Abstract
The bacterium Vibrio natriegens can double with a generation time of less than 10 min (R. G. Eagon, J. Bacteriol. 83:736-737, 1962), a growth rate that requires an extremely high rate of protein synthesis. We show here that V. natriegens' high potential for protein synthesis results from an increase in ribosome numbers with increasing growth rate, as has been found for other bacteria. We show that V. natriegens contains a large number of rRNA operons, and its rRNA promoters are extremely strong. The V. natriegens rRNA core promoters are at least as active in vitro as Escherichia coli rRNA core promoters with either E. coli RNA polymerase (RNAP) or V. natriegens RNAP, and they are activated by UP elements, as in E. coli. In addition, the E. coli transcription factor Fis activated V. natriegens rrn P1 promoters in vitro. We conclude that the high capacity for ribosome synthesis in V. natriegens results from a high capacity for rRNA transcription, and the high capacity for rRNA transcription results, at least in part, from the same factors that contribute most to high rates of rRNA transcription in E. coli, i.e., high gene dose and strong activation by UP elements and Fis.
Generation times depend not only on the genetic constitution of a bacterial species but also on the environment in which that species is growing. In the laboratory, changing the nutritional environment can vary the growth rate of a culture. Since faster-growing cells make more protein per unit time than slower-growing cells, it has long been appreciated that faster-growing cells either would have to contain ribosomes that translate faster or would need an increased number of ribosomes (40).
Some bacteria, including the pathogens Vibrio parahaemolyticus, an important causative agent of food poisoning, and Clostridium perfringens, which causes food poisoning and gas gangrene, can grow with a generation time as short as 10 min (10, 32, 34, 43, 52, 55). We reasoned that an understanding of the mechanisms responsible for the high protein synthesis rate in extremely fast-growing bacteria might be relevant to an understanding of their virulence. As an example of a fast-growing bacterium, we chose to study Vibrio natriegens, a nonpathogenic, gram-negative species isolated from salt mud marshes that has been reported to have a maximal generation time of about 10 min (15).
In the cases examined to date, cells regulate ribosome synthesis rather than ribosome activity. Since making ribosomes is an energy-intensive process requiring the synthesis of 16S, 23S, and 5S rRNAs as well as over 50 ribosomal proteins (25), cells devote energy to ribosome synthesis only when needed. Thus, studies of enteric bacteria, including Escherichia coli and Salmonella enterica serovar Typhimurium, have shown that the ribosome synthesis rate increases proportionally to the square of the growth rate (40). rRNA transcription is also proportional to the square of the growth rate and is the rate-limiting step in ribosome synthesis (42).
The regulation of rRNA transcription has been characterized best in E. coli, where there are seven rRNA operons (36), each with two promoters, P1 and P2 (12, 26), recognized by the RNA polymerase holoenzyme containing σ70 (Eσ70 RNAP; subunit composition, α2ββ"σ70ω). These promoters contain three RNAP recognition elements, two (referred to as the −10 and −35 hexamers) that interact with σ70 and one (referred to as the UP element), positioned upstream of the −35 hexamer, that interacts with the two α subunits (48). Consensus sequences have been derived for each of the recognition elements: 5"-TATAAT-3" and 5"-TTGACA-3" (28) for the −10 and −35 hexamers, respectively, and 5"-AAA(A/T)(A/T)T(A/T)TTTTAAAAAA-3" for the UP element (−57 to −41 with respect to the transcription start site) (16, 17). The −10 and −35 hexamers in rRNA promoters are separated by 16 bp, 1 bp less than the 17-bp consensus. UP elements stimulate transcription 20- to 50-fold from each of the 7 rrn P1 promoters in vivo (31, 46, 48). In addition, the transcription factor Fis stimulates transcription another three- to eightfold by binding to sites upstream of the UP element at all 7 rrn P1 promoters (31, 49). Thus, the unusual strength of rrn P1 promoters in E. coli is attributable primarily to the presence of UP elements and Fis.
V. natriegens has been reported to have the remarkable doubling time of 9.8 min (15). Here, we report that, as is the case for the better-studied but slower-growing E. coli, V. natriegens increases its number of ribosomes with the growth rate in order to achieve its extraordinarily high rate of protein synthesis. We show that multiple mechanisms contribute to this high ribosome synthesis efficiency, including high rRNA gene copy number; strong promoters that contain near-consensus −10, −35, and UP elements; and activation by the transcription factor Fis. In addition, V. natriegens rRNA promoters exhibit the relatively short-lived open-complex characteristic of rRNA promoters in E. coli, potentially contributing to the regulation of these promoters in vivo.
MATERIALS AND METHODS
Strains, plasmids, and cloning of V. natriegens rrn promoter regions.
The strains and plasmids used in this study are listed in Table 1. All DNA sequences are presented 5" to 3" for the nontemplate strand.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype | Source |
|---|---|---|
| Strains | ||
| RLG3654 | V. natriegens ATCC 14048 | 15 |
| RLG3499 | E. coli VH1000 = MG1655 lacZ lacI pyrE+ | 19 |
| λ System I lysogens | ||
| RLG 5104 | VH1000 λ V. natriegens rrnA P1-P2 (−248 to +8)-lacZ | This work |
| RLG 5105 | VH1000 λ V. natriegens rrnA P1-P2 (−248 to +1)-lacZ | This work |
| RLG 5106 | VH1000 λ V. natriegens rrnA P1-P2 (−148 to +8)-lacZ | This work |
| RLG 3871 | VH1000 λ E. coli rrnB P1-P2 (−600 to +2)-lacZ | This work |
| Plasmids | ||
| pRLG3748 | PRLG770 with −35 con promoter (−54 to +16) | 22 |
| pRLG5100 | M13mp19 with V. natriegens rrnA P1-P2 from −460 to start of 16S rRNA | This work |
| pRLG5101 | pRLG770 with V. natriegens rrnA P1-P2 (−248 to +8) | This work |
| pRLG5102 | pRLG770 with V. natriegens rrnA P1 (−248 to +1) | This work |
| pRLG5103 | pRLG770 with V. natriegens rrnA P2 (−148 to +8) | This work |
| pRLG6096 | pRLG770 with V. natriegens rrnA P2 (−41 to +8) | This work |
| pRLG6097 | pRLG770 with V. natriegens rrnA P2 (−66 to +8) | This work |
| pRLG6098 | pRLG770 with V. natriegens rrnA P1 (−41 to +1) | This work |
| pRLG6099 | pRLG770 with V. natriegens rrnA P1 (−66 to +1) | This work |
| pRLG5111 | M13mp19 with V. natriegens rrnB P1-P2 from −400 to start of 16S rRNA | This work |
| pRLG5117 | pRLG770 with V. natriegens rrnB P1-P2 | This work |
| pRLG5114 | pRLG770 with V. natriegens rrnC P2 promoter | This work |
| pRLG5112 | M13mp19 with V. natriegens rrnD P2 from −103 to start of 16S rRNA | This work |
| pRLG5113 | M13mp19 with V. natriegens rrnE P2 from −100 to start of 16S rRNA | This work |
| pRLG3858 | pRLG770 with E. coli rrnB P1-P2 (−152 to +7) | This work |
| pRLG862 | pRLG770 with E. coli rrnB P1 (−88 to +1) | 49 |
| pRLG5944 | pRLG770 with E. coli rrnB P1 (−61 to +1) | This work |
| pWR55 | pRLG770 with E. coli rrnB P1 (−41 to +1) | 48 |
Because the DNA sequences upstream of the V. natriegens 16S rRNA genes were not known, we used the Vectorette II system (Genosys, The Woodlands, Tex.) to obtain the rRNA regulatory regions from several operons. Briefly, V. natriegens chromosomal DNA was isolated; digested with either EcoRI, HindIII, ClaI, or BamHI; and ligated to Vectorette II DNA. Vectorette II DNA consists of partially double-stranded DNA fragments of known sequence with ends compatible with the fragments generated by digestion of the chromosomal DNA. PCR from the Vectorette DNA and a highly conserved sequence within the 16S rRNA genes, using the primer RLG3121 (5"-GGCCTGAATTCCCAGCGTTCAATCTGAGCCATGATC-3"), which hybridizes to the DNA region corresponding to the 5" end of the 16S rRNA, and the Vectorette II primer (5"-GCAGGAGAACCCCATGAAGCTTGAATTCGGATCCACGTTG-3"), which hybridizes to the complement of the single-stranded region of the Vectorette DNA ligated to the chromosomal DNA fragments, allowed amplification of the promoter regions. The PCR products were cloned into M13mp19 for sequencing (using the RLG3121 primer [see above]) and designated rrnA to rrnE, and promoter fragments with the endpoints listed in Table 1 were subcloned into pRLG770 (49) for in vitro transcription assays.
For numbering purposes in the V. natriegens sequences, in operons where the start sites have not been determined, the P1 start site was defined as the G 9 bp downstream from the −10 hexamer and the P2 start site was defined as the C 7 bp downstream from the −10 hexamer.
Determination of RNA/protein ratios.
V. natriegens ATCC 14048 and E. coli VH1000 were grown at 37°C in different media: brain heart infusion (Difco), Luria broth (LB), or M9 minimal medium (41) supplemented with 0.4% glucose or glycerol and 10 μg of thiamine/ml. V. natriegens cultures were supplemented with 1.5% Instant Ocean sea salts (Aquarium Systems, Inc.) (44, 56). The cultures were inoculated to an A600 of approximately 0.01, grown for ≈5 generations, and then cooled on ice. Aliquots (20 ml) were pelleted by centrifugation and resuspended in 20 ml of Tris-Cl (pH 7.9)-4 mM EDTA, the A600 was measured, 2-ml aliquots of each culture were sonicated, and the sonicated cells were then used to determine the RNA/protein ratio in duplicate using the orcinol (51) and Bradford reagents (Bio-Rad), respectively, as described previously (20).
Southern blotting.
Chromosomal DNA from V. natriegens and E. coli was isolated as described previously (57). DNA (0.5 μg) was digested with 20 U of restriction enzymes (NE Biolabs) (see Fig. 2), electrophoresed on 0.8% agarose gels in 1× Tris-borate-EDTA buffer at 30 V for 16 h, and blotted to GeneScreen Plus membranes (NEN Research Products) using the protocol provided by the manufacturer. Either of two 32P-labeled oligonucleotides hybridizing to the nontemplate strand of very highly conserved regions of the 16S rRNA gene was used as a probe: RLG3133 (5"-CCAGCGTTCAATCTGAGCC-3") hybridizes to the 5" end of the 16S rRNA gene, and probe RLG1492r (5"-GGYTACCTTGTTACGACTT-3") (38) hybridizes to the 3" end of the 16S rRNA gene.
FIG. 2.
Southern hybridization analysis of rrn operons from V. natriegens. Lane 1, 32P-labeled λ DNA cleaved with HindIII (control). Lane 2, E. coli chromosomal DNA cleaved with PstI plus BglII (control). Lanes 3 to 8, V. natriegens DNA cleaved with the indicated restriction enzymes. Probe RLG3133 was used for lanes 2 to 7 and is complementary to the beginning of the 16S rRNA gene, and probe RLG1492r was used for lane 8 and is complementary to the 16S rRNA gene (38).
β-Galactosidase determination.
E. coli strains with λ prophages containing promoter-lacZ fusions were grown on LB agar at 30°C, and then fresh colonies were inoculated into LB medium to an A600 of approximately 0.02. The cultures were grown for 4 generations at 30°C to an A600 of 0.4, harvested, and lysed, and β-galactosidase activity was measured (41).
Purification of V. natriegens RNAP.
V. natriegens was grown at 30°C in 1 liter of brain heart infusion medium supplemented with 1.5% sea salts. Cells were harvested and stored overnight at −20°C; 3 g of cells was thawed, resuspended in 10 ml of lysis buffer (50 mM HEPES [pH 7.9], 2 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, and 5% glycerol), and sonicated with five bursts of 20 s each to lyse the cells and shear the DNA; and the sonicate was centrifuged at 44,000 × g to obtain a clear but viscous lysate. Crushed ammonium sulfate (2.26 g; 40% final concentration) was added to 10 ml of cleared lysate, stirred for 1 h, and centrifuged at 8,000 × g for 50 min. The pellet was resuspended in a solution of 10 ml of 10 mM Tris (pH 7.9), 5% glycerol, 0.1 mM EDTA, 1 mM DTT, and 100 mM NaOAc and then centrifuged at 21,000 × g for 15 min. The 10-ml solution (10 mg/ml) was filtered, and 1 ml was loaded on a 75-ml Superdex 200 (Pharmacia) size exclusion column and washed with a solution of 160 ml of 10 mM Tris (pH 7.9), 5% glycerol, 0.1 mM EDTA, 0.1 mM DTT, and 100 mM NaOAc. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and those containing RNAP were pooled and loaded on a MonoQ column (Pharmacia) equilibrated in buffer A with 100 mM NaCl (buffer A is 10 mM Tris [pH 7.9], 5% glycerol, 0.1 mM EDTA, and 0.1 mM DTT), and RNAP was eluted at 1 ml/min using a 100 mM-to-1 M NaCl gradient in buffer A. One-milliliter fractions were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis; V. natriegens RNAP eluted in fractions 22 to 24. RNAP was further purified and concentrated using Microcon-100 filters (Millipore), and the concentrated RNAP was dialyzed against storage buffer containing 10 mM Tris (pH 7.9), 50% glycerol, 0.1 mM EDTA, 0.1 mM DTT, and 100 mM NaOAc. The protein concentration was determined as described above. The RNAP preparation had a specific activity ≈1/12 of that of our standard E. coli preparation, derived in part from a higher level of contamination with other proteins and in part from a higher proportion of inactive RNAP in the preparation. The concentrations of V. natriegens RNAP used for in vitro transcription were adjusted accordingly to compensate for the lower specific activity.
Primer extension from RNA produced in vitro.
Transcripts for primer extension mapping of start sites were synthesized in 120-μl reaction mixtures containing 0.2 nM supercoiled rrnA P1-P2 template (pRLG5104); 10 mM HEPES; 100 mM NaOAc; 10 mM MgCl2; 0.1 mM EDTA (pH 8.0); 1 mM DTT; 100 μM (each) GTP, CTP, ATP, and UTP; and 24 nM V. natriegens RNAP. The reaction was incubated for 15 min at 22°C and then extracted with phenol-choloroform and precipitated with 2 volumes of ethanol and 20 μg of glycogen. The precipitated RNA was resuspended in 12 μl of 10 mM Tris-Cl (pH 7.9).
RLG1620 primer (5"-GCGCTACGGCGTTTCACTTC-3") was phosphorylated using polynucleotide kinase and [γ-32P]ATP. One microliter (≈1.0 pmol) of primer (≈105 cpm) was mixed with 12 μl (≈0.1 pmol) of RNA template and 4 μl of 5× Moloney murine leukemia virus buffer (Promega, Madison, Wis.). The 1620 primer hybridizes to the pRLG770 vector template strand ≈40 bp downstream of the site of promoter insertion. The primer was extended with 1 μl (200 U) of Moloney murine leukemia virus reverse transcriptase and 2.0 μl of 5 mM deoxynucleoside triphosphates, and the reaction was incubated at 45°C for 30 min and stopped by addition of 0.3 volume of NaOAc, 5 μg of glycogen, and 2 volumes of 100% ethanol.
Lifetime of RNAP-promoter complexes.
Open-complex half-lives were measured as described previously (5, 7) with minor modifications. Supercoiled templates (specified in the figure legends) containing either V. natriegens rrnA P1-P2 or E. coli rrnB P1-P2 were incubated with 24 nM V. natriegens RNAP or 2 nM E. coli RNAP, respectively, for 15 min at 30°C in 40 mM Tris-Cl (pH 7.9), 10 mM MgCl2, 60 mM NaCl, 1 mM DTT, and 0.1 μg of bovine serum albumin/μl. Aliquots (10 μl) were withdrawn into tubes containing 1.5 μl of nucleoside triphosphates (NTPs; final concentrations, 100 μM GTP, 100 μM CTP, 100 μM ATP, and 10 μM [α-32P]UTP [5 μCi]) at different times after the addition of heparin (final concentration, 10 μg/ml). Transcription reactions were stopped after 15 min and analyzed on 5% acrylamide-8 M urea gels. The fractions of open complexes remaining at different times were plotted as described previously (5, 7). Open-complex half-life is not affected by the specific activity of the RNAP preparation, since the rate of open-complex decay is not RNAP concentration dependent.
In vitro transcription.
Multiple-round transcription was started by the addition of 24 nM V. natriegens RNAP (or 2 nM E. coli RNAP) to 0.2 nM supercoiled plasmid templates (specified in the figure legends). Transcription assays to measure nucleotide dependence of the V. natriegens rrnA P1 promoter (pRLG5104) were performed at 22°C in 40 mM Tris-Cl (pH 7.9), 130 mM KCl, 10 mM MgCl2, 1 mM DTT, 100 μg of bovine serum albumin/ml, 100 μM CTP, 100 μM ATP, 10 μM UTP, and 5 μCi of [α-32P]UTP (NEN), and 15 to 2,000 μM GTP. Fis-dependent transcription was measured in the same buffer but containing 100 mM NaCl instead of 130 mM KCl and 100 μM GTP, CTP, and ATP and 10 μM UTP instead of the concentrations given above. Transcription was stopped with an equal volume of formamide loading buffer 15 min after the addition of RNAP. Samples were electrophoresed on 5% polyacrylamide-8 M urea gels, and the dried gels were visualized and quantified by phosphorimaging (ImageQuant Software; Molecular Dynamics).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences determined in this study are as follows: AF417938 for rrnA (119 bp upstream of the P1 start site to 191 bp downstream of the P2 start site), AF417939 for rrnB (119 bp upstream of the P1 start site to 192 bp downstream of the P2 start site), AF417940 for rrnC (44 bp upstream of the P1 start site to 173 bp downstream of the P2 start site), AF417941 for rrnD (101 bp upstream of the P2 start site to 147 bp downstream of the P2 start site), and AF417942 for rrnE (56 bp upstream of the P2 start site to 168 bp downstream of the P2 start site).
RESULTS AND DISCUSSION
RNA/protein ratios in V. natriegens increase with growth rate.
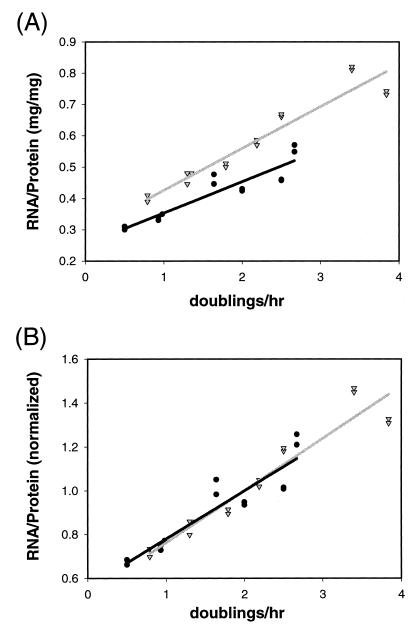
We determined the RNA (which is predominantly rRNA) and protein concentrations from both V. natriegens and E. coli at 37°C at different growth rates (Fig. 1). V. natriegens doubled in as little as ≈15 min (μ (doublings/h) = 4), while E. coli doubled in as little as ≈26 min (μ = 2.3 generations per h). In both species, the RNA/protein ratios increased proportionally with the growth rate (Fig. 1A). The V. natriegens RNA/protein ratio was slightly higher than that for E. coli at the same growth rate, but when the RNA/protein ratios were normalized to give the same value at a common growth rate (μ = 2.0 generations per h) to facilitate visual comparison of the slopes (Fig. 1B), the slopes of the two curves were identical within error.
FIG. 1.
RNA/protein ratios as a function of growth rate in V. natriegens ATCC 14048 and E. coli VH1000. (A) RNA/protein ratios from cultures grown in media supporting different growth rates at 37°C (see Materials and Methods). (B) Curves were normalized to the same value at a growth rate of 2.0 doublings/h as described previously (14). Triangles, V. natriegens; circles, E. coli.
It has been estimated that E. coli produces approximately 70,000 ribosomes/cell when doubling with a generation time of 25 min (40), which would extrapolate to about 90,000 ribosomes/cell if E. coli were able to grow at ≈4 doublings/h like V. natriegens. Figure 1 suggests that V. natriegens produces about 115,000 ribosomes/cell at ≈4 doublings/h.
Since RNA/protein ratios are higher in V. natriegens than in E. coli for a given growth rate (i.e., the overall protein synthesis rate), V. natriegens produces less protein per unit of rRNA than E. coli. Thus, it takes more ribosomes to make the same amount of protein in this bacterium than in E. coli. These results indicate that V. natriegens does not meet its need for a rate of protein synthesis higher than that of E. coli by making ribosomes that translate faster. Rather, V. natriegens increases the ribosome number to a level even higher than that of E. coli to provide the high rate of protein synthesis required for short generation times. In the remainder of this report, we investigate potential mechanisms to account for the extremely high rate of rRNA transcription required to achieve such a high ribosome synthesis rate.
V. natriegens contains about 13 rrn operons.
rRNA transcription in V. natriegens could be increased by having an increased rRNA gene dose. Southern blot analysis was carried out to determine the number of rRNA operons in V. natriegens. Genomic DNA was cleaved with a variety of restriction endonucleases, separated by gel electrophoresis, blotted, and hybridized to 32P-labeled probes. Similar results were obtained with two different 19-mer probes, one complementary to the highly conserved 5" end of the 16S rRNA gene and the other complementary to the highly conserved 3" end of the 16S rRNA gene (see Materials and Methods).
Bands of different lengths depend primarily on restriction site heterogeneity outside of the rRNA operons themselves, since rRNA sequences in different operons within an organism are typically highly conserved. Blots were quantified by phosphorimaging. Some bands contained two- or threefold more radioactivity than other bands of similar size, indicating that hybridizing fragments from two or three operons probably migrated to similar positions in the gel.
Some digests contained as many as 10 hybridizing bands of different sizes (e.g., Fig. 2, lanes 7 and 8). When multiple bands migrating to the same gel position were taken into account in these and other digests, the analysis suggested that there are at least 13 rRNA operons in V. natriegens. This should be considered a minimal estimate, since some restriction digests could have resulted in fragments that were too small to be resolved on the gel or retained on the nitrocellulose membrane.
Different Vibrio species have been reported to contain different numbers of rRNA operons, including 8 for Vibrio cholerae (29) and at least 9 for V. parahaemolyticus (4), while another fast-growing bacterium, C. perfringens, was reported to have 10 rRNA operons (23). The rRNA operon number correlates with the ability to respond to upshifts (11, 37, 50, 53) and generally (but not perfectly) with the maximal growth rate (11, 37, 50, 53). Although inactivation of a single E. coli rRNA operon does not decrease the growth rate significantly, inactivation of several operons does result in slower growth (3). It has also been reported that an excess number of rRNA operons can be a burden to cells growing at low growth rates (53). On balance, it seems likely that the high number of rRNA genes in V. natriegens contributes to the potential for a high rRNA synthesis rate.
Nucleotide sequences of V. natriegens rRNA promoters.
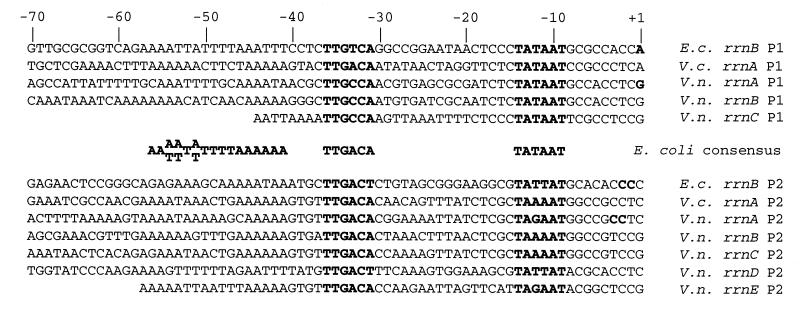
Since rRNA synthesis is limited primarily by the rate of transcription initiation, we identified the DNA sequences from regulatory regions preceding several of the V. natriegens rRNA operons. Since the V. natriegens genome sequence has not been determined, rRNA promoter regions were cloned using an asymmetric PCR strategy (Vectorette [described in Materials and Methods]). Intact or partial sequences for the regulatory regions from five operons (including three rrn P1 and five rrn P2 promoters) were obtained (arbitrarily designated rrnA to rrnE [Fig. 3]; see Materials and Methods for sequence endpoints and GenBank accession numbers). The V. cholerae genome sequence has recently been reported and contains eight rRNA operons (29). The sequences of the promoter region from one V. cholerae rRNA operon and from the E. coli rrnB operon are provided for comparison.
FIG. 3.
DNA sequences from the core promoters and A+T-rich upstream sequences of V. natriegens (V.n.) rrn P1 and P2 promoters. The −35 and −10 hexamers for P1 and P2 are shown in boldface. E. coli (E.c.) rrnB and V. cholerae (V.c.) rrnA sequences are shown for comparison. The E. coli consensus sequences for the −35 and −10 hexamer (28), as well as for the UP element (16, 17), are also indicated. See Materials and Methods for GenBank accession numbers. Start sites (in boldface) have been determined experimentally only for the seven E. coli rrn promoters and for the V. natriegens rrnA promoter.
Sequences corresponding to the −10 and −35 RNAP recognition hexamers were easily identified by inspection, despite the fact that the distances between the rrn P1 and rrn P2 promoters were different from those in E. coli and varied in the different V. natriegens rRNA operons (see below). As in the E. coli rrnB P1 promoter, each of the three cloned V. natriegens rrn P1 promoters contained a perfect E. coli −10 consensus, a −35 hexamer differing at only one position from the E. coli consensus, and 16-bp spacers between the two recognition hexamers. The −10 and −35 hexamers in the V. natriegens rrn P2 promoters either matched or differed at one position from the E. coli consensus (28), and the spacing between the two hexamers was again 16 bp. Both rrn P1 and P2 promoters contained A+T-rich sequences upstream of −40 that likely function as UP elements (see below). In each operon, the sequences between the −10 hexamer and the transcription start site are G+C rich in both promoters, as in E. coli. In E. coli, this sequence is known as the “discriminator” region, where mutations alter regulation of transcription initiation (6, 33, 35).
The V. cholerae rrn P1 and P2 promoters, like those from E. coli and V. natriegens, contain near-consensus −35 and −10 hexamers separated by 16 bp and are preceded by UP element-like A+T-rich elements (1, 27, 31, 46, 48). Interestingly, while the distance separating the P1 and P2 promoters in E. coli varies in different operons, from 105 bp at rrnD, rrnE, and rrnH to 120 bp at rrnB, in V. natriegens the distance between the two promoters varied from ≈115 bp to as much as 148 bp, while in V. cholerae the P1-P2 spacer varied from ≈70 to 100 bp.
Relative strengths of V. natriegens and E. coli rRNA promoters in E. coli.
Since no genetic system has been developed for introducing cloned promoters into V. natriegens and measuring their activities in vivo, we constructed lacZ fusions to the V. natriegens rrnA rRNA promoters on bacteriophage λ and measured their activities in E. coli strains lysogenic for these λ phages. Even in this heterologous system, the V. natriegens promoters were extremely strong (Table 2); the V. natriegens promoter-lacZ fusion containing both the rrnA P1 and P2 promoters (V. natriegens sequences from 248 bp upstream of the P1 start site to 8 bp downstream from the P2 start site) produced more β-galactosidase than a construct containing the E. coli rrnB P1 and P2 promoters (−600 to +2). The V. natriegens rrnA P1 promoter was about twofold stronger than the V. natriegens rrnA P2 promoter. These experiments indicate that the V. natriegens rRNA promoters are extremely active in E. coli, indicative of their likely high activities in V. natriegens. Furthermore, as is the case for the E. coli rrn P1 and P2 promoters, it appears that V. natriegens rrnA P1 is stronger than P2 in rich medium (at least in E. coli).
TABLE 2.
Comparison of V. natriegens rrnA and E. coli promoter activities in E. coli
| Strain | Promotera | Activityb |
|---|---|---|
| RLG 5104 | V. natriegens P1-P2 (−248 to +8) | 6,840 ± 55 |
| RLG 5105 | V. natriegens P1 (−248 to +1) | 5,893 ± 36 |
| RLG 5106 | V. natriegens P2 (−148 to +8) | 2,915 ± 22 |
| RLG 4295 | E. coli P1-P2 (−600 to +2) | 5,890 ± 49 |
Numbers in parentheses refer to the sequence endpoints of the promoter fragments used for constructing lacZ fusions. The V. natriegens and E. coli P1-P2 fusions contain 248 or 600 bp upstream of the P1 start site, respectively, and 8 or 2 bp downstream of the P2 start site, respectively. V. natriegens P1 contains 148 bp upstream of the P1 transcription start site. V. natriegens P2 contains 148 bp upstream and 8 bp downstream of the P2 transcription start site.
Activities were measured in LB and are given in Miller units of β-galactosidase activity. The values are the averages of three experiments carried out in duplicate.
Mapping of the rrnA P1 and P2 start sites in vitro.
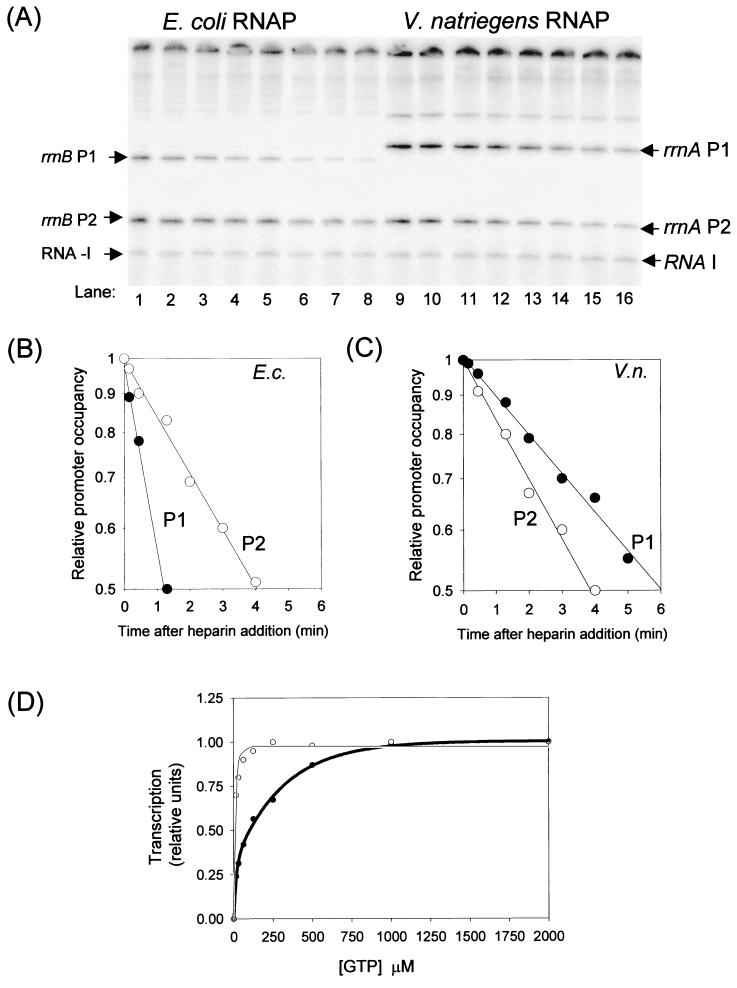
In order to characterize the properties of the V. natriegens rRNA promoters directly, we purified RNA polymerase from V. natriegens (see Materials and Methods) and conducted transcription experiments in vitro. Start sites were mapped for the rrnA P1 and P2 promoters by primer extension from RNAs synthesized in vitro using a primer hybridizing ≈50 bp downstream from the rrnA P2 −10 hexamer (Fig. 4). The transcripts initiate from the regions predicted from the sequence analysis (Fig. 3). There appear to be three start sites for rrnA P1, corresponding to +7U, +8C, and +9G with respect to the −10 hexamer, with G at the +9 position being by far the most prominent start site. The rrnA P2 start sites are 6 to 8 bp downstream from the −10 hexamer, corresponding to +6C, +7C, and +8U with respect to the −10 hexamer, with the C start sites favored over the U start site. The E. coli rrn P1 promoters also start with a purine 9 bp downstream from the −10 hexamer, and the P2 promoters also start with pyrimidines 6 to 8 bp downstream from the −10 hexamer (35; H. Murray and R. L. Gourse, unpublished data).
FIG. 4.
Determination of rrnA P1 and P2 transcription start sites from V. natriegens. In vitro transcription reactions and primer extension were carried out as described in Materials and Methods. The primer extension products were subjected to electrophoresis on an 8% acrylamide-8 M urea gel. DNA sequencing ladders (T, C, G, and A) are shown in the four lanes on the left and in the one lane on the right of the extension reaction and were generated with the same plasmid used for the in vitro transcription reaction. (A) Both P1 and P2 start sites are shown. (B) Gels from the same reaction shown in panel A but electrophoresed longer to improve resolution.
Activation of V. natriegens rrnA P1 and rrnA P2 in vitro.
E. coli rRNA promoters owe much of their strength to the presence of UP elements (A+T-rich sequences that bind the RNAP α subunit) in both the rrn P1 and P2 promoters and to binding sites for the transcription factor Fis upstream of the P1 promoter (31, 49). In vitro transcription assays were used to determine if V. natriegens rRNA promoters were stimulated by the presence of UP elements and Fis.
rrnA P1 promoter fragments with (promoter endpoints, −66 to +1) or without (promoter endpoints, −41 to +1) putative UP elements were inserted upstream of transcription terminators on supercoiled plasmid templates. A second set of templates contained the rrnA P2 promoter with (promoter endpoints, −66 to +8) or without (promoter endpoints, −41 to +8) upstream sequences. Both sets of plasmids also contained the RNA I promoter, which served as a control, since it does not contain an UP element. Transcription reactions were carried out using V. natriegens RNAP (Fig. 5A). The A+T-rich sequences upstream of the −35 regions in both V. natriegens rrnA P1 and rrnA P2 each stimulated transcription 14-fold (lanes 3 to 6). We conclude that these A+T-rich DNA sequences make major contributions to V. natriegens rrnA P1 and P2 promoter activity in the absence of trans-acting factors. For comparison, the E. coli rrnB P1 UP element activated its core promoter 10-fold under the same buffer conditions when transcribed by E. coli RNAP (lanes 1 and 2).
FIG. 5.
Transcription from plasmids containing the V. natriegens rRNA promoters in vitro. (A) UP element-dependent transcription. Lanes 1 and 2, E. coli (E.c.) RNAP. Lanes 3 to 6, V. natriegens (V.n.) RNAP. Lane 1, E. coli rrnB P1 promoter lacking (−) the UP element (pWR55; −41 to +1). Lane 2, E. coli rrnB P1 promoter including (+) the UP element (pRLG5944; −61 to +1). Lane 3, V. natriegens rrnA P1 promoter lacking upstream sequences (pRLG6098; −41 to +1). Lane 4, V. natriegens rrnA P1 promoter including upstream sequences (pRLG6099; −66 to +1). Lane 5, V. natriegens rrnA P2 promoter lacking upstream sequences (pRLG6096; −41 to +8). Lane 6, V. natriegens rrnA P2 promoter lacking upstream sequences (pRLG6097; −66 to +8). (B) Transcription with wild-type E. coli RNAP (WT; lanes 1, 3, and 5) or αΔ235 RNAP (Δ; lanes 2, 4, and 6). Lanes 1 and 2, E. coli rrnB P1 promoter (pRLG862). Lanes 3 and 4, V. natriegens rrnA P1 promoter (pRLG5102). Lanes 5 and 6, V. natriegens rrnA P2 promoter (pRLG5103). (C) Activation by Fis. Lanes 1 and 2, E. coli RNAP and E. coli rrnB P1-P2 (pRLG3858; −152 to +7). Lanes 3 and 4, V. natriegens RNAP and V. natriegens rrnA P1-P2 (pRLG5101; −248 to +8). Lanes 1 and 3, no Fis. Lanes 2 and 4, 100 nM Fis.
Previous studies have shown that UP elements recruit RNAP through interactions with the C-terminal domain of the α subunit (αCTD) (47, 48) and that E. coli RNAP lacking the αCTD is defective for UP element-dependent transcription (46, 48). The V. cholerae αCTD is identical to the E. coli αCTD except for the C-terminal two amino acids, and the seven amino acid residues most important for DNA binding are 100% conserved in the two species (21, 29). Since neither V. cholerae nor V. natriegens α mutant RNAP is available, we tested whether the V. natriegens rrnA P1 and P2 promoters are transcribed by E. coli RNAP and whether this transcription is dependent on the presence of the αCTD.
Transcription from E. coli rrnB P1 was sevenfold more efficient with RNAP containing the αCTD than with RNAP containing the truncated α (Fig. 5B, lanes 1 and 2). The V. natriegens rrnA P1 and rrnA P2 promoters were transcribed efficiently by E. coli RNAP in vitro, and transcription was reduced 10-fold and 3-fold, respectively, with the RNAP lacking the αCTD (lanes 4 and 6). We conclude that the V. natriegens rrnA P1 and P2 A+T-rich sequences upstream of their respective −35 hexamers interact with αCTD and that these UP elements contribute to the extraordinary activity of rRNA promoters in V. natriegens.
The rrn P1 transcription activator Fis is the most abundant nucleoid-associated protein in E. coli during exponential growth (18, 54), and a Fis homolog is present in most enterics, including V. cholerae (7, 29). V. cholerae Fis is extremely similar to E. coli Fis (83% identity; 91% similarity), and the residues in Fis that interact with αCTD and the amino acid residues in αCTD that interact with Fis are 100% conserved (2, 9). Likewise, the amino acid residues in the helix-turn-helix DNA binding motif of V. cholerae Fis are conserved. Therefore, we tested whether E. coli Fis would activate V. natriegens P1 promoters in vitro. Fis increased transcription from V. natriegens rrnA P1 by V. natriegens RNAP 6-fold (Fig. 5C, lanes 3 and 4) under conditions where it increased transcription from E. coli rrnB P1 by E. coli RNAP 10-fold (Fig. 5C, lanes 1 and 2). Therefore, although we have not explored whether there is a V. natriegens fis gene, our data suggest that a Fis homolog contributes to the strength of V. natriegens rRNA transcription.
V. natriegens promoters form short-lived open complexes.
During the multistep process of transcription initiation, RNAP first binds to the promoter to form a short-lived closed complex, which then isomerizes through at least one intermediate to form an open complex in which the DNA in the −10 hexamer and start site region is locally unwound (13, 30, 45). E. coli rRNA promoters form open complexes with half-lives of only a few minutes or less under solution conditions where most other E. coli promoters make open complexes with lifetimes of several hours (5, 24, 39). We have proposed that the short lifetime of the open complex is crucial for regulation of rRNA transcription, because it makes the promoters subject to regulation by changing concentrations of guanosine 3"-diphosphate 5"-diphosphate (ppGpp) and the initiating NTP (5, 6, 19; D. A. Schneider, H. Murray, and R. L. Gourse, unpublished data).
Because V. natriegens rRNA promoters clearly have some features in common with E. coli rRNA promoters, we tested whether the V. natriegens rrnA P1 and rrnA P2 promoters form short-lived open complexes. Under conditions where neither V. natriegens nor E. coli RNAP dissociated detectably from the RNA I promoter during the time course of the experiment (24 min) (Fig. 6A and data not shown), the half-lives of the V. natriegens rrnA P1 and P2 open complexes containing V. natriegens RNAP were ≈6 and 3.5 min, respectively (Fig. 6C). Under the same conditions, the half-lives of the E. coli rrnB P1 and P2 open complexes (containing E. coli RNAP) were ≈1 and 4 min, respectively (Fig. 6B), consistent with values obtained previously (5; Murray and Gourse, unpublished). Thus, V. natriegens rRNA promoters form relatively short-lived open complexes.
FIG. 6.
Characteristics of rrn P1 and P2 open complexes. (A) Open-complex half-lives. Transcription from rrn P1 and P2 promoters was measured following heparin addition. A representative gel is shown (see Materials and Methods). Lanes 1 to 8, E. coli RNAP and E. coli rrnB P1-P2 (pRLG3858). Lanes 9 to 16, V. natriegens RNAP and V. natriegens rrnA P1-P2 (pRLG5101). (B and C) Quantitation by phosphorimager analysis of lanes 1 to 8 (B) and lanes 9 to 16 (C). Solid circles, P1; open circles, P2. (D) Initiating NTP concentration dependence of V. natriegens rrnA P1 promoter in vitro. pRLG5101 was transcribed using 24 nM V. natriegens RNAP in 130 mM KCl transcription buffer (see Materials and Methods) in the presence of increasing GTP concentrations (solid circles). The −35 con promoter (22) was transcribed under the same conditions (open circles). E.c., E. coli; V.n., V. natriegens.
We next tested whether transcription from V. natriegens rrnA P1 by V. natriegens RNAP would require high concentrations of the initiating NTP in vitro. The V. natriegens rrnA P1 promoter required relatively high concentrations of GTP (but not ATP, CTP, or UTP) for maximal transcription, consistent with the observation that this promoter initiates primarily with GTP (Fig. 6D and data not shown). The concentration of GTP required for half-maximal transcription from rrnA P1 was at least 10-fold greater than that required for transcription from a control promoter that also starts with GTP, −35 con (22), under the same conditions (Fig. 6D). These data suggest that transcription from the V. natriegens rrnA P1 promoter might be regulated by the concentration of the initiating NTP in vivo, as has been proposed for the E. coli rrnD P1 promoter (5, 6, 19; D. A. Schneider and R. L. Gourse, unpublished data). Since V. natriegens rrnA P2 forms even shorter-lived open complexes than V. natriegens rrnA P1 (see above), we speculate that rrnA P2 might also be regulated by the concentration of its initiating NTP.
Homologs for relA and spoT, the genes responsible for ppGpp synthesis, are present in the V. cholerae genome sequence (29). The short-lived open complexes formed by V. natriegens rRNA promoters with RNAP most likely would make them sensitive to inhibition by high concentrations of ppGpp, as in E. coli (5). Therefore, we speculate that V. natriegens rRNA promoters are also regulated by ppGpp.
Conclusion.
We have determined that V. natriegens amplifies ribosome numbers to establish the high rate of protein synthesis required for its extremely short generation times. Our investigations have revealed multiple mechanisms that contribute to V. natriegens' high capacity for ribosome synthesis. These include a high rRNA gene dose and stimulation of promoter activity by UP elements and Fis. Furthermore, we expect that V. natriegens rRNA transcription might be regulated by at least some of the same mechanisms used in E. coli, since V. natriegens rRNA promoters share sequence and kinetic features crucial for E. coli rRNA regulation.
There are likely to be additional strategies that contribute to the high rate of rRNA synthesis in V. natriegens that have not been investigated here. For example, we would expect that V. natriegens has a mechanism similar to that in E. coli for preventing premature termination of rRNA transcription (12). Furthermore, the location of rRNA operons relatively near the origin of replication contributes to the high rate of rRNA synthesis in E. coli, since this part of the chromosome is present in multiple copies per cell at high growth rates. While it is tempting to ascribe the high capacity for rRNA synthesis in V. natriegens to its high rRNA gene dose, without knowing the effect of each contributing mechanism relative to the effect of the comparable mechanism in E. coli, it is not possible to ascribe the high rate of rRNA transcription to any one factor.
Acknowledgments
We thank Larry Anthony for help with V. natriegens RNAP purification, Angela Arndt for help with sequencing the V. natriegens promoter regions, Heath Murray for constructs containing E. coli rrnB P2 promoters, and Wilma Ross, Melanie Barker, and other members of the Gourse laboratory for helpful comments.
This work was supported by grant GM37048 from the National Institutes of Health and a Hatch grant from the U.S. Department of Agriculture to R.L.G.
REFERENCES
- 1.Aiyar, S. E., R. L. Gourse, and W. Ross. 1998. Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase alpha subunit. Proc. Natl. Acad. Sci. USA 95:14652-14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar, S. E., S. M. McLeod, W. Ross, C. A. Hirvonen, M. S. Thomas, R. C. Johnson, and R. L. Gourse. 2002. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J. Mol. Biol., in press. [DOI] [PubMed]
- 3.Asai, T., C. Codon, J. Voulgaris, D. Zaporojets, B. Shen, M. Al-Omar, C. Squires, and C. L. Squires. 1999. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 181:3803-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bag, P. K., S. Nandi, R. K. Bhadra, T. Ramamurthy, S. K. Bhattacharya, M. Nishibuchi, T. Hamabata, S. Yamasaki, Y. Takeda, and G. B. Nair. 1999. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J. Clin. Microbiol. 37:2354-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 6.Barker, M. M., and R. L. Gourse. 2001. Regulation of rRNA transcription correlates with NTP sensing. J. Bacteriol. 183:6315-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartlett, M. S., T. Gaal, W. Ross, and R. L. Gourse. 1998. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J. Mol. Biol. 279:331-345. [DOI] [PubMed] [Google Scholar]
- 8.Beach, M. B., and R. Osuna. 1998. Identification and characterization of the fis operon in enteric bacteria. J. Bacteriol. 180:5932-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bokal, A. J., W. Ross, T. Gaal, R. C. Johnson, and R. L. Gourse. 1997. Molecular anatomy of a transcription activation patch: FIS-RNA polymerase interactions at the Escherichia coli rrnB P1 promoter. EMBO J. 16:154-162.0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant, A. E., and D. L. Stevens. 1997. The pathogenesis of gas gangrene, p. 185-196. In J. I. Rood (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, San Diego, Calif.
- 11.Condon, C., D. Liveris, C. Squires, I. Schwartz, and C. L. Squires. 1995. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177:4152-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Condon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59:623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.deHaseth, P. L., M. L. Zupancic, and M. T. Record, Jr. 1998. RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 180:3019-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson, R. R., T. Gaal, H. A. deBoer, P. L. deHaseth, and R. L. Gourse. 1989. Identification of promoter mutants defective in growth-rate-dependent regulation of rRNA transcription in Escherichia coli. J. Bacteriol. 171:4862-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eagon, R. G. 1962. Pseudomonas natriegens: a marine bacterium with a generation time of less than 10 minutes. J. Bacteriol. 83:736-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estrem, S. T., T. Gaal, W. Ross, and R. L. Gourse. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA 95:9761-9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estrem, S. T., W. Ross, T. Gaal, Z. W. S. Chen, W. Niu, R. H. Ebright, and R. L. Gourse. 1999. Bacterial promoter architecture: subsite structure of UP elements and interactions with the C-terminal domain of the RNA polymerase alpha subunit. Genes Dev. 13:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finkel, S. E., and R. C. Johnson. 1992. The Fis protein: it's not just for DNA inversion anymore. Mol. Microbiol. 6:3257-3265. [DOI] [PubMed] [Google Scholar]
- 19.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092-2097. [DOI] [PubMed] [Google Scholar]
- 20.Gaal, T., and R. L. Gourse. 1990. Guanosine 3"-diphosphate 5"-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5533-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaal, T., W. Ross, E. E. Blatter, H. Tang, X. Jia, V. V. Krishnan, N. Assa-Munt, R. H. Ebright, and R. L. Gourse. 1996. DNA-binding determinants of the alpha subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 10:16-26. [DOI] [PubMed] [Google Scholar]
- 22.Gaal, T., W. Ross, S. T. Estrem, L. H. Nguyen, R. R. Burgess, and R. L. Gourse. 2001. Promoter recognition and discrimination by Eσs RNA polymerase. Mol. Microbiol. 42:939-954. [DOI] [PubMed] [Google Scholar]
- 23.Garnier, T. B., B. Canard, and S. T. Cole. 1991. Cloning, mapping, and molecular characterization of the rRNA operons of Clostridium perfringens. J. Bacteriol. 173:5431-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gourse, R. L. 1988. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 16:9789-9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gourse, R. L., T. Gaal, S. E. Aiyar, M. M. Barker, S. T. Estrem, C. A. Hirvonen, and W. Ross. 1998. Strength and regulation without transcription factors: lessons from bacterial rRNA promoters. Cold Spring Harbor Symp. Quant. Biol. 63:131-139. [DOI] [PubMed] [Google Scholar]
- 26.Gourse, R. L., T. Gaal, M. S. Bartlett, J. A. Appleman, and W. Ross. 1996. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50:645-677. [DOI] [PubMed] [Google Scholar]
- 27.Gourse, R. L., W. Ross, and T. Gaal. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37:687-695. [DOI] [PubMed] [Google Scholar]
- 28.Hawley, D., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helmann, J. D., and P. L. deHaseth. 1999. Protein-nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry 38:5959-5967. [DOI] [PubMed] [Google Scholar]
- 31.Hirvonen, C. A., W. Ross, C. E. Wozniak, E. Marasco, J. R. Anthony, S. E. Aiyar, V. Newburn, and R. L. Gourse. 2001. Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J. Bacteriol. 183:6305-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, E. A. 1990. Clostridium perfringens food poisoning, p. 229-240. In D. O. Cliver (ed.), Foodborne diseases. Academic Press, Inc., San Diego, Calif.
- 33.Josaitis, C. A., T. Gaal, and R. L. Gourse. 1995. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc. Natl. Acad. Sci. USA 92:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joseph, S. W., R. R. Colwell, and J. B. Kaper. 1982. Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 10:77-124. [DOI] [PubMed] [Google Scholar]
- 35.Keener, J., and M. Nomura. 1996. Regulation of ribosome synthesis, p. 1417-1428. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 36.Kiss, A., B. Sain, and P. Venetianer. 1977. The number of rRNA genes in Escherichia coli. FEBS Lett. 79:77-79. [DOI] [PubMed] [Google Scholar]
- 37.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Sequencing and hybridization techniques in bacterial systems. Wiley, Chichester, United Kingdom.
- 39.Leirmo, S., and R. L. Gourse. 1991. Factor-independent activation of Escherichia coli rRNA transcription. I. Kinetic analysis of the roles of the upstream activator region and supercoiling on transcription of the rrnB P1 promoter in vitro. J. Mol. Biol. 220:555-568. [DOI] [PubMed] [Google Scholar]
- 40.Maaloe, O., and N. O. Kjeldgaard. 1966. Control of macromolecular synthesis: a study of DNA, RNA, and protein synthesis in bacteria. Benjamin, New York, N.Y.
- 41.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Miura, A., J. H. Krueger, S. Itoh, H. A. deBoer, and M. Nomura. 1981. Growth rate-dependent regulation of ribosome synthesis in E. coli: expression of the lacZ and galK genes fused to ribosomal promoters. Cell 25:773-782. [DOI] [PubMed] [Google Scholar]
- 43.Okabe, S. 1974. Statistical review of food poisoning in Japan—especially that by Vibrio parahaemolyticus, p. 5-8. In T. Fujino, G. Sakaguchi, R. Sakazaki, and Y. Takeda (ed.), International symposium on Vibrio parahaemolyticus. Saikon Publishing Co., Ltd., Tokyo, Japan.
- 44.Payne, W. J. 1960. Effects of sodium and potassium ions on growth and substrate penetration of a marine pseudomonad. J. Bacteriol. 80:696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Record, M. T., Jr., W. S. Reznikoff, M. L. Craig, K. L. McQuade, and P. J. Schlax. 1996. Escherichia coli RNA polymerase (Eσ70), promoters, and the kinetics of the steps of transcription initiation, p. 792-820. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 46.Ross, W., S. E. Aiyar, J. Salomon, and R. L. Gourse. 1998. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J. Bacteriol. 180:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross, W., A. Ernst, and R. L. Gourse. 2001. Fine structure of E. coli RNA polymerase-promoter interactions: alpha subunit binding to the UP element minor groove. Genes Dev. 15:491-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 49.Ross, W., J. F. Thompson, J. T. Newlands, and R. L. Gourse. 1990. E. coli FIS protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 9:3733-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt, T. M. 1997. Multiplicity of rRNA operons in prokaryotic genomes, p. 221-229. In F. J. deBruijn, J. R. Lupski, and G. M. Weinstock (ed.), Bacterial genomes: physical structure and analysis. Chapman and Hall, New York, N.Y.
- 51.Schneider, W. C. 1955. Determination of nucleic acids in tissues by pentose analysis. Methods Enzymol. 3:680-684. [Google Scholar]
- 52.Smith, L. D. S. 1992. The genus Clostridium—medical, p. 1867-1878. In A. Balows, H. G. Truper, M. Dorkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 53.Stevenson, B. S., and T. M. Schmidt. 1997. Growth rate-dependent expression of RNA from plasmid-borne rRNA operons in Escherichia coli. J. Bacteriol. 180:1970-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talukder, A. A., and A. Ishihama. 1999. Twelve species of the nucleoid-associated protein from Escherichia coli. J. Biol. Chem. 274:33105-33113. [DOI] [PubMed] [Google Scholar]
- 55.Twedt, R. M., and R. M. E. Novelli. 1971. Modified selective and differential isolation medium for Vibrio parahaemolyticus. Appl. Microbiol. 22:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webb, C. D., and W. J. Payne. 1971. Influence of Na+ ion on synthesis of macromolecules by a marine bacterium. Appl. Microbiol. 21:1080-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson, K. 1994. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. I. John Wiley and Sons, New York, N.Y. [Google Scholar]