Abstract
The Salmonella pathogenicity island 2 (SPI-2) type III secretion system is expressed by intracellular bacteria and translocates effector proteins across the vacuolar membrane. Signals sensed by Salmonella enterica serovar Typhimurium in the intracellular compartment activate SPI-2 gene expression through the two-component regulatory system SsrAB. The effects of environmental and genetic signals on expression of the SsrAB-regulated gene sspH2 were examined. SsrAB-dependent activation of sspH2 was detected in the presence of both low and moderate concentrations of magnesium or calcium and at acidic and neutral pHs. The levels of expression were comparable to those detected in bacteria recovered from cultured macrophages. The induction in media at alkaline pHs (pH 7.5 and 8.0) was greatly reduced compared to the induction observed at pH 7.0 or at a lower pH, suggesting that alkaline pH represses SsrAB activation. In addition, the PhoPQ two-component system, which is also activated intracellularly, was not required for activation of SsrAB.
Salmonella enterica serovar Typhimurium virulence requires two type III secretion systems (TTSS) encoded in large blocks of horizontally acquired DNA known as Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2, respectively) (6). The SPI-1 TTSS is expressed in the extracellular milieu and translocates effector proteins upon contact with the plasma membrane of host cells (8, 13). In contrast, the SPI-2 TTSS is expressed in the intracellular environment after phagocytosis by macrophages or invasion of epithelial cells (2). The SPI-2 TTSS translocates several effector proteins, which facilitate intracellular replication of bacteria, across the vacuolar membrane. The two-component regulatory system SsrAB encoded in SPI-2 controls the expression of SPI-2 genes encoding the TTSS and effectors. SsrAB also activates a regulon encoded outside SPI-2, including at least five SPI-2 TTSS effectors (sspH2, sseI, sseJ, sifA, and sifB) (1, 9, 15).
Studies in which expression of SPI-2 genes in defined culture media has been examined have begun to elucidate the environmental signals sensed by SsrAB (3, 7). Expression can be achieved by growth in minimal media having various compositions. Deiwick et al. concluded that SPI-2 gene expression is induced in defined media whose magnesium, calcium, or phosphate contents are limited, and they presented evidence that suggested that this induction is dependent on both the SsrAB two-component system encoded in SPI-2 and the PhoPQ two-component system (3). In contrast, Lee et al. (7) presented data which suggested that the magnesium concentration did not affect SPI-2 gene expression; rather, media which were acidic (pH 4.5) at the beginning of growth induced SsrAB activation to express SPI-2 genes, while media which were neutral (pH 7) at the beginning of growth did not induce activation. In addition, the OmpR-EnvZ two-component system was found to exhibit transcriptional regulation of expression of ssrAB by direct binding to the ssrAB promoter, suggesting that osmolarity may also play a role in SPI-2 gene expression (7).
In this work we further defined the environmental and genetic signals required for activation of SsrAB by performing a detailed analysis of the effects of medium pH, magnesium concentration, and PhoP-PhoQ on SsrAB-dependent gene transcription.
Magnesium limitation is not required for SsrAB activation.
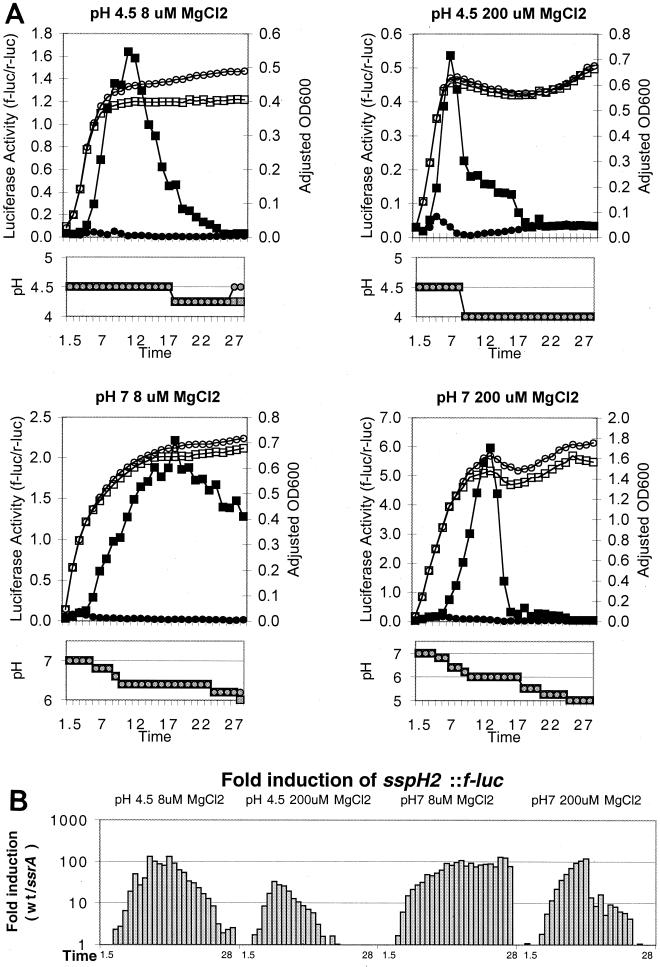
Previous studies demonstrated that SsrAB is activated in magnesium-limited medium (3). However, in these studies the researchers examined single time points and therefore may not have detected expression earlier or later in the bacterial growth cycle. β-Galactosidase is a commonly used reporter for transcription in bacteria. Because this enzyme has a long half-life, cumulative gene expression can be determined over several hours, and it is sometimes possible to use single end point measurements to assay transcription. In contrast, the half-life of firefly luciferase, which was used by Deiwick et al. and in this work, is approximately 15 min in Salmonella serovar Typhimurium (data not shown). Thus, a thorough examination of expression at many time points is required to determine whether a luciferase fusion is expressed. In order to more rigorously examine the effects of magnesium and acidic pH on SsrAB activation, we examined expression of sspH2, which encodes an SPI-2 TTSS translocated effector whose transcription is highly regulated by SsrAB (10). Salmonella serovar Typhimurium expressing a single copy of a transcriptional fusion of sspH2 to the firefly luciferase (f-luc) encoded in the pGPLFR03 suicide plasmid (strain EM207 and derivatives of this strain [10]) was grown in N minimal media [5 mM KCl, 7.5 mM (NH4)2SO4 0.5 mM K2SO4, 1 mM KH2PO4, 0.1 M Tris HCl, 0.1% Casamino Acids, 38 mM glycerol, 200 μM bis-Tris] containing magnesium at levels which Deiwick et al. previously found to be either inducing (8 μM MgCl2) or noninducing (200 μM MgCl2). The pHs of media containing magnesium at these two concentrations were adjusted to either 7 or 4.5; these pHs were found to be noninducing or inducing by Lee et al. Firefly luciferase activity was normalized for renilla luciferase (r-luc) activity, which is expressed from a constitutive promoter in pGPLFR03.
When organisms were grown at an initial pH of 4.5 or 7.0 in the presence of either a high magnesium concentration or a low magnesium concentration, sspH2::f-luc was found to be highly induced in wild-type bacteria but not in strains carrying an ssrA::mTn5 (5) mutation. The maximum expression of sspH2::f-luc was 100- to 130-fold higher in wild-type bacteria than in ssrA null strains, except in pH 4.5 medium containing 200 μM MgCl2, in which 33-fold induction was observed (Fig. 1). Interestingly, although the levels of induction of sspH2::f-luc were similar in three of the media analyzed, the gross f-luc/r-luc activity in the media with an initial pH of 7.0 containing 200 μM MgCl2 was approximately threefold higher than the activity observed in pH 4.5 or 7.0 media containing 8 μM MgCl2. SsrAB-dependent induction of sspH2::f-luc expression began in the late logarithmic phase of growth and continued through the early stationary phase in all four media. The levels of expression observed in these media are comparable to the levels previously observed in bacteria in cