Abstract
A 1H nuclear magnetic resonance (1H NMR) assay was used to study the enzymatic transformation of cis-dienelactone, a central intermediate in the degradation of chloroaromatics. It was shown that the product of the cis-dienelactone hydrolase reaction is maleylacetate, in which there is no evidence for the formation of 3-hydroxymuconate. Under acidic conditions, the product structure was 4-carboxymethyl-4-hydroxybut-2-en-4-olide. Maleylacetate was transformed by maleylacetate reductase into 3-oxoadipate, a reaction competing with spontaneous decarboxylation into cis-acetylacrylate. One-dimensional 1H NMR in 1H2O could thus be shown to be an excellent noninvasive tool for monitoring enzyme activities and assessing the solution structure of substrates and products.
A major route for mineralization of chloroaromatic compounds by microorganisms is their transformation into chlorocatechols and their further metabolism by enzymes of the chlorocatechol pathway (24). In this metabolic pathway, chlorocatechols are subject to intradiol cleavage to form the respective chloromuconates, which are converted by chloromuconate cycloisomerases into cis- or trans-dienelactone. The dienelactones undergo hydrolysis by dienelactone hydrolase. The hydrolysis product formed during the metabolism of 4-chlorocatechol was tentatively identified as “maleylacetate” based on its absorption characteristics (λmax = 243 nm in aqueous alkali) by Evans et al. (11). Similarly, Tiedje et al. (31) postulated maleylacetate and chloromaleylacetate as products formed during the metabolism of 4-chloro- and 3,5-dichlorocatechol, respectively. Those authors noted, however, that UV absorption was essentially quenched upon acidification, a behavior resembling keto-enol tautomerism, raising the question of the actual solution structure of maleylacetate. More recently, Seibert et al. claimed that the enol form (3-hydroxy-2,4-hexadienedioate, 3-hydroxymuconate) is thermodynamically favored under physiological conditions and exhibits an absorption maximum at 243 nm (28). The disappearance of this absorption under acidic conditions was believed to be due to the presence of the keto form, 3-oxo-cis-4-hexenedioate (maleylacetate in the strict sense), under these conditions. Despite the authors' assumption that 3-hydroxymuconate was the actual substrate of the purified reductase, the enzyme was termed “maleylacetate” reductase. Later, Prucha et al. (23) showed that the hydrolysis product of 3-methyldienelactone, supposedly 3-methylmaleylacetate or 3-hydroxy-4-methylmuconate, has a cyclic structure under acidic conditions (4-carboxymethylene-4-hydroxy-3-methylbut-2-en-4-olide, 4-hy-droxy-3-methylmuconolactone), while no indication of the configuration under physiological conditions was given. Thus, although the structure of the decarboxylation product has been published (27), the actual solution structure of “maleylacetate” remained uncertain, presumably due to its reportedly high instability.
“Maleylacetate” is an intermediate not only in the degradation of chlorocatechols via the chlorocatechol ortho-cleavage pathway but also in the degradation of more highly substituted aromatics, such as 2,4,5-trichlorophenoxyacetate (1, 7), 2,4,6-trichlorophenol (16, 20), pentachlorophenol (19, 33), and lindane (18), even though different reaction sequences yielding maleylacetate have been proposed. Moreover, “maleylacetate” is assumed to be an intermediate in the degradation of resorcinolic compounds (5). Thus, “maleylacetate” is a central intermediate in the metabolism of chloroaromatic as well as natural aromatic compounds. Intermediates similar in structure to “maleylacetate” have been found in various other pathways involved in aromatic degradation. 2-Hydroxymuconate (2-hydroxy-2,4-hexadienedioate) was reported to be an intermediate in the degradation of catechols after extradiol cleavage (14, 25). In aqueous solution, this compound is in equilibrium with 2-oxo-4-hexenedioate, a structural isomer of “maleylacetate” (3-oxo-cis-4-hexenedioate), and 4-oxalocrotonate tautomerase catalyzes the otherwise slow transformation of 2-oxo-4-hexenedioate to 2-oxo-3-hexenedioate (32). However, the actual solution structures of such intermediates have been investigated in only a few cases. Thus, knowledge of the identity of the substrates for the following reaction and the actual enzyme mechanism involved is limited. Previously, Guthrie (13) showed that acetopyruvate exists in three major forms in aqueous solution and Pokorny et al. (22) proved that the enol form is the actual substrate for hydrolysis by acetopyruvate hydrolase.
To unequivocally identify the structure of the dienelactone hydrolysis product under physiological conditions, we have characterized its structure by in situ 1H nuclear magnetic resonance (1H NMR) analysis, a method recently shown to be a powerful tool for investigating the solution structures of such intermediates (2).
Dienelactone hydrolase and maleylacetate reductase in Ralstonia eutropha JMP222(pBBR1M-1)
In order to characterize the structure of the dienelactone hydrolysis product under physiological conditions, R. eutropha JMP222(pBBR1M-1) was grown on 3-chlorobenzoate as recently described (21). R. eutropha JMP222 is a derivative of R. eutropha JMP134 (9) cured of plasmid pJP4. pJP4 was shown to contain two chlorocatechol gene clusters, namely, module I (8) and module II (17), which both encode a “maleylacetate” reductase (15, 28). Only module I was present in JMP222(pBBR1M-I). Enzyme activities were estimated in cell extracts by methods described previously (21). R. eutropha JMP222(pBBR1M-I) expresses high levels of dienelactone hydrolase (1,200 U/g of protein with 80 μM cis-dienelactone as the substrate) during growth on 3-chlorobenzoate. In contrast to recent observations (21), we were also able to show the existence of a significant level of maleylacetate reductase activity in cell extracts (up to 700 U/g of protein). We could prove that the respective enzyme was extremely unstable under different conditions; e.g., only a small percentage of activity (usually less than 20%) was recovered when a cell extract was subjected to purification by fast protein liquid chromatography using a MonoQ HR5/5 column with Tris-HCl as the eluent system (data not shown). However, high activities (always higher than 60%) were recovered when phosphate buffer was the eluent. Such a buffer system was necessary for the later 1H NMR analysis to avoid interference of signals originating from any protein fraction added to the in situ transformation NMR assay. A cell extract of strain JMP222(pBBR1M-I) was therefore prepared in phosphate buffer, and a total of 5 to 8 mg of protein was loaded onto a MonoQ HR5/5 column and eluted with a linear gradient of 0 to 0.5 M NaCl in 50 mM phosphate buffer (pH 7.4) at a flow rate of 0.5 ml/min. Fractions of 0.5 ml were collected and analyzed for dienelactone hydrolase activity as described previously (21). Under these conditions, dienelactone hydrolase eluted at a NaCl concentration of 0.03 mM, whereas the bulk of the maleylacetate reductase activity (approximately 60 to 70% of the applied activity) eluted at a NaCl concentration of 0.35 mM.
The transformation of cis-dienelactone (0.08 mM) was analyzed spectrophotometrically as described previously (28) with aliquots of fractions containing dienelactone hydrolase activity, and the formation of a product exhibiting an absorption maximum at 243 nm under physiological conditions was confirmed. This maximum disappeared upon acidification as described previously (28). Thus, the transformation product can be assumed to be identical to that formed by homogeneous maleylacetate reductase (28). Higher concentrations of cis-dienelactone (2 mM) were transformed at rates nearly five times those calculated for substrate concentrations of 0.08 mM, as expected for an enzyme with a low affinity (Km value of approximately 0.14 mM) for cis-dienelactone (26). Complete turnover of cis-dienelactone was confirmed by high-pressure liquid chromatography analysis using a Shimadzu high-pressure liquid chromatography system equipped with a SC125/Lichrospher 5-μm (Bischoff, Leonberg, Germany) column and an aqueous solvent system (flow rate, 1 ml/min) containing 0.1% (vol/vol) H3PO4 (87%), and 20% (vol/vol) methanol.
