anti-PduA antibody (Fig. 3D).
FIG. 3.
Localization of diol dehydratase and the PduA protein by immunoelectron microscopy. Cells were grown on succinate minimal medium supplemented with 1,2-propanediol to induce expression of the pdu operon. For each panel the arrows point to gold particles that indicate the location of either diol dehydratase or the PduA protein. The strain and protein labeled in each panel are as follows. (A) S. enterica, diol dehydratase. (B) BE182 (ΔpduA), diol dehydratase. (C) S. enterica, PduA. (D) BE182 (ΔpduA), PduA. Bars at lower right-hand corners are 100 nm in length.
Localization of diol dehydratase in a pduA mutant.
Labeling of wild-type and BE182 (ΔpduA) cells with anti-diol dehydratase antibodies illustrated the effect of a nonpolar pduA deletion mutation on the localization of diol dehydratase (Fig. 3A and B). In the wild-type strain, diol dehydratase localized to the polyhedral organelles (Fig. 3A), whereas in strain BE182 (ΔpduA), which is unable to synthesize polyhedra, diol dehydratase was found in large diffuse aggregates at the poles of the cell (Fig. 3B). This is consistent with the role of the PduA protein as a component of a shell that encases diol dehydratase.
The pduA gene is required for the formation of polyhedral organelles.
The effect of a pduA::TPOP1 insertion mutation on polyhedral organelle formation was investigated. TPOP1 insertions are polar in the absence of tetracycline but nonpolar when growth media are supplemented with this antibiotic (27). Strain BE33 (pduA672::TPOP1) exhibited a polyhedron-negative phenotype and failed to grow on 1,2 propanediol-CN-B12 minimal medium in the absence of tetracycline. When tetracycline was used to induce downstream expression from the TPOP1 element, BE33 was still unable to synthesize polyhedra, but the ability to grow on 1,2-propanediol minimal medium was restored. This indicated that the pduA gene was required for organelle formation but not for growth on 1,2-propanediol. Two additional nonpolar pduA mutations were also shown to prevent organelle formation but allow growth on 1,2-propanediol-CN-B12 minimal medium. They were BE182 (ΔpduA) and BE39 (pduA::TPOP1 fragment). These results provided further evidence that the pduA gene was required for the formation of polyhedral organelles but not for growth on 1,2-propanediol.
Complementation of the polyhedral organelle-negative phenotype of a ΔpduA mutant strain.
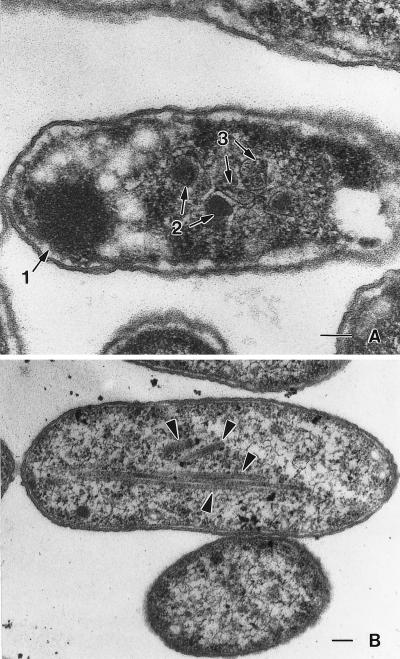
Strain BE228 contains a nonpolar deletion of the pduA coding sequence as well as plasmid pTA749, which allows expression of the PduA protein in response to IPTG. To determine whether the pduA gene present on the plasmid could complement the ΔpduA mutation for polyhedral body formation, strain BE228 was grown on 1,2-propanediol succinate minimal medium using several IPTG concentrations and then examined by electron microscopy. Strain BE228 synthesized both normal and abnormal polyhedra at 0.01 mM IPTG (Fig. 4A). In the absence of IPTG, no normally shaped organelles were formed and aberrantly shaped structures were rare. The finding that BE228 (ΔpduA/pTA749) produced apparently normal organelles in the presence of IPTG, but not in its absence, showed that the organelle-negative phenotype of strain BE182 was a consequence of the ΔpduA mutation. The apparent explanation for the formation of aberrant organelles is that proper organelle assembly required a specific ratio of the PduA protein to other organelle proteins; the prevalence of aberrant polyhedra increased with increasing IPTG concentration (increasing levels of the PduA protein), and at concentrations of ≥0.1 mM, primarily aberrant structures were observed (Fig. 4B). The fact that higher PduA expression levels alters organelle shape supports a structural role for the PduA protein in the formation of the structures.
FIG. 4.
Complementation of a ΔpduA mutation for formation of polyhedral organelles. Strain BE228 (ΔpduA/pTA749-PduA expression plasmid) was grown on minimal succinate medium supplemented with 1,2-propanediol and IPTG and then examined by transmission electron microscopy. Strain BE228 was grown with 0.01 (A) and 0.1 (B) mM IPTG, respectively. (A) The numbered arrows indicate the locations of the following structures: 1, polar inclusion body; 2, polyhedral organelles; 3, abnormally shaped organelles. (B) The arrows indicate the aberrant rod-like structures observed in some cells. Bars at the lower right-hand corners are 100 nm in length.
Strains with pduA mutations show a period of interrupted growth when cultured on 1,2-propanediol-CN-B12 minimal medium.
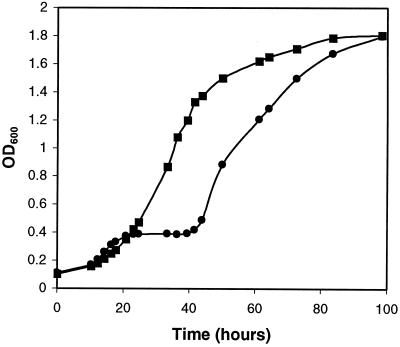
During aerobic growth on 1,2-propanediol-CN-B12 minimal medium, strain BE182 (ΔpduA) grew similarly to the wild-type strain except for a period of interrupted growth that initiated at 20 h and persisted until the 40-h mark (Fig. 5). Before the onset of interrupted growth, BE182 (ΔpduA) grew with a generation time of 6.8 h, which was slightly faster than the 8.4-h generation time of the wild type-strain. Strain BE182 (ΔpduA) also grew slightly faster than the wild type after the period of interrupted growth; its generation time was 7.7 h, compared to a 9-h generation time for the wild-type strain. The maximum optical density reached by both the wild-type strain and BE182 (ΔpduA) was 1.8 at 600 nm. Viable cell counts of the mutant and wild-type strains gave results similar to optical density measurements. The period of interrupted growth was observed in four separate experiments. This phenomenon suggested that a toxic compound was accumulating and inhibiting the growth of S. enterica mutants unable to form polyhedral organelles during growth on 1,2-propanediol. Presumably, growth resumed after the induction of genes that mitigated the toxicity problem. Alternatively, interrupted growth could have resulted from the depletion of an essential nutrient. However, this seems unlikely, since interrupted growth was dependent on the concentrations of 1,2-propanediol and CN-B12 present in the growth medium (see below).
FIG. 5.
Growth of the wild-type strain and a pduA mutant on 1,2-propanediol-CN-B12 minimal medium. Results for the wild type, S. enterica serovar Typhimurium LT2 (▪), and BE182 (nonpolar ΔpduA) (•) are shown. Cells were cultured as described in Materials and Methods.
Effects of 1,2-propanediol concentration on the growth rates of wild-type and pduA mutant strains.
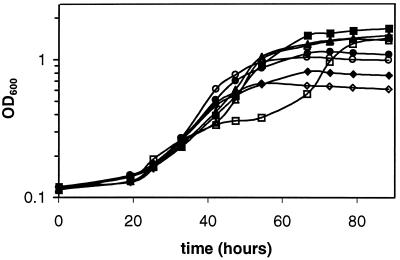
The effects of the 1,2-propanediol concentration on the growth of the wild type and a strain containing a nonpolar pduA deletion mutation (BE182) were examined (Fig. 6). At 1,2-propanediol concentrations of 0.01 and 0.05%, the wild-type strain and BE182 grew similarly. The generation times of the wild-type were 10.8 and 11.9 h, respectively, whereas the generation times of BE182 (ΔpduA) were 11.7 and 10.7 h, respectively. At higher 1,2-propanediol concentrations, some distinct differences between the wild type and strain BE182 were noted. At 1,2-propanediol concentrations of 0.2 and 0.4%, the wild-type strain grew with generation times of 10.2 and 12.6 h. At similar 1,2-propanediol concentrations, strain BE182 grew with generation times of 14.5 and 20.3 h followed by a period of interrupted growth and then much shorter generation times of 8 and 10.2 h. In strain BE182 (ΔpduA), the duration of interrupted growth was shorter at a 1,2-propanediol concentration of 0.2% (10 h) than at 0.4% (20 h). Thus, interrupted growth was observed at higher concentrations but not at lower concentrations of 1,2-propanediol. This suggested that interrupted growth resulted from the accumulation of a toxic compound derived from 1,2-propanediol. Propionaldehyde was a likely candidate, since this compound is an intermediate of 1,2-propanediol degradation and aldehydes are well known to have toxic effects on cells. Amino acid supplementation was not used in this experiment, as very slight growth occurred on the amino acids alone. Consequently, the generation times observed are longer than those shown in Fig. 5.
FIG. 6.
Effects of various 1,2-propanediol concentrations on the growth of strain BE182 (ΔpduA) and the wild-type strain. Cells were cultured on 1,2-propanediol-CN-B12 minimal media having the following 1,2-propanediol concentrations: 0.4% (▪, □), 0.2% (▴, ▵), 0.1% (•, ○), and 0.05% (⧫, ◊). Closed symbols, wild type, S. enterica serovar Typhimurium LT2. Open symbols, BE182 (ΔpduA).
Effects of CN-B12 concentration on the growth rates of wild-type and pduA null mutant strains.
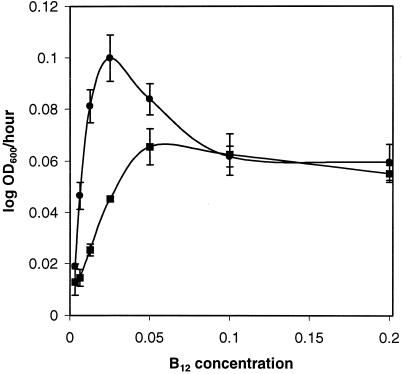
The effects of the CN-B12 concentration on the growth of the wild type and a strain containing a nonpolar pduA deletion mutation (BE182) were examined (Fig. 7). Strain BE182 (ΔpduA) grew faster than the wild type at concentrations of CN-B12 from 0.003125 to 0.05 μg/ml; the largest difference was seen at a concentration of 0.00625 μg/ml, where the generation times were 47.5 h for the wild-type strain and 14.9 h for BE182 (ΔpduA). At CN-B12 concentrations of 0.1 and 0.2 μg/ml, BE182 (ΔpduA) reached a maximal growth rate similar to that of the wild-type strain but exhibited the aforementioned interrupted growth (see above). These findings are consistent with a model in which an intermediate of 1,2-propanediol degradation has a toxic effect that interrupts growth. At higher CN-B12 concentrations, 1,2-propanediol would be catabolized at a faster rate and the toxic compound would accumulate to higher levels. This would result in a more pronounced interruption of growth. Subsequently, genes that lessen the effects of the toxic compound would be induced, and the growth rate would increase. In addition, the above results indicate that the polyhedral organelles act as a barrier to B12. The possible significance of this finding is addressed in the Discussion section. In this experiment, amino acid supplementation was not used, as slight growth occurs on the amino acids alone.
FIG. 7.
Effects of various CN-B12 concentrations on the growth of strain BE182 (ΔpduA) and the wild-type strain (S. enterica). Cells were cultured on 1,2-propanediol minimal medium supplemented with CN-B12 at the concentrations indicated. ▪, wild-type; •, BE182 (ΔpduA). Error bars represent one standard deviation.
DISCUSSION
Previously we showed that S. enterica formed polyhedral organelles during growth on 1,2-propanediol-CN-B12 minimal medium (6). Based on electron microscopy studies and analogy with carboxysomes, we proposed that these organelles consisted of a proteinaceous shell that encased coenzyme B12-dependent diol dehydratase and perhaps other enzymes (6). Here we present genetic and biochemical evidence that PduA is a shell protein of the polyhedral organelles of S. enterica. Immunolabeling experiments showed that the PduA protein localized to the periphery of the organelles (Fig. 3C). Strains having nonpolar pduA mutations were unable to synthesize organelles (Fig. 3D), and expression of the PduA protein from a plasmid vector resulted in aberrantly shaped organelles at higher expression levels (Fig. 4). Thus, three independent lines of evidence indicate that the PduA protein is a component of the shell of the polyhedral organelles of S. enterica.
The results reported here are also consistent with the localization of diol dehydratase within the shell of the organelle. A ΔpduA strain (organelle negative) grew faster than the wild type at limiting B12 concentrations, suggesting that the organelles act as barriers to B12. In strains with pduA mutations, diol dehydratase was no longer associated with the organelles but was found in large inclusion bodies located at the poles of the cell (Fig. 3A and B). Prior immunogold labeling studies showed that diol dehydratase localized primarily to the interior of the organelles (6). Hence, several findings indicate that diol dehydratase localizes within the shell of the organelles. However, the possibility that diol dehydratase is also a component of the organelle shell cannot be excluded at this time.
The shell of the S. enterica polyhedral organelles is likely to be composed of several proteins in addition to the PduA protein. There are four carboxysome shell protein homologues encoded within the pdu operon, PduAJKT, and previous studies showed that polar insertion mutations downstream of pduH caused the formation of aberrant organelles (6). Furthermore, the shells of carboxysomes are known to be composed of multiple proteins (34, 35), and the occurrence of multiple shell gene homologues within an operon has been observed previously in Halothiobacillus (2, 3), Synechococcus (17, 18, 22, 24), and Synechocystis (39).
The function of the Salmonella organelles is currently unknown, but it seems likely that the shells of these organelles act as permeability barriers. Some models propose that carboxysomes concentrate CO2 for RuBisCO, which is encased within the shell of the organelle (15, 25, 34, 35). Similarly, the S. enterica organelles might be used to concentrate 1,2-propanediol (the substrate), or Ado-B12 (the required cofactor) for diol dehydratase. However, the growth studies reported here showed that a pduA mutant unable to form organelles grew similarly to or better than the wild type on lower concentrations of 1,2-propanediol and CN-B12 (a precursor of Ado-B12). These results indicated that the S. enterica organelles are not involved in the concentration of 1,2-propanediol or Ado-B12.
Previously it was suggested that the polyhedral organelles of S. enterica were used to sequester toxic aldehydes (36). S. enterica forms polyhedral organelles during B12-dependent growth on both 1,2-propanediol and ethanolamine, and aldehyde intermediates are a common feature of both degradative pathways (13, 28). The studies reported here support a model in which the polyhedral organelles of S. enterica function to minimize aldehyde toxicity. The finding that ΔpduA mutants undergo a period of interrupted growth at higher 1,2-propanediol concentrations but not at lower 1,2-propanediol concentrations is consistent with formation of a toxic compound derived from 1,2-propanediol. In the first step of 1,2-propanediol degradation, coenzyme B12-dependent diol dehydratase catalyzes the formation of propionaldehyde from 1,2-propanediol. Presumably, higher concentrations of 1,2-propanediol would result in a higher rate of propionaldehyde formation and in greater toxicity. In addition, pduA mutants showed interrupted growth on 1,2-propanediol minimal medium supplemented with higher levels of CN-B12 but not on similar medium supplemented with lower levels of CN-B12. S. enterica converts CN-B12 to Ado-B12, the required cofactor for diol dehydratase. Hence, at higher CN-B12 concentrations, it is expected that propionaldehyde would be formed faster and greater toxicity would result. Following the period of interrupted growth, mutants unable to form organelles resumed growth at a rate similar to that of the wild-type strain. Presumably, growth resumed after the induction of appropriate stress-response genes.
Results also showed that mutant strains unable to produce polyhedral organelles grew substantially faster than the wild-type strain on 1,2-propanediol minimal medium supplemented with lower levels of CN-B12. This result indicated that the S. enterica organelles present a barrier to Ado-B12. Although this would result in slower growth at lower CN-B12 (Ado-B12) concentrations, it could help minimize propionaldehyde toxicity at higher CN-B12 concentrations. Interestingly, this suggests a model in which the polyhedral organelles of S. enterica function to minimize aldehyde toxicity by moderating the rate of propionaldehyde production through the control of Ado-B12 availability.
Acknowledgments
This work was supported by grant GM59486 from the National Institutes of Health and by the Florida Agricultural Experiment Station.
We thank H. C. Aldrich and D. S. Williams for their invaluable assistance with the electron microscopy studies and Tetsuo Toraya for providing the anti-diol dehydratase antibody used in the immunolabeling studies.
Footnotes
Florida Agricultural Experiment Station Journal Series no. R-08598.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baker, S. H., S. C. Lorbach, M. Rodriguez-Buey, D. S. Williams, H. C. Aldrich, and J. M. Shively. 1999. The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in Thiobacillus neapolitanus. Arch. Microbiol. 172:233-239. [DOI] [PubMed] [Google Scholar]
- 3.Baker, S. H., D. S. Williams, H. C. Aldrich, A. C. Gambrell, and J. M. Shively. 2000. Identification and localization of the carboxysome peptide Csos3 and its corresponding gene in Thiobacillus neapolitanus. Arch. Microbiol. 173:278-283. [DOI] [PubMed] [Google Scholar]
- 4.Berkowitz, D., J. M. Hushon, H. J. Whitfield, Jr., J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobik, T. A., M. E. Ailion, and J. R. Roth. 1992. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J. Bacteriol. 174:2253-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conner, C. P., D. M. Heithoff, S. M. Julio, R. L. Sinsheimer, and M. J. Mahan. 1998. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl. Acad. Sci. USA 95:4641-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, R. W., D. Botstein, J. R. Roth, and Cold Spring Harbor Laboratory. 1980. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 10.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Heithoff, D. M., C. P. Conner, U. Hentschel, F. Govantes, P. C. Hanna, and M. J. Mahan. 1999. Coordinate intracellular expression of Salmonella genes induced during infection. J. Bacteriol. 181:799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horswill, A. R., and J. C. Escalante-Semerena. 1997. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J. Bacteriol. 179:928-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeter, R. M. 1990. Cobalamin-dependent 1,2-propanediol utilization by Salmonella typhimurium. J. Gen. Microbiol. 136:887-896. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, C. L., E. Pechonick, S. D. Park, G. D. Havemann, N. A. Leal, and T. A. Bobik. 2001. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J. Bacteriol. 183:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan, A., and L. Reinhold. 1999. CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:539-570. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence, J. G., and J. R. Roth. 1996. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics 142:11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludwig, M., D. Sultemeyer, and G. D. Price. 2000. Isolation of ccmKLMN genes from the marine cyanobacterium, Synechococcus sp. PCC7002 (Cyanophyceae), and evidence that CcmM is essential for carboxysome assembly. J. Phycol. 36:1109-1118. [Google Scholar]
- 18.Marco, E., I. Martinez, M. Ronem Tarazi, M. I. Orus, and A. Kaplan. 1994. Inactivation of ccmO in Synechococcus sp. strain PCC 7942 results in a mutant requiring high levels of CO2. Appl. Environ. Microbiol. 60:1018-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marco, E., and M. I. Orus. 1993. Trichlorfon-induced inhibition of nitrate and ammonium uptake in cyanobacteria. J. Exp. Bot. 44:501-508. [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Obradors, N., J. Badia, L. Baldoma, and J. Aguilar. 1988. Anaerobic metabolism of the L-rhamnose fermentation product 1,2-propanediol in Salmonella typhimurium. J. Bacteriol. 170:2159-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa, T., D. Amichay, and M. Gurevitz. 1994. Isolation and characterization of the ccmM gene required by the cyanobacterium Synechocystis PCC6803 for inorganic carbon utilization. Photosynth. Res. 39:183-190. [DOI] [PubMed] [Google Scholar]
- 23.Price, G. D., and M. R. Badger. 1991. Evidence for the role of carboxysomes in the cyanobacterial CO2-concentrating mechanism. Can. J. Bot. 69:963-973. [Google Scholar]
- 24.Price, G. D., S. M. Howitt, K. Harrison, and M. R. Badger. 1993. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J. Bacteriol. 175:2871-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price, G. D., D. Sultemeyer, B. Klughammer, M. Ludwig, and R. Badger Murray. 1998. The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins, and recent advances. Can. J. Bot. 76:973-1002. [Google Scholar]
- 26.Price-Carter, M., J. Tingey, T. A. Bobik, and J. R. Roth. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rappleye, C. A., and J. R. Roth. 1997. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J. Bacteriol. 179:5827-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roof, D. M., and J. R. Roth. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth, J. R., J. G. Lawrence, and T. A. Bobik. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50:137-181. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Schmieger H. 1971. Method for detection of phage mutants with altered transducing ability. Mol. Gen. Genet. 110:378-381. [DOI] [PubMed] [Google Scholar]
- 32.Shively, J. M., F. L. Ball, and B. W. Kline. 1973. Electron microscopy of the carboxysomes (polyhedral bodies) of Thiobacillus neapolitanus. J. Bacteriol. 116:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shively, J. M., C. E. Bradburne, H. C. Aldrich, T. A. Bobik, J. L. Mehiman, S. Jin, and S. H. Baker. 1998. Sequence homologs of the carboxysomal polypeptide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboxysomes-polyhedral bodies. Can. J. Bot. 76:906-916. [Google Scholar]
- 34.Shively, J. M., and R. S. English. 1991. The carboxysome, a prokaryotic organelle: a mini-review. Can. J. Bot. 69:957-962. [Google Scholar]
- 35.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 36.Stojiljkovic, I., A. J. Baumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium LT2: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toraya, T., S. Honda, and S. Fukui. 1979. Fermentation of 1,2-propanediol with 1,2-ethanediol by some genera of Enterobacteriaceae, involving coenzyme B12-dependent diol dehydratase. J. Bacteriol. 139:39-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 39.Wu Tian, F., D. Liu Yong, and R. Song Li. 2000. Selection and ultrastructural observation of a high-CO2-requiring mutant of cyanobacterium Synechococcus sp. PCC7942. Acta Bot. Sin. 42:116-121. [Google Scholar]