Abstract
Charged amino acids in the predicted transmembrane portion of PcaK, a permease from Pseudomonas putida that transports 4-hydroxybenzoate (4-HBA), were required for 4-HBA transport, and they were also required for P. putida to have a chemotactic response to 4-HBA. An essential amino acid motif (DGXD) containing aspartate residues is located in the first transmembrane segment of PcaK and is conserved in the aromatic acid/H+ symporter family of the major facilitator superfamily of transporters.
The major facilitator superfamily (MFS) of transporters is the largest group of electrochemical-potential-driven permeases found in living organisms (18, 22, 23). The superfamily presently consists of 37 families (http://www.biology.ucsd.edu/∼msaier/transport/2__A__1.html) that generally group according to substrate specificity. Although the MFS is huge, only two of its members; LacY, the lactose permease of Escherichia coli (family 5), and TetA, a tetracycline efflux protein encoded by transposon Tn 10 (family 3 ) have been studied in detail from a functional point of view.
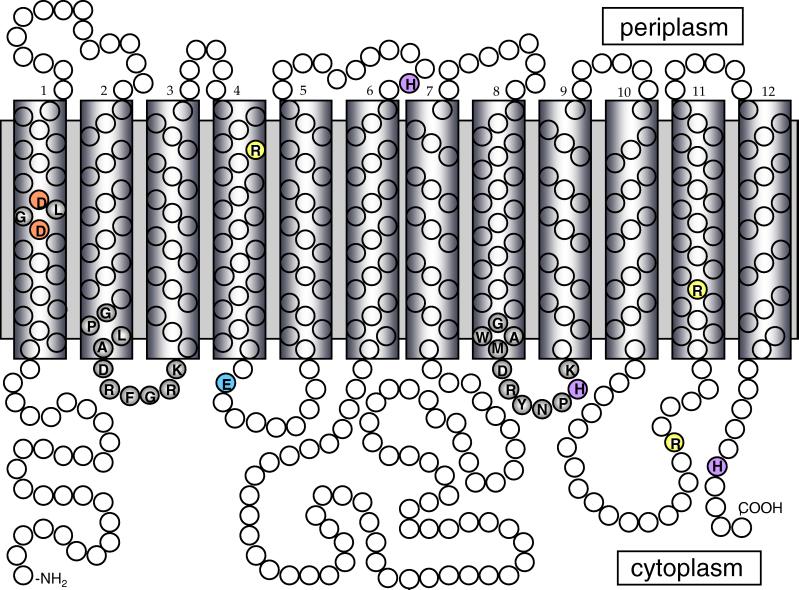
PcaK is a 4-hydroxybenzoate (4-HBA) permease from the bacterium Pseudomonas putida that is unusual in that it also mediates chemotaxis of P. putida to 4-HBA (9, 17). PcaK is the founding member of the aromatic acid/H+ symporter family (AAHS) (family 15) of the MFS (18). Under the recently proposed transporter classification (TC) system, PcaK has been assigned the TC number 2.A.1.15.1 (22). Other members of family 15 include transport systems for the herbicide 2,4-dichlorophenoxyacetate; the lignin monomer vanillate; the aromatic acids 3-hydroxyphenylpropionate and benzoate; and cis,cis-muconate, a dicarboxylic acid that is an intermediate in benzoate degradation (2, 3, 6, 14, 28). As is typical of MFS transporters, PcaK has 12 predicted membrane spanning regions and two conserved amino acid sequences in the hydrophilic regions between the second and third transmembrane regions and between the eighth and ninth transmembrane regions that are required for substrate accumulation (5) (Fig. 1). It also has four charged amino acids in predicted transmembrane regions that are conserved in all members of the AAHS family but not in other families of the MFS (Table 1). These include two aspartate residues in transmembrane segment 1 (TM1) that are part of a conserved DXGD motif, as well as two arginine residues, one in TM4 and the other in TM11 (Table 1). PcaK also has a glutamate at position 144 and an aspartate at position 386, which do not clearly lie within transmembrane regions but are conserved in all AAHS family members.
FIG. 1.
Predicted membrane topology of PcaK. The model is based upon the algorithm of Jones et al. (11). Other algorithms yielded an almost identical topology. Predicted hydrophobic transmembrane segments are enclosed in boxes. The amino acid residues of PcaK that were mutagenized in this study are shaded various colors. The two conserved amino acid sequences in the 2-3 and 8-9 cytoplasmic loops are shaded grey.
TABLE 1.
Conserved charged amino acids located within transmembrane regions of AAHS family members of MFS
| Proteina | Organism | Substrate | % Identityc | DGXDd | Presence of amino acide
|
|||
|---|---|---|---|---|---|---|---|---|
| R124 | R398 | E144 | R386 | |||||
| PcaK | Pseudomonas putida | 4-HBA | 100 | DGLD | + | + | + | + |
| PcaK | Pseudomonas aeruginosa | 4-HBA | 82 | DGLD | + | + | + | + |
| PcaK | Acinetobacter calcoaceticus | 4-HBA | 64 | DGID | + | + | + | + |
| BenK | Acinetobacter calcoaceticus | Benzoate | 33 | DGYD | + | + | + | + |
| FcbT | Arthrobacter sp. strain SU | 4-Chlorobenzoateb | 31 | DGMD | + | + | + | + |
| TfdK | Ralstonia eutropha | 2,4-Dichlorophenoxyacetate | 33 | DGYD | + | K | + | + |
| MmlH | Ralstonia eutropha | 4-Methylmuconolactone | 25 | DSFD | + | K | + | + |
| HppK | Rhodococcus globerulus | 3-Hydroxyphenylpropionateb | 29 | DGFE | + | + | + | + |
| MhpT | Escherichia coli | 3-Hydroxyphenylpropionateb | 39 | EGLD | + | + | + | + |
| MucK | Acinetobacter calcoaceticus | cis-cis muconate | 20 | DGAD | + | + | + | + |
| VanK | Acinetobacter calcoaceticus | Vanillate | 29 | DGFD | + | + | + | + |
References or GenBank accession numbers for represented AAHS family members are as follows: P. putida: PcaK, Q51955. P. aeruginosa: PcaK, G83616. A. calcoaceticus: PcaK, Q43975; BenK, AAF63452; MucK, AAC27117; VanK, AAC27108. Arthrobacter sp. strain SU: FcbT, AAD25166. R. eutropha: TfdK, AAC44725; MmlH, CAA67957. R. globerulus: HppK, AAB81315. E. coli: MhpT, P77589.
The compound listed is a putative substrate for the respective protein.
Percents are relative to 100% amino acid identity for P. putida PcaK based upon GAP analysis (16).
DGXD, the four-amino-acid motif located within the first transmembrane region of AAHS family members.
Amino acid residue numbers are based upon the P. putida PcaK sequence. +, amino acid residue is conserved in GAP alignments; K, lysine residue at position 398.
As a step towards defining amino acid sequences that have a major functional role in members of the AAHS family of the MFS, we mutated the conserved charged amino acids in the PcaK permease and examined the effects of the mutations on rates of 4-HBA transport. Several histidines that are less well conserved in the AAHS family were also mutated. We examined the effects of all of the mutations on the chemotactic response of P. putida to 4-HBA.
Construction of pcaK mutants
Plasmid pHJD193, which contains a 1,656-bp segment of DNA encompassing the pcaK gene and its native promoter (5), was used as template for the generation of site-directed mutants of pcaK by the incorporation of a phosphorylated oligonucleotide during PCR amplification (15). 5"-Phosphorylated mutagenic primers were designed to incorporate one codon change and a silent restriction enzyme recognition site to allow the screening of mutated PCR products. The outside primers and PCR conditions were as described previously (5). The nucleotide sequences of all mutant pcaK genes were verified by DNA sequencing. The PCR products were cloned into the gentamicin resistance broad-host-range vector pBBR1MCS-5 (13). E. coli cells were transformed with plasmid DNA by the method described by Hanahan (8). Plasmids carrying gentamicin resistance genes were mobilized from E. coli DH5α into the P. putida pcaK mutant PRS4085 by triparental matings with E. coli HB101 (pRK2013) (4). P. putida cells were grown at 30°C in a defined mineral medium with 4-HBA or succinate as described previously (9). For P. putida, the antibiotics gentamicin and kanamycin were used at final concentrations of 5 and 100 μg/ml, respectively; for E. coli, ampicillin, gentamicin, and kanamycin were used at final concentrations of 100, 20, and 100 μg/ml, respectively.
4-HBA transport is abolished by mutations that insert uncharged amino acids into transmembrane regions of PcaK
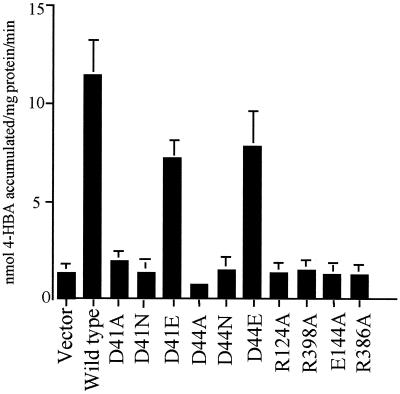
The two aspartates in TM1 of PcaK (D41 and D44) were changed to alanines (D41A and D44A), asparagines (D41N and D44N), and glutamates (D41E and D44E). Within TM4 and TM11, R124 and R328 were each changed to alanine residues. The wild-type PcaK protein and each mutant PcaK protein was expressed in the P. putida pcaK null mutant strain PRS4085 under the control of the native pcaK promoter. The rate of [14C]4-HBA transport was measured as described previously (5). The D41A, D41N, D44A, and D44N mutant PcaK proteins were unable to catalyze any measurable transport of 4-HBA (Fig. 2). When the negative charge at amino acid positions 41 and 44 was restored (D41E and D44E changes), 4-HBA transport was 63 and 67% of the wild-type rate. The R124A and R398A amino acid substitutions resulted in mutant PcaK proteins that were completely defective in their ability to catalyze 4-HBA transport (Fig. 2). Charged amino acid residues that are not located within transmembrane regions of PcaK are also involved in 4-HBA transport. E144 and R386, which are positioned in the hydrophilic cytoplasmic loop between the fourth and fifth transmembrane segments and 10th and 11th transmembrane segments of PcaK, respectively, are both required for 4-HBA transport. When these residues were each changed to alanines, 4-HBA transport was reduced to background levels. Site-directed amino acid changes in the three histidine residues of PcaK resulted in mutant proteins that facilitated the transport of 4-HBA from slightly above background to wild-type levels (Table 2).
FIG. 2.
Transport of 4-HBA by P. putida strain PRS4085 expressing wild-type and mutant PcaK proteins. Proteins were expressed in P. putida cells from the broad-host range vector pBBR1MCS-5. Data are the average levels of 4-HBA accumulation for at least four separate experiments, each done in duplicate. Standard deviations are represented by error bars.
TABLE 2.
Transport and chemotaxis phenotypes of pcaK histidine site-directed mutants
| Mutation | % 4-HBA accumulationa | % 4-HBA chemotaxisb |
|---|---|---|
| Wild type | 100 | 100 |
| H183A | 46 | 54 |
| H328A | 52 | 62 |
| H328R | 15 | 0 |
| H444A | 105 | 100 |
100% accumulation is the amount of 4-HBA accumulated in one min by the pcaK mutant strain PRS4085 complemented with a wild-type pcaK gene from plasmid pHJD193. This value is 12 nmol of accumulated 4-HBA/mg of protein. Transport values are the averages of least four separate experiments, each done in duplicate.
Chemotaxis was measured with 4-HBA swarm plates. A value of 100% means that the average swarm diameter of strain PRS4085 was complemented with the wild-type pcaK gene. Swarm plates were incubated at 30°C for 20 h.
pcaK mutations also affect chemotaxis
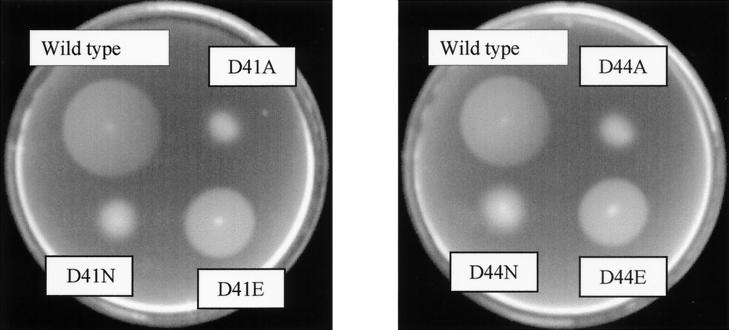
Chemotaxis to 4-HBA was assessed by a soft agar swarm plate assay. PcaK mutant (strain PRS4085) cells carrying either wild-type pcaK or mutant pcaK genes in trans on plasmid pBBR1MCS-5 were stabbed into the centers of minimal medium plates that had been solidified with 0.3% Nobel agar and that contained either 0.5 mM 4-HBA or 1.0 mM succinate. A wild-type chemotactic response is indicated by a sharp, rapidly growing ring that forms in response to the gradient of 4-HBA that is created as cells metabolize the carbon source. The ability of mutant PcaK proteins to mediate chemotaxis to 4-HBA was determined by comparing the diameter of the swarm ring formed by each mutant to the diameter of the swarm ring formed by cells expressing wild-type PcaK protein. In general, the 4-HBA chemotaxis phenotypes of each PcaK mutant matched in severity the 4-HBA transport phenotype. The PcaK mutant proteins that were completely defective in catalyzing 4-HBA transport were also completely defective in mediating a chemotactic response to 4-HBA (data not shown). Cells expressing mutant PcaK proteins had a wild-type response to the chemoattractant succinate. The mutant proteins that catalyzed partial levels of 4-HBA transport could also partially complement the 4-HBA chemotaxis minus phenotype of the pcaK mutant strain PRS4085 (Fig. 3; Table 2).
FIG. 3.
Chemotaxis phenotypes of pcaK mutants with D41 and D44 site-directed changes. Chemotaxis was measured with 4-HBA swarm plates. The wild type is strain PRS4085 complemented with the wild-type pcaK gene. Swarm plates were incubated at 30°C for 20 h.
Mutant PcaK proteins did not have defects in membrane localization
Each PcaK mutant protein resembled the wild-type protein in its level of expression and its cellular localization. To test this, wild-type and mutant PcaK proteins were labeled with l-[35S]methionine during expression from a T7 promoter in E. coli BL21 (DE3) (26). We have shown previously that, when expressed in E. coli, PcaK catalyzes 4-HBA transport at a rate similar to that seen in P. putida (17). Mutant pcaK genes were amplified by PCR using the pBBR1MCS-5 clones as templates, cloned into pT7-5 (27), and expressed as described previously (5). Each mutant PcaK protein localized to the cell membrane at levels that were similar to those of the wild-type PcaK (data not shown). The cytoplasmic fraction contained no labeled protein.
Conclusions
MFS transporters are widely distributed in archea, bacteria, and eukaryotes. This is an ancient family of transporters and individual members have diverged to the point where it can be difficult to identify them as belonging to the MFS without using advanced computational methods (21). All MFS members share, however, a common molecular architecture that consists of 12 or 14 transmembrane helices that are thought to be packed to form the perimeter of a pore through which the transported substrate crosses the cell membrane. Movement of the solute is often accompanied by the translocation of a proton or other ion. Of the 448 amino acids of PcaK, 48 are charged: 28 positive and 20 negative. The four charged amino acids that are located within transmembrane regions of the protein (D41, D44, R124, and R328) are each required for 4-hydroxybenzoate transport. Each is also conserved in all members of the aromatic acid/H+ symporter family of the MFS. In TetA, a histidine residue and aspartate residues located in transmembrane domains have been identified as being required for the exchange of tetracycline and a H+ (25, 29, 30). Extensive studies on the lactose permease have identified pairs of charged amino acid residues (a negatively charged side chain paired with a positively charged side chain) in transmembrane helices that are involved in substrate translocation, H+ translocation, or helix packing (7, 12, 20). It is possible that transmembrane aspartate and arginine residues are charge paired in PcaK and intimately involved in one or more of these three processes, although more detailed studies will be required to prove this. Hydrophobicity plots predict that E144 and H183 of PcaK lie very close to the membrane, and it is possible that in the native protein these amino acids are in fact buried in the cytoplasmic membrane. This is probably not the case for R386, and it is unclear what this particular amino acid might contribute to the transport function of PcaK. The H328 residue of PcaK is located within the second of two conserved stretches of amino acids that are critical for substrate translocation in MFS permeases by acting as a gate for channel opening and closing (10). Therefore, the effects of the H328A and H328R changes on transport are likely due to structural changes within the 8-9 cytoplasmic loop of PcaK.
Mutants that are completely defective in PcaK expression are unable to sense and respond to 4-HBA in chemotaxis assays (9). 4-HBA diffuses across the cell membrane and into P. putida at rates sufficient to support wild-type rates of growth under the conditions in which 4-HBA chemotaxis is measured (9). Thus, simple accumulation of this aromatic acid within cells is not sufficient for chemotaxis to occur; PcaK must be present. This and previous work (5) shows that pcaK mutants that are partially defective in transport have chemotaxis defects of equivalent severity.
Transporters for aromatic acids have been identified that belong to MFS families other than the AAHS family (1, 19, 24). So, membership in the AAHS is not an obligatory characteristic of all MFS permeases that recognize aromatic compounds as a substrate.
Acknowledgments
This work was supported by Public Health Service grant GM56665 from the National Institute of General Medical Science.
REFERENCES
- 1.Chang, H. K., and G. J. Zylstra. 1999. Characterization of the phthalate permease OphD from Burkholderia cepacia ATCC 17616. J. Bacteriol. 181:6197-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collier, L. S., N. N. Nichols, and E. L. Neidle. 1997. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J. Bacteriol. 179:5943-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Argenio, D. A., A. Segura, W. M. Coco, P. V. Bunz, and L. N. Ornston. 1999. The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK. J. Bacteriol. 181:3505-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ditty, J. L., and C. S. Harwood. 1999. Conserved cytoplasmic loops are important for both the transport and chemotaxis functions of PcaK, a protein from Pseudomonas putida with 12 membrane-spanning regions. J. Bacteriol. 181:5068-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrandez, A., J. L. Garcia, and E. Diaz. 1997. Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl)propionate catabolic pathway of Escherichia coli K-12. J. Bacteriol. 179:2573-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frillingos, S., M. Sahin-Toth, J. Wu, and H. R. Kaback. 1998. Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. Faseb J 12:1281-1299. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 9.Harwood, C. S., N. N. Nichols, M.-K. Kim, J. L. Ditty, and R. E. Parales. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jessen-Marshall, A. E., N. J. Paul, and R. J. Brooker. 1995. The conserved motif, GXXX(D/E)(R/K)XG[X](R/K)(R/K), in hydrophilic loop 2/3 of the lactose permease. J. Biol. Chem. 270:16251-16257. [DOI] [PubMed] [Google Scholar]
- 11.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 12.Kaback, H. R., and J. Wu. 1999. What to do while awaiting crystals of a membrane transport protein and thereafter. Acc. Chem. Res. 32:805-813. [Google Scholar]
- 13.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad host range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 14.Leveau, J. H., A. J. Zehnder, and J. R. van der Meer. 1998. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 180:2237-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michael, S. F. 1994. Mutagenesis by incorporation of a phosphorylated oligo during PCR amplification. BioTechniques 16:411-412. [PubMed] [Google Scholar]
- 16.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 17.Nichols, N. N., and C. S. Harwood. 1997. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prieto, M. A., and J. L. Garcia. 1997. Identification of the 4-hydroxyphenylacetate transport gene of Escherichia coli W: construction of a highly sensitive cellular biosensor. FEBS Lett. 414:293-297. [DOI] [PubMed] [Google Scholar]
- 20.Sahin-Toth, M., A. Karlin, and H. R. Kaback. 2000. Unraveling the mechanism of the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:10729-10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saier, M. H., Jr. 2000. Families of transmembrane sugar transport proteins. Mol. Microbiol. 35:699-710. [DOI] [PubMed] [Google Scholar]
- 22.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saier, M. H., Jr., J. T. Beatty, A. Goffeau, K. T. Harley, W. H. Heijne, S. C. Huang, D. L. Jack, P. S. Jahn, K. Lew, J. Liu, S. S. Pao, I. T. Paulsen, T. T. Tseng, and P. S. Virk. 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1:257-279. [PubMed] [Google Scholar]
- 24.Saint, C. P., and P. Romas. 1996. 4-Methylphthalate catabolism in Burkholderia (Pseudomonas) cepacia Pc701: a gene encoding a phthalate-specific permease forms part of a novel gene cluster. Microbiology 142:2407-2418. [DOI] [PubMed] [Google Scholar]
- 25.Someya, Y., A. Niwa, T. Sawai, and A. Yamaguchi. 1995. Site-specificity of the second-site suppressor mutation of the Asp-285-Asn mutant of metal-tetracycline/H+ antiporter of Escherichia coli and the effects of amino acid substitutions at the first and second sites. Biochemistry 34:7-12. [DOI] [PubMed] [Google Scholar]
- 26.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 27.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams, P. A., and L. E. Shaw. 1997. MucK, a gene in Acinetobacter calcoaceticus ADP1 (BD413) encodes the ability to grow on exogeneous cis,cis-muconate as sole carbon source. J. Bacteriol. 179:5935-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi, A., K. Adachi, T. Akasaka, N. Ono, and T. Sawai. 1991. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. Histidine 257 plays an essential role in H+ translocation. J. Biol. Chem. 266:6045-6051. [PubMed] [Google Scholar]
- 30.Yamaguchi, A., T. Akasaka, N. Ono, Y. Someya, M. Nakatani, and T. Sawai. 1991. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. Roles of the aspartyl residues located in the putative transmembrane helices. J. Biol. Chem. 267:7490-7498. [PubMed] [Google Scholar]