Solution structure of metabolites formed from cis-dienelactone by dienelactone hydrolase as determined by 1H NMR
In order to characterize the substrate structural changes occurring under the various solution conditions described above, the transformation of cis-dienelactone was monitored by 1H NMR spectroscopy using an NMR sample containing 30 mM sodium phosphate buffer (pH 7.2), 2 mM cis-dienelactone, and 0.14 ml of D2O in a total volume of 0.7 ml. The one-dimensional 1H NMR spectra were recorded at 300 K on a Bruker Avance DMX 600 NMR spectrometer locked to the deuterium resonance of D2O in the solution. Spectra were recorded by using the standard Bruker one-dimensional nuclear Overhauser effect spectroscopy suppression sequence with 280 scans, each with a 1.8-s acquisition time and a 1.3-s relaxation delay. The center of the suppressed water signal was used as an internal reference (δ = 4.80 ppm).
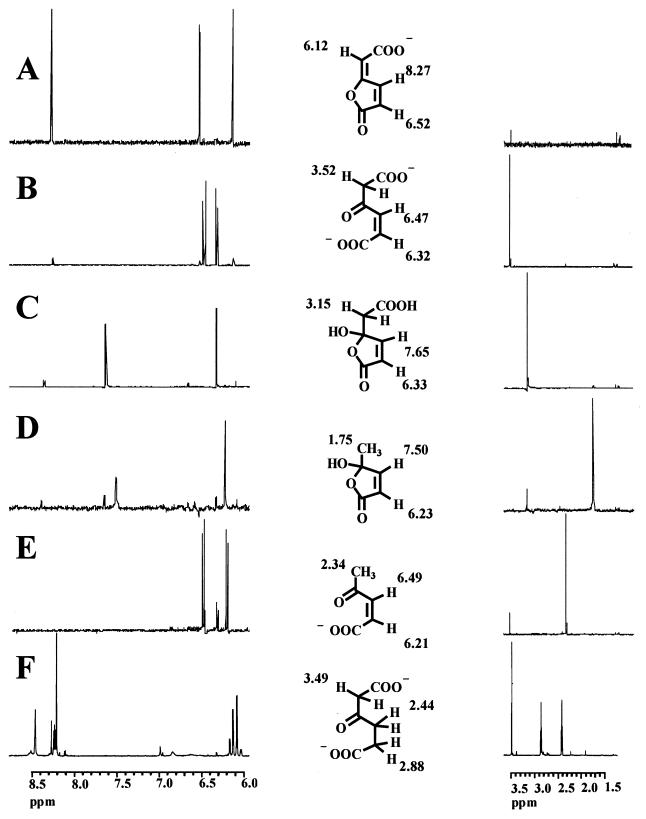
After the in situ 1H spectrum of cis-dienelactone had been recorded (Fig. 1A), 10 μl of dienelactone hydrolase (corresponding to an activity of 30 mU) was added. Figure 1B shows the spectrum after complete transformation of cis-dienelactone. The sample was then adjusted to a pH of 2.5 by the addition of HCl, and a spectrum was recorded under acidic conditions (Fig. 1C). The spectrum of the sample was then recorded at appropriate time intervals to monitor the expected decarboxylation reaction, which should yield cis-acetylacrylate acylal (Fig. 1D), the 1H NMR spectrum of which has already been reported (30). After complete decarboxylation, the sample was neutralized by addition of NaOH to give a final pH of 8, and the structure of the product was analyzed under neutral conditions (Fig. 1E). In all cases, the structures of the various products were evident from the intensity of the 1H signals, their characteristic chemical shifts, and the magnitude of the coupling constants (Fig. 1).
FIG. 1.
1H NMR spectra of cis-dienelactone and transformation products. cis-Dienelactone (2 mM) in 50 mM phosphate buffer (pH 7.4) (A) was incubated for 1 h with partially purified dienelactone hydrolase (43 mU/ml) (B), acidified to pH 2.5 (C), and kept under acidic conditions for 48 h (D), and the resulting solution was neutralized (E). (F) 1H NMR spectrum of cis-dienelactone after 1 h of incubation with partially purified dienelactone hydrolase (29 mU/ml) plus partially purified maleylacetate reductase (71 mU/ml). The proposed solution structures cis-dienelactone (A), 3-oxo-cis-4-hexenedioate (maleylacetate) (B), 4-carboxymethyl-4-hydroxybut-2-en-4-olide (4-hydroxymuconolactone) (C), cis-acetylacrylate acylal (D), cis-acetylacrylate (E), and 3-oxoadipate (F) are indicated.
Only two olefinic protons (δ = 6.32 and 6.47 ppm) were present in the product formed from cis-dienelactone by dienelactone hydrolase. Furthermore, the presence of a methylene function in the product (δ = 3.52 ppm) excluded its identity with 3-hydroxymuconate (Fig. 1B). The vicinal coupling of 12 Hz observed for the olefinic systems is characteristic of their cis configuration in an open-chain system and thus indicates the identity of the product with maleylacetate in the strict sense. Acidification resulted in a downfield shift of the resonance line of the methylene function (δ = 3.15 ppm) and a high-field shift of the resonance lines of one olefinic proton (two olefinic protons resonate at 6.33 and 7.65 ppm). The small coupling constant of 5.8 Hz indicates the presence of the olefinic system in a closed five-membered ring system (4). These data (Fig. 1C) indicate that maleylacetate, under acidic conditions, is 4-carboxymethyl-4-hydroxybut-2-en-4-olide (4-hydroxymuconolactone). There were no indications of the presence of 3-hydroxymuconate under either physiological or acidic conditions. However, a mechanism of dienelactone hydrolase catalysis involving a nucleophilic attack by a cysteine residue has been proposed (6). The enolate anion generated after collapse of the first tetrahedral intermediate is supposed to abstract a proton from a nearby water molecule, leaving a hydroxyl which is appropriately placed for attack of the acyl carbon in the subsequent deacylation step, which should generate 3-hydroxymuconate. Obviously, in contrast to 2-hydroxymuconate (32), 3-hydroxymuconate belongs to the class of fast-reacting enols (10), giving rise in milliseconds to the corresponding keto-derivative, maleylacetate, and thus, the initial dienelactone hydrolysis product could not be observed. Even when cis-dienelactone was transformed in the NMR tube with 200 mU of enzyme, generating a 0.3 mM concentration of product/min, no signals corresponding to the formation of 3-hydroxymuconate were visible. However, the presence of such a fast equilibrium step and the production of 3-hydroxymuconate as the initial hydrolysis product remain to be proven.
Under acidic conditions, a slow decarboxylation was observed with cis-acetylacrylate acylal as the product (Fig. 1D), as reported by Schlömann (27). The small coupling constant of 5.8 Hz between the two olefinic protons resonating at 6.23 and 7.50 ppm indicates their presence in a closed-ring system. Three protons of a methyl group resonate at 1.75 ppm. These values are in accordance with those previously reported for cis-acetylacrylate acylal (30).
At neutral pH (Fig. 1E), the compound is present in the open-ring form, as evidenced by the high coupling constant of 12.3 Hz between the two olefinic protons (6.21 and 6.49 Hz), the downfield shift of the proton neighboring the oxygen-substituted carbon atom compared to the closed form, and the respective upfield shift of the signal of the methyl group (2.34 ppm). These values are again very similar to those reported previously for cis-acetylacrylate (30).
Transformation of maleylacetate by maleylacetate reductase as determined by 1H NMR
Maleylacetate prepared in situ from cis-dienelactone by partially purified dienelactone hydrolase is further transformed by maleylacetate reductase, supposedly yielding 3-oxoadipate by simultaneous oxidation of NADH. This reaction was monitored by NMR analysis as follows. In a first reaction, cis-dienelactone (2 mM) was transformed by partially purified dienelactone hydrolase (corresponding to 20 mU) as described above, and after complete transformation to maleylacetate, the reaction mixture was supplemented with NADH (3 mM) and partially purified maleylacetate reductase (corresponding to 50 mU). In addition to the reaction product, the instability of maleylacetate under neutral conditions (27) led to cis-acetylacrylate through spontaneous decarboxylation of maleylacetate. In a second experiment, cis-dienelactone (2 mM) was incubated with dienelactone hydrolase (corresponding to 20 mU) and an excess of maleylacetate reductase (corresponding to 50 mU) in the presence of NADH (3 mM), thus avoiding the intermediate accumulation of maleylacetate. Besides the appearance of signals from the oxidation of NADH to NAD, the formation of a single reaction product which was identical to the one formed during successive transformation of cis-dienelactone via maleylacetate was clearly visible. Its identity with 3-oxoadipate (Fig. 1F) is evident from the presence of three methylene functions (2.44 and 2.88 ppm [vicinal coupling constant = 7.0 Hz] and 3.49 ppm). These signals were clearly separated from those originating from NAD and residual NADH.
3-Oxo-cis-4-hexenedioate as a stable intermediate in the degradation of chloroaromatics
Monitoring biocatalyzed reactions and metabolic pathways using NMR spectroscopy is of growing interest (2). The method is noninvasive, and analyses can be performed in 1H2O or appropriate buffer systems, which allows one to monitor transformations under physiological conditions. It allows characterization of substrates and products without any manipulation, thus avoiding structural changes during extraction or derivatization procedures. The 1H NMR analysis also allows the identification of solution structures of substrates and intermediates, as shown by Pokorny et al. (22). In the present study, we showed that maleylacetate exists in two stable solution structures, maleylacetate in the strict sense under physiological conditions and its cyclic lactone structure, 4-hydroxymuconolactone, under acidic conditions. The enol form, 3-hydroxymuconate, was not observed in the present study. This is in contrast to previous assumptions (28) of an equilibrium between the enol form (supposed to be the major solution structure under physiological conditions) and maleylacetate (supposed to be the major solution structure under acidic conditions).
The observed absorption maximum at 243 nm is evidently due to maleylacetate. All related keto acids, the solution structures of which have been elucidated, show similar absorption maxima, e.g., 2-oxo-3-hexendioate (λmax = 236 nm [32]), 4,6-dioxo-2-heptenoate (maleylacetone, λmax = 243 nm [29]), and cis-acetylacrylate (λmax = 240 nm [30]), due to their conjugated double-bond system, whereas enol tautomers, such as 2-hydroxymuconate (λmax = 295 nm [32]), 4-hydroxy-6-oxo-2,4-heptadieneoate (λmax = 312 nm [29]), and 4-hydroxy-6-oxo-2-methyl-2-4-heptadienoate (λmax = 325 nm [3]), absorb at significantly higher wavelengths due to extended conjugation.
In contrast to maleylacetate which, depending on the pH, exists as the keto and the lactol forms, different solution structures have been reported for maleylacetone, methylmaleylacetone, and acetopyruvate, including an enol tautomer, which under physiological conditions accounts for approximately 50% of the solution structure mixture (3, 12, 22, 29). The only apparent difference between maleylacetate and (methyl) maleylacetone is the higher acidity of the methylene protons in the latter case, favoring formation of the enol tautomer.
Beside maleylacetate, 2-chloromaleylacetate has been proposed as an intermediate in various chloroaromatic-degradative pathways. However, the identification of 2-chloromaleylacetate as an intermediate in the degradation of chloroaromatic compounds via hydroquinones as intermediates is as yet only tentative and based on mass spectrometric data of presumably 2-chloromaleylacetate or further reaction products (18, 19, 33). Moreover, new unidentified reaction products were recently observed during the enzyme-catalyzed transformation of 3,5-dichlorocatechol via 2-chloromaleylacetate. At least some of these products were assumed to be formed by spontaneous reactions of 2-chloromaleylacetate. We propose that in situ NMR is an appropriate tool for monitoring such reactions and for characterizing products formed by enzyme-catalyzed and spontaneous reactions. Experiments to monitor the metabolic fate of 2-chloromaleylacetate are being performed.
Acknowledgments
We are indebted to W. Reineke and P. Rapp for stimulating discussions. We thank I. Plumeier, B. Jaschok-Kentner, and C. Kakoschke for technical assistance.
The work was supported by EC grant EVK1-CT-1999-00023. P.N. was supported by the DFG-European Graduate College 653. D.H.P. and B.G. acknowledge support from the collaborative grant BMBF-IB/CONICYT-Chile and FONDECYT 8990004.
REFERENCES
- 1.Benvenuti, M., F. Briganti, A. Scozzafava, L. Golovleva, V. M. Travkin, and S. Mangani. 1999. Crystallization and preliminary crystallographic analysis of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E: a novel dioxygenase involved in the biodegradation of polychlorinated aromatic compounds. Acta Crystallogr. D 55:901-903. [DOI] [PubMed] [Google Scholar]
- 2.Brecker, L., and D. W. Ribbons. 2000. Biotransformations monitored in situ by proton nuclear magnetic resonance spectroscopy. Trends Biotechnol. 18:197-202. [DOI] [PubMed] [Google Scholar]
- 3.Cain, R. B., P. Fortnagel, S. Hebenbrock, G. W. Kirby, H. J. S. McLenaghan, G. V. Rao, and S. Schmidt. 1997. Biosynthesis of a cyclic tautomer of (3-methylmaleyl)acetone from 4-hydroxy-3,5-dimethylbenzoate by Pseudomonas sp. HH35 but not by Rhodococcus rhodochrous N75. Biochem. Biophys. Res. Commun. 238:197-201. [DOI] [PubMed] [Google Scholar]
- 4.Chapman, O. L. 1963. Variation of vinyl proton coupling constant with ring-size in cis-cyclic olefins. J. Am. Chem. Soc. 85:2014-2016. [Google Scholar]
- 5.Chapman, P. J., and D. W. Ribbons. 1976. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J. Bacteriol. 125:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheah, E., G. W. Ashley, J. Gary, and D. Ollis. 1993. Catalysis by dienelactone hydrolase: a variation on the protease mechanism. Proteins 16:64-78. [DOI] [PubMed] [Google Scholar]
- 7.Daubaras, D. L., K. Saido, and A. M. Chakrabarty. 1996. Purification of hydroxyquinol 1,2-dioxygenase and maleylacetate reductase: the lower pathway of 2,4,5-trichlorophenoxyacetic acid metabolism by Burkholderia cepacia AC1100. Appl. Environ. Microbiol. 62:4276-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Don, R. H., and J. M. Pemberton. 1985. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pJP4. J. Bacteriol. 161:466-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duhaime, R. M., and A. C. Weedon. 1987. Direct measurement of the rates of reketonization of dienolates produced by photochemical enolization of β-alkyl α,β-unsaturated ketones in aqueous basic solution. J. Am. Chem. Soc. 109:2479-2483. [Google Scholar]
- 11.Evans, W. C., B. S. W. Smith, P. Moss, and H. N. Fernley. 1971. Bacterial metabolism of 4-chlorophenoxyacetate. Biochem. J. 122:509-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fowler, J., and S. Seltzer. 1970. The synthesis of model compounds for maleylacetoacetic acid. Maleylacetone. J. Org. Chem. 35:3529-3532. [Google Scholar]
- 13.Guthrie, J. 1972. Acetopyruvic acid: rate and equilibrium constants for hydration and enolization. J. Am. Chem. Soc. 94:7020-7024. [Google Scholar]
- 14.Harayama, S., M. Rekik, K.-L. Ngai, and L. N. Ornston. 1989. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J. Bacteriol. 171:6251-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasberg, T., D. L. Daubaras, A. M. Chakrabarty, D. Kinzelt, and W. Reineke. 1995. Evidence that operons tcb, tfd, and clc encode maleylacetate reductase, the fourth enzyme of the modified ortho pathway. J. Bacteriol. 177:3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latus, M., H.-G. Seitz, J. Eberspächer, and F. Lingens. 1995. Purification and characterization of hydroquinol 1,2-dioxygenase from Azotobacter sp. strain GP1. Appl. Environ. Microbiol. 61:2453-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leveau, J. H. J., F. Konig, H. Fuchslin, C. Werlen, and, J. R. van der Meer. 1999. Dynamics of multigene expression during catabolic adaptation of Ralstonia eutropha JMP134 (pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Mol. Microbiol. 33:396-406. [DOI] [PubMed] [Google Scholar]
- 18.Miyauchi, K., Y. Adachi, Y. Nagata, and M. Takagi. 1999. Cloning and sequencing of a novel meta-cleavage dioxygenase gene whose product is involved in degradation of gamma-hexachlorocyclohexane in Sphingomonas paucimobilis. J. Bacteriol. 181:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohtsubo, Y., K. Miyauchi, K. Kanda, T. Hatta, H. Kiyohara, T. Senda, Y. Nagata, Y. Mitsui, and M. Takagi. 1999. PcpA, which is involved in the degradation of pentachlorophenol in Sphingomonas chlorophenolica ATCC39723, is a novel type of ring-cleavage dioxygenase. FEBS Lett. 459:395-398. [DOI] [PubMed] [Google Scholar]
- 20.Padilla, L., V. Matus, P. Zenteno, and B. Gonzalez. 2000. Degradation of 2,4,6-trichlorophenol via chlorohydroxyquinol in Ralstonia eutropha JMP134 and JMP222. J. Basic Microbiol. 40:243-249. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Pantoja, D., L. Guzman, M. Manzano, D. H. Pieper, and B. Gonzalez. 2000. Role of tfdCIDIEIFI and tfdDIICIIEIIFII gene modules in catabolism of 3-chlorobenzoate by Ralstonia eutropha JMP134(pJP4). Appl. Environ. Microbiol. 66:1602-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pokorny, D., L. Brecker, M. Pogorevc, W. Steiner, H. Griengl, T. Kappe, and D. W. Ribbons. 1999. Proton-nuclear magnetic resonance analyses of the substrate specificity of a β-ketolase from Pseudomonas putida, acetopyruvate hydrolase. J. Bacteriol. 181:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prucha, M., V. Wray, and D. H. Pieper. 1996. Metabolism of 5-chlorosubstituted muconolactones. Eur. J. Biochem. 237:357-366. [DOI] [PubMed] [Google Scholar]
- 24.Reineke, W. 1998. Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu. Rev. Microbiol. 52:287-331. [DOI] [PubMed] [Google Scholar]
- 25.Sala-Trepat, J. M., and D. C. Evans. 1971. The meta-cleavage of catechol by Azotobacter species: 4-oxalocrotonate pathway. Eur. J. Biochem. 20:400-413. [DOI] [PubMed] [Google Scholar]
- 26.Schlömann, M. 1994. Evolution of chlorocatechol catabolic pathways. Biodegradation 5:301-321. [DOI] [PubMed] [Google Scholar]
- 27.Schlömann, M., P. Fischer, E. Schmidt, and H.-J. Knackmuss. 1990. Enzymatic formation, stability, and spontaneous reactions of 4-fluoromuconolactone, a metabolite of the bacterial degradation of 4-fluorobenzoate. J. Bacteriol. 172:5119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seibert, V., K. Stadler-Fritzsche, and M. Schlömann. 1993. Purification and characterization of maleylacetate reductase from Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 175:6745-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seltzer, S. 1981. Nuclear magnetic resonance, paramagnetic ion induced relaxation method to differentiate between 1,3-diketo and 1,3-keto-enol isomers. J. Org. Chem. 46:2643-2650. [Google Scholar]
- 30.Seltzer, S., and K. D. Stevens. 1967. The preparation of cis-β-acetylacrylic acid. J. Org. Chem. 33:2708-2711. [Google Scholar]
- 31.Tiedje, J. M., J. M. Duxbury, M. Alexander, and J. E. Dawson. 1969. 2,4-D metabolism: pathway of degradation of chlorocatechols by Arthrobacter sp. J. Agric. Food Chem. 17:1021-1026. [DOI] [PubMed] [Google Scholar]
- 32.Whitman, C. P., B. A. Aird, W. R. Gillespie, and N. J. Stolowich. 1991. Chemical and enzymatic ketonization of 2-hydroxymuconate, a conjugated enol. J. Am. Chem. Soc. 113:3154-3162. [Google Scholar]
- 33.Xu, L., K. Resing, S. L. Lawson, P. C. Babbitt, and S. D. Copley. 1999. Evidence that pcpA encodes 2,6-dichlorohydroquinone dioxygenase, the ring cleavage enzyme required for pentachlorophenol degradation in Sphingomonas chlorophenolica strain ATCC 39723. Biochemistry 38:7659-7669. [DOI] [PubMed] [Google Scholar]