Abstract
Nickel acquisition is necessary for urease activity, a major virulence factor of the human gastric pathogen Helicobacter pylori. The nickel permease NixA of H. pylori is a member of the single-component nickel-cobalt transporter family. To identify functionally relevant amino acids of NixA, single-site exchanges were introduced into NixA via PCR-based mutagenesis. This study investigated one of the recognition motifs for this family in transmembrane segment III and other conserved amino acids, mostly with possible nickel-binding capacities. The mutant alleles were expressed in Escherichia coli, and activity of the altered permeases was analyzed by measuring nickel accumulation and urease activity. Expression was checked by immunoblotting after sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a NixA-specific antibody. Replacement of Phe-75 and His-79—both part of the characteristic sequence motif—and of Asn-127, Thr-195, and Ser-197 with alanine abolished nickel uptake in the E. coli system. The results were unchanged if these amino acids were replaced with residues more similar to the original amino acid. The phenotype of the null mutants was independent of the culture medium. Mutation of Val-82, Tyr-242, Thr-260, His-181, and His-15 strongly affected uptake activity under nickel limitation on complex Luria-Bertani medium but had little effect in minimal medium. Eight other conserved amino acids (Ser-80, Ser-81, Phe-119, Trp-180, Tyr-183, Trp-244, Pro-249, and Asn-256) were found to be dispensable for the function of NixA. These results show that atypical nickel-binding amino acids play an important function in nickel uptake and that most of the essential amino acids are clustered in conserved motifs.
The transition metal nickel is an essential trace element of at least five biological processes (14, 31): (i) hydrolysis of urea, (ii) oxidation and evolution of molecular hydrogen, (iii) carbon monoxide dehydrogenase-mediated acetate metabolism under anaerobic conditions, (iv) reduction of methyl coenzyme M to methane, and (v) detoxification of superoxide anion radicals. Uptake of nickel is a prerequisite for those organisms which catalyze nickel-dependent reactions. Ni2+—the most prevalent form—is taken up by nonspecific Mg2+ transport systems and high-affinity systems specific for the transport of nickel (see reference 8 for a review).
In general, two types of nickel-specific uptake systems have been identified so far: (i) the multiple-component ATP binding cassette system called Nik, which was thought for a long time to be unique to Escherichia coli (39), until homologous systems were identified and characterized in Brucella suis (17) and Vibrio parahaemolyticus (28); and (ii) the nickel-cobalt transporter family, comprising homologous single polypeptides in a variety of microorganisms (8, 32)—Helicobacter pylori (25), Ralstonia eutropha (7), Bradyrhizobium japonicum (11), Rhodococcus rhodochrous (18), and the thermophilic Bacillus species strain TB-90 (21)—which have all been characterized biochemically, or at least physiologically. During database searches, related sequences have been identified in the genomes of Mycobacterium tuberculosis and Mycobacterium avium; Salmonella enterica serovar Paratyphi, Salmonella enterica serovar Typhi, and Salmonella enterica serovar Typhimurium; Staphylococcus aureus; Yersinia pestis; and the fission yeast Schizosaccharomyces pombe (8).
Members of the second family share two recognition sequences within their common topology of eight transmembranehelices (TMs): NH2-Arg/Lys-His-Ala-Xaa-Asp-Ala-Asp-His-Ile/Leu-COOH in TM II and NH2-Gly-(Xaa)2-Phe-(Xaa)2-Gly-His-Ser/Thr-Ser/Thr-Val/Ile-Val-COOH in TM III (32). Besides these two conserved motifs, 48 other conserved amino acids scattered through the protein could be detected after sequence alignment. Recently, two other motifs have been proposed (9): NH2-Leu-Gly-Xaa-Asp/Glu-Thr-Ala/Ser-Thr/Ser-Glu-COOH in TM V and NH2-Gly-Met-(Xaa)3-Asp-Thr/Ser-Xaa-Asp-COOH in TM VI.
The high-affinity nickel transport protein NixA of the human pathogen H. pylori was discovered when a gene bank clone of strain ATCC 43504 was found to enhance the coexpressed urease activity in E. coli (25). The urease—an important virulence factor of H. pylori—is a major sink of nickel in this organism, representing up to 6% of the soluble cell protein (16). It converts urea to ammonia and carbamate, the latter decomposing spontaneously to carbon dioxide and ammonia. The released ammonia has been postulated to allow the survival of H. pylori and its colonization of the low-pH environment of the gastric mucosa, which causes Type B gastritis as well as gastric and duodenal ulceration (1, 4, 19, 23, 24). Persistent infection is strongly associated with the development of gastric carcinoma and MALT lymphoma (10, 29, 34).
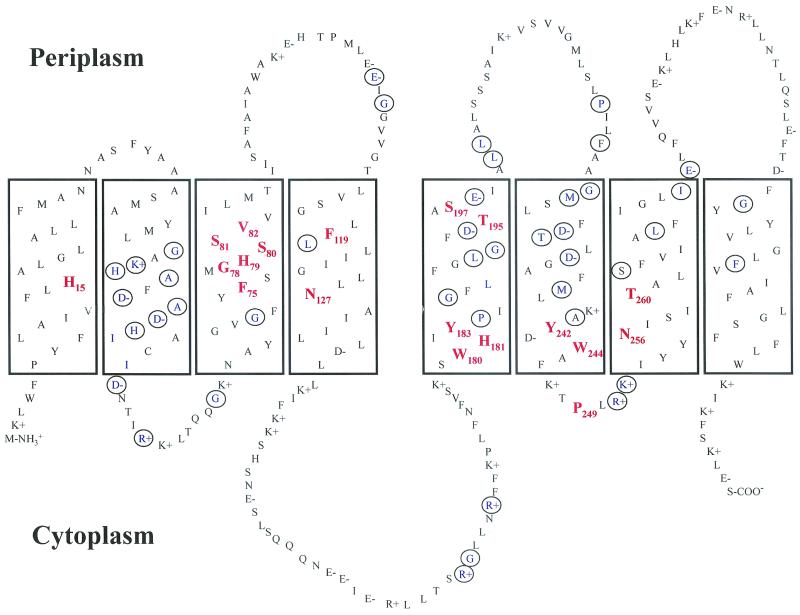
The high affinity of NixA is well suited for effective scavenging by H. pylori of the extremely low levels of nickel from the human body, which are estimated to be in the range of 2 to 11 nM (36). A first clue to which amino acids might be important for recognizing and transporting nickel could come from a search for conserved amino acids among homologous permeases. The location of the two characteristic sequences in TMs II and III and of the other conserved amino acids is shown in Fig. 1, representing a recently proposed topological model of NixA (13).
FIG. 1.
Proposed two-dimensional model of NixA, the nickel permease of H. pylori. The eight boxes represent the TMs connected by four periplasmic (above the TMs) and three cytoplasmic (below the TMs) loops. The N and C termini reside in the cytoplasm. Amino acids are given in the one-letter code; the ones exchanged in this study are shown in red and numbered. Other conserved amino acids that have not been exchanged in this study are circled.
In two previous papers, the consequences of amino acid replacements in high-affinity nickel transporters were studied: one concentrated on the conserved motif in TM II of the NixA homologue HoxN (7), and the other focused on conserved histidine, aspartate, and glutamate residues of NixA (12), well known to bind nickel in vitro. It is unknown if nickel binding is also the key role of these amino acids in vivo.
In this study, we report the role of conserved amino acids with low nickel-binding constants in vitro, harboring hydroxyl, amide, and aromatic groups in their side chains. We focused on the signature sequence of TM III of NixA and other conserved residues closer to the C terminus. The intracellular nickel contents of strains expressing the mutant proteins were assessed by measurement of nickel accumulation and urease activity.
MATERIALS AND METHODS
Materials.
63NiCl2 (24.7 TBq/mol) and [35S]dATPαS were from Amersham. Zinsser Aquasafe 300 Plus was used for liquid scintillation analyses in a Packard 1900 TR counter. Biochemicals were purchased from Boehringer Mannheim-Roche Molecular Biochemicals, Amersham, Life Technologies, and Stratagene. For detection of NixA-anti-NixA antibody complexes in Western immunoblots, an alkaline phosphatase-coupled goat anti-rabbit antibody (Sigma) was used. Nitrocellulose blotting membranes were from Schleicher & Schuell.
Bacterial strains and plasmids.
E. coli strain CC118 [araD139 Δ(ara leu)7697 ΔlacX74 phoAΔ20 galE thi rpsE rpoB argE(Am) recA1] (22) was used as the host strain for recombinant plasmids. Plasmid pUEF202 contains the nixA gene of H. pylori encoding the nickel permease NixA (25). Plasmid pHP808 harbors the whole urease operon of H. pylori (15). Both plasmids were a gift from H. L. T. Mobley (University of Maryland, Baltimore).
Construction of a nixA deletion mutant.
Plasmid pUEF202 was digested with the single-cutting enzyme AflII and treated with T4 DNA polymerase. After precipitation, the linearized plasmid was digested with MluNI, which cuts only once in plasmid pUEF202 and generates blunt ends. The larger fragment was isolated from an agarose gel, ligated, and transformed into E. coli. The resulting clone (internal deletion of 237 amino acids out of 331) was sequenced.
Site-directed mutagenesis.
Mutagenesis was carried out by the PCR-based method of Chen and Przybyla (2), which in most cases requires two rounds of PCR but only a single mutagenesis primer. The mutagenic primers used in this study are available on request. Pfu polymerase was used for DNA amplification, with plasmid pUEF202 as a template. The products of the second PCR round were digested with appropriate restriction enzymes, purified on agarose gels, and used to replace the respective fragments of plasmid pUEF202. The complete sequence of synthetic DNAs was verified by the dideoxy chain termination method of Sanger et al. (33).
Nickel accumulation.
The nickel accumulation assay was adopted from Wolfram et al. (38). Overnight cultures of E. coli CC118 (pUEF202 or its site-directed derivatives) were used to inoculate (0.6%) fresh Luria-Bertani (LB) medium containing ampicillin (60 μg/ml) and 500 nM 63NiCl2 (24.7 TBq/mol). The cultures were incubated at 37°C with vigorous shaking for 6 h, washed twice with buffer A (50 mM Tris-HCl, 10 mM MgCl2 [pH 7.5]), and concentrated 10-fold in the same buffer. An aliquot of the suspension (50 μl) was subjected to liquid scintillation analysis. The cellular nickel content was expressed as picomoles per milligram of protein.
Urease assay.
E. coli CC118 (pHP808; pUEF202 or its site-directed vaiants) was grown aerobically overnight in LB medium in the presence of ampicillin (60 μg/ml), chloramphenicol (25 μg/ml), and 10 μM NiCl2 at 37°C. The cells were washed once with 50 mM KPO4 (pH 7.0) and concentrated 10-fold in the same buffer. Urease activity was measured with permeabilized cells, quantifying the rate of ammonium ion released from urea by the formation of indophenol (26). The assay mix consisted of the cell suspension in 50 mM KPO4 (pH 7.0), 35 mM NaCl, and 0.14 mM N-cetyl-N,N,N-trimethylammoniumbromide. The reactions were initiated by the addition of a urea solution to a final concentration of 10 mM, and the release of ammonium ion was determined in timed aliquots. The aliquots were added to a phenol-containing reagent mix. After the development of an indophenol dye from the ammonia-containing sample, the absorbance was measured at 546 nm and the amount of ammonia was calculated from a calibration curve. One unit of enzyme is defined as the amount of enzyme required to form 2 μmol of ammonium ion per min at 37°C.
Isolation of crude bacterial membranes.
E. coli CC118 (pUEF202 or its site-directed derivatives) was grown overnight in LB medium supplemented with ampicillin (60 μg/ml) and 500 nM NiCl2, washed once with buffer B (50 mM Tris-HCl, 100 mM NaCl [pH 7.5]), and concentrated 10-fold in the same buffer with a mix of protease inhibitors (Complete Mini; Boehringer Mannheim-Roche Molecular Biochemicals). The cells were then disrupted by three passages in a French pressure cell at 20,000 lb/in2, and the lysate was cleared by centrifugation (5,000 × g; 15 min; 4°C). The supernatant was ultracentrifuged (100,000 × g; 60 min; 4°C). The resulting pellet containing the membrane proteins was resuspended in 60 μl of buffer B with the protease inhibitor mix and adjusted with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer to a protein concentration of 2.5 mg/ml and stored at −80°C.
Western blot analysis.
Membrane proteins (30 μg) were separated by SDS-PAGE (10% polyacrylamide) and electroblotted on nitrocellulose membranes. NixA and its site-directed mutant derivatives were detected by antibodies to NixA raised in rabbits (a gift from H. L. T. Mobley).
Protein determination.
Protein was determined by the method of Lowry et al. (20).
RESULTS
Selection of conserved amino acids for site-directed mutagenesis.
Sequence alignment of the 10 members of the nickel-cobalt transporter family identified two recognition motifs (32) and 48 other conserved amino acids (9, 12, 18). In this study, we concentrated on amino acids of the motif Phe-75-Xaa-Xaa-Gly-78-His-79-Ser-80-Ser-81-Val-82 in TM III of the nickel permease NixA and Phe-119, Asn-127, Trp-180, His-181, Tyr-183, Thr-195, Ser-197, Tyr-242, Trp-244, Pro-249, Asn-256, and Thr-260 (Fig. 1), which have not been analyzed for their functional relevance in previous studies. The 10 underlined amino acids out of 18 were strictly conserved in all 10 members of the protein family, and 5 (Phe-119, His-181, Tyr-183, Trp-244, and Asn-256) were conserved in 9 of the members. Ser-81 and Ser-197—only conserved in four and three family members, respectively—were otherwise exclusively replaced by threonine. Val-82 was present in eight homologous nickel permeases. Additionally, His-15 in TM I, which is conserved in five transporters among the NixA homologues, was substituted. Conserved lipophilic amino acids were mostly neglected, as well as conserved positively charged amino acids. The latter have been shown to be without effect on nickel transport and urease activity (12). Substitution by alanine was chosen because of its small size and its inability to bind nickel, except for Gly-78, which was replaced by the more bulky isoleucine, and for Val-82, which was replaced by the hydrophilic serine.
Effect of substitution of the conserved amino acids on nickel accumulation and urease activity.
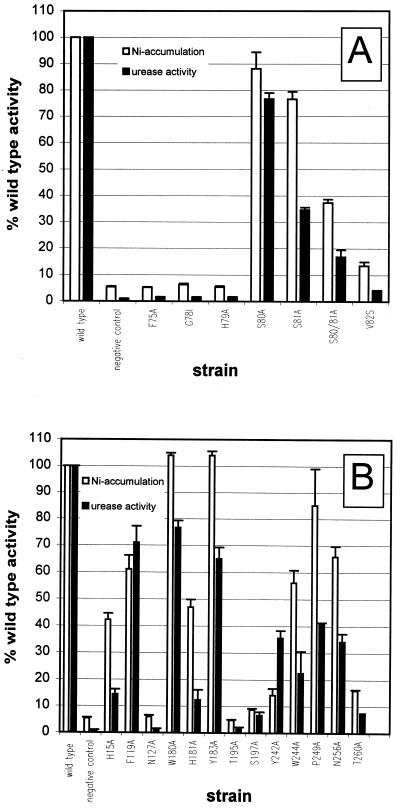
The functional contribution of the amino acids of the conserved motif in TM III of NixA is shown in Fig. 2A, and those of all other amino acids are shown in Fig. 2B. Nickel accumulation and urease activity were completely abolished in 6 of the 20 mutants: F75A, G78I, H79A, N127A, T195A, and S197A. These mutants were considered biologically relevant, since they reduced the measured activities to the level of the negative control. In four mutants (S80A, W180A, Y183A, and P249A), neither nickel accumulation nor urease activity was changed compared to the wild type. In nine others (H15A, S81A, V82S, F119A, H181A, Y242A, W244A, N256A, and T260A), nickel uptake and urease activity ranged between 15 and 80% of those of the wild type. These effects were not considered biologically relevant even for the drastically reduced H15A, V82S, H181A, Y242A, and T260A mutants, since the phenotypic effects were inconsistent: on minimal medium, urease activity was closer to wild-type levels than on the complex LB medium. This is probably due to the fact that the free nickel concentration is lower in LB medium because of its higher complexation capacity.
FIG. 2.
Effects of exchanged amino acids on nickel uptake. The bars represent nickel uptake measured either as nickel accumulation or urease activity and are the mean values of at least three independent measurements (+ standard deviations). Activity is given as a percentage of wild-type activity; 100% urease activity corresponds to a mean value of 124 mU/mg of protein (ranging from 100 to 147 mU/mg of protein), and 100% nickel accumulation corresponds to a mean value of 25.7 pmol/mg of protein (ranging from 19.4 to 37.8 pmol/mg). (A) Effects of exchanges in the conserved motif in TM III; (B) effects of exchanges in other conserved amino acids scattered through NixA.
The mutants S80A and S81A, surprisingly, did not show a strong phenotype, although in all of the homologous nickel permeases, hydroxyl-containing amino acids are strictly conserved at these positions. Therefore, a mutant was constructed in which both hydroxyl moieties were concomitantly changed. This double mutant still retained significant activity (Fig. 2A). In contrast to the S197A mutant, which showed nickel accumulation and urease activity less than 10% of the wild type levels, Ser-80 and Ser-81 seemed to be irrelevant.
In general, it was observed that the percentage of nickel accumulation was higher than the urease activity in comparison to the wild type, which may mean that not all the intracellular nickel was incorporated into urease. Nevertheless, nickel accumulation correlated significantly with the urease activity observed in the mutants (R2 = 0.77; P = 0.0001).
Effect of replacing amino acid on the mutant phenotype.
In three cases (Phe-75, His-79, and Thr-195), additional mutations were introduced using amino acids with physicochemical properties closer to the original amino acid than alanine. His-79 was replaced by serine, which could not functionally substitute at this position. Phe-75 was additionally replaced by methionine and tyrosine. Methionine at this position was poorly compensating, as was alanine, and tyrosine led to a considerable increase in nickel accumulation (more than 20% of the wild-type phenotype) but only a very weak increase in urease activity under standard conditions (about 8% of wild-type activity). A NixA mutant harboring a serine at position 195 instead of the original threonine showed a residual activity of about 12 to 13% of the NixA wild type in both assays.
During the course of the experiments, it was observed that replacement with isoleucine reduced nickel uptake more strongly than replacement by alanine at the same position (tested for His-181, Glu-198, and Asp-234), although both amino acids are hydrophobic. Therefore, alanine seemed to be a better choice for substitution to judge the functional relevance of the original amino acid.
Expression of the mutants.
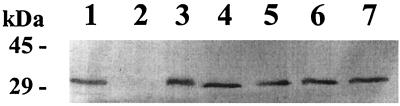
In order to check whether the reduction of nickel-dependent activities in the NixA mutants was due to a lack of expression in E. coli, crude membrane extracts were isolated and subjected to Western blot analysis with a NixA-specific antibody. Figure 3 shows a Western blot for the mutants with the strongest effects on nickel accumulation: the mutant proteins of NixA derivatives H79A, F75A, N127A, T195A, and S197A were synthesized in approximately the same amounts as the NixA wild type. G78I gave a nonreproducible weak signal, which renders evaluation as an essential amino acid uncertain.
FIG. 3.
Expression of NixA mutant proteins. A Western blot of crude membrane extracts of E. coli strains after separation by SDS-PAGE is shown. On the left, the molecular mass of the marker is given in kilodaltons. Lanes: 1, wild type; 2, negative control (nixA deletion mutant); 3, F75A; 4, H79A; 5, N127A; 6, T195A; 7, S197A.
DISCUSSION
The present report describes the functional role of conserved amino acids of the nickel permease NixA of H. pylori. The study was focused on the amino acids of a recognition motif in TM III and on 13 other amino acids scattered through the protein. We found five amino acids to be crucial for nickel accumulation and urease activity: Phe-75 and His-79, located in the recognition motif, and Asn-127, Thr-195, and Ser-197. The results show that the recognition motif in TM III of the nickel-cobalt permease family is as important as the one in TM II and that amino acids with low in vitro affinity to nickel are as essential for the function of NixA as those with high nickel affinity.
Based on our results and the results of Fulkerson et al. (12), a total of 12 amino acids have been shown to perform an essential function in the NixA-mediated uptake of nickel: Asp-47, Asp-49, His-44, and His-50 in TM II; Asp-55 in the cytoplasm following TM II; Phe-75 and His-79 in TM III; Asn-127 in TM IV; Asp-194, Thr-195, and Ser-197 in TM V; and Asp-231 in TM VI. Evidently, a great portion of the functionally relevant amino acids are histidine and aspartate residues; they rank among the best nickel binders, directly after cysteine, according to their affinities (27). Interestingly, four amino acids identified in this study have low nickel-binding constants in vitro; indeed, this is the first identification of this kind of amino acid as being essential for the nickel uptake function in a nickel-specific transporter. In contrast, positively charged conserved amino acids were dispensable for the function of NixA (12). It cannot be assumed that all the amino acids important for transport activity directly interact with the substrate. They could participate as well in stabilization of the overall protein structure or of the nickel interaction domains or be involved in conformational changes between substrate binding and release. This applies even more to amino acids like phenylalanine, serine, asparagine, and threonine, found to be necessary in this study, with nickel-binding affinities about 10-fold lower than the one for aspartate (27).
A hint about the role of the amino acids in the transport process might come from kinetic studies of single-site mutant transporters and the determination of the characteristic parameters Km and Vmax. A change in the affinity (Km) of CorA mutants in S. enterica serovar Typhimurium was interpreted as a loss of a substrate binding site (35, 37). CorA has a high capacity and a low affinity for nickel, facilitating reliable kinetic analysis. However, the members of the nickel-cobalt transporter family have high affinity but low capacity for nickel. Kinetic analyses are difficult (6, 7) and were not performed in any of the cited studies. Our results are based on nickel accumulation experiments. Nevertheless, we performed kinetic studies with our mutants; all five mutants with less than 10% of the wild-type nickel accumulation showed a massive decrease in nickel transport capacity; three of them (N127A, T195A, and S197A) were indistinguishable from the negative control. In the other two mutants (F75A and H79A), affinity for nickel was decreased 8- and 14-fold, respectively, with an almost unchanged maximal transport rate. Urease activity correlated best with nickel accumulation, which was therefore chosen as the more relevant parameter to evaluate the function of NixA.
Replacement of the original amino acids by conservative residues might allow us to analyze whether the size or the chemical property is more important for the function of a specific amino acid. Replacement of Phe-75 with alanine and methionine abolished nickel uptake, while tyrosine at this position could partially restore uptake. Aromaticity might play a role at this position, yet clearly phenylalanine was preferred. Thr-195 could be partly replaced by serine, showing that a hydroxyl moiety is necessary at this position.
According to the topology model, it is evident that most of the essential amino acids are located in TMs and clustered in or near motifs (9, 32) (Fig. 1). An exception is Asn-127 in TM IV; however, it could well be that in the three-dimensional arrangement Asn-127 was placed near other functionally important residues. Phe-75 in the topology model presented is situated in the middle of TM III (13) (Fig. 1). Others have proposed a position near the transition of the membrane and the cytoplasm (5). In the latter case, Phe-75 might be involved in stabilization of the membrane segment at the interface of the bilayer and the cytoplasm, as was proposed for conserved aromatic residues of CorA (35). On the other hand, Phe-75 might interact with nickel even at this location, since it is known in the case of the well-characterized melibiose and lactose carrier of E. coli that amino acids located in the cytoplasmic half of TMs or even in the cytoplasm could be responsible for substrate specificity or cation coupling (30).
In respect to substrate specificity, it was interesting that His-15 in TM I is part of a sequence motif (NH2-Val-Xaa-Leu-His-Val-Leu-Gly-Xaa-Ala-Leu-COOH) which was identified in NhlF of R. rhodochrous (18) among the NixA homologues and in the nonhomologous COT1 protein of Saccharomyces cerevisiae (3). Both proteins are involved in cobalt transport. Since NixA harbors this motif as well, it would be worthwhile to discover whether cobalt is also a substrate for NixA.
Mutagenesis studies are important steps in characterizing the nickel transport function of NixA. It would be interesting to analyze the virulence of H. pylori strains expressing mutant NixA derivatives in an animal model, since the nickel-dependent urease activity plays a key role in the successful colonization of the stomach mucosa. However, this must await the advent of suitable genetic tools to introduce and express genes with point mutations in H. pylori.
Acknowledgments
We thank Harry L. T. Mobley for the plasmids pHP808, containing the entire urease operon of H. pylori, and pUEF202, harboring the nixA gene of H. pylori, as well as for the NixA-specific antibody.
This work was supported by the Swiss National Fund (32-45998-95).
REFERENCES
- 1.Blaser, M. J. 1990. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J. Infect. Dis. 161:626-633. [DOI] [PubMed] [Google Scholar]
- 2.Chen, B., and A. E. Przybyla. 1994. An efficient site-directed mutagenesis method based on PCR. BioTechniques 17:657-659. [PubMed] [Google Scholar]
- 3.Conklin, D. S., J. A. McMaster, M. R. Culbertson, and C. Kung. 1992. COT1, a gene involved in cobalt accumulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:3678-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon, M. F. 1991. Helicobacter pylori and peptic ulceration: histopathological aspects. J. Gastroenterol. Hepatol. 6:125-130. [DOI] [PubMed] [Google Scholar]
- 5.Eitinger, T., and B. Friedrich. 1994. A topological model for the high-affinity nickel transporter of Alcaligenes eutrophus. Mol. Microbiol. 12:1025-1032. [DOI] [PubMed] [Google Scholar]
- 6.Eitinger, T., and B. Friedrich. 1997. Microbial nickel transport and incorporation into hydrogenases, p. 235-256. In G. Winkelmann and C. J. Carrano (ed.), Transition metals in microbial metabolism. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 7.Eitinger, T., L. Wolfram, O. Degen, and C. Anthon. 1997. A Ni2+ binding motif is the basis of high affinity transport of the Alcaligenes eutrophus nickel permease. J. Biol. Chem. 272:17139-17144. [DOI] [PubMed] [Google Scholar]
- 8.Eitinger, T., and M.-A. Mandrand-Berthelot. 2000. Nickel transport systems in microorganisms. Arch. Microbiol. 173:1-9. [DOI] [PubMed] [Google Scholar]
- 9.Eitinger, T., O. Degen, U. Böhnke, and M. Müller. 2000. Nicp1, a relative of bacterial transition metal permeases in Schizosaccharomyces pombe, provides nickel ion for urease biosynthesis. J. Biol. Chem. 275:18029-18033. [DOI] [PubMed] [Google Scholar]
- 10.Forman, D., D. G. Newell, F. Fullerton, J. W. G. Yarnell, A. R. Stacey, N. Wald, and F. Sitas. 1991. Association between Helicobacter pylori infection and risk of gastric cancer: evidence from a prospective study. Br. Med. J. 302:1302-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, C., S. Javedan, F. Moshiri, and R. J. Maier. 1994. Bacterial genes involved in incorporation of nickel into hydrogenase enzyme. Proc. Natl. Acad. Sci. 91:5099-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulkerson, J. F., Jr., R. M. Garner, and H. L. T. Mobley. 1999. Conserved residues and motifs in the NixA protein of Helicobacter pylori are critical for the high affinity transport of nickel ions. J. Biol. Chem. 273:235-241. [DOI] [PubMed] [Google Scholar]
- 13.Fulkerson, J. F., Jr., and H. L. T. Mobley. 2000. Membrane topology of the NixA nickel transporter of Helicobacter pylori: two nickel transport-specific motifs within transmembrane helices II and III. J. Bacteriol. 182:1722-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hausinger, R. P. 1997. Metallocenter assembly in nickel-containing enzymes. J. Biol. Inorg. Chem. 2:279-286. [Google Scholar]
- 15.Hu, L.-T., P. F. Foxall, R. Russell, and H. L. T. Mobley. 1992. Purification of recombinant Helicobacter pylori urease apoenzyme encoded by ureA and ureB. Infect. Immun. 60:2657-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, L.-T., and H. L. T. Mobley. 1990. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect. Immun. 58:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jubier-Maurin, V., A. Rodrigue, S. Ouahrani-Bettache, M. Layssac, M.-A. Mandrand-Berthelot, S. Köhler, and J.-P. Liautard. 2001. Identification of the nik gene cluster of Brucella suis: regulation and contribution to urease activity. J. Bacteriol. 183:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komeda, H., M. Kobayashi, and S. Shimizu. 1997. A novel transporter involved in cobalt uptake. Proc. Natl. Acad. Sci. 94:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, A., J. Fox, and S. Hazell. 1993. Pathogenicity of Helicobacter pylori: a perspective. Infect. Immun. 61:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowry, O. H., N. J. Roseborough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 183:265-275. [PubMed] [Google Scholar]
- 21.Maeda, M., M. Hidaka, A. Nakamura, H. Masaki, and T. Uozumi. 1994. Cloning, sequencing, and expression of thermophilic Bacillus sp. strain TB-90 urease gene complex in Escherichia coli. J. Bacteriol. 176:432-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manoil, C., and J. Beckwith. 1985. Tn phoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall, B. J., D. B. McGechie, P. A. Rogers, and R. J. Glancy. 1985. Pyloric Campylobacter infection and gastroduodenal disease. Med. J. Australia 142:439-444. [DOI] [PubMed] [Google Scholar]
- 24.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet i:1311-1315. [PubMed]
- 25.Mobley, H. L. T., R. M. Garner, and P. Bauerfeind. 1995. Helicobacter pylori nickel transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol. Microbiol. 16:97-109. [DOI] [PubMed] [Google Scholar]
- 26.Mobley, H. L. T., and R. P. Hausinger. 1989. Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53:85-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nriagu, J. O. 1980. Global cycle and properties of nickel, p. 1-26. In J. O. Nriagu (ed.), Nickel in the environment. Wiley & Sons, Inc., New York, N.Y.
- 28.Park, K.-S., T. Iida, Y. Yamaichi, T. Oyagi, K. Yamamoto, and T. Honda. 2000. Genetic characterization of DNA region containing the trh and ure genes of Vibrio parahaemolyticus. Infect. Immun. 68:5742-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsonnet, J., S. Hansen, L. Rodriguez, A. Gelb, R. Warnke, E. Jellum, N. Orentreich, J. Vogelman, and G. Friedman. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 330:1267-1271. [DOI] [PubMed] [Google Scholar]
- 30.Poolman, B., and W. N. Konings. 1993. Secondary solute transport in bacteria. Biochim. Biophys. Acta 1183:5-39. [DOI] [PubMed] [Google Scholar]
- 31.Ragsdale, S. W. 1998. Nickel biochemistry. Curr. Opin. Chem. Biol. 2:208-215. [DOI] [PubMed] [Google Scholar]
- 32.Saier, M. H., Jr., B. H. Eng, S. Fard, J. Garg, D. A. Haggerty, W. J. Hutchinson, D. L. Jack, E. C. Lai, H. J. Liu, D. P. Nusinew, A. M. Omar, S. S. Pao, I. T. Paulson, J. A. Quan, M. Sliwinski, T.-T. Tseng, S. Wachi, and G. B. Young. 1999. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim. Biophys. Acta 1422:1-56. [DOI] [PubMed] [Google Scholar]
- 33.Sanger, F., S. Micklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sipponen, P., V. J. Kosuneutu, M. Riikela, and K. Seppala. 1992. Infection and chronic gastritis in gastric cancer. J. Clin. Pathol. 45:318-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, R. L., M. A. Szegedy, L. M. Kucharski, C. Walker, R. M. Wiet, A. Redpath, M. T. Kaczmarek, and M. E. Maguire. 1998. The CorA Mg2+ transport protein of Salmonella typhimurium—mutagenesis of conserved residues in the third membrane domain identifies a Mg2+ pore. J. Biol. Chem. 273:28663-28669. [DOI] [PubMed] [Google Scholar]
- 36.Sundermann, F. W., Jr. 1993. Biological monitoring of nickel in humans. Scand. J. Work Environ. Health 19(Suppl.):34-38. [PubMed] [Google Scholar]
- 37.Szegedy, M. A., and M. E. Maguire. 1999. The CorA Mg2+ transport protein of Salmonella typhimurium—mutagenesis of conserved residues in the second membrane domain. J. Biol. Chem. 274:36973-36979. [DOI] [PubMed] [Google Scholar]
- 38.Wolfram, L., B. Friedrich, and T. Eitinger. 1995. The Alcaligenes eutrophus protein HoxN mediates nickel transport in Escherichia coli. J. Bacteriol. 177:1836-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, L.-F., C. Navarro, and M.-A. Mandrand-Berthelot. 1991. The hydC region contains a multicistronic operon (nik) involved in nickel transport in Escherichia coli. Gene 107:37-42. [DOI] [PubMed] [Google Scholar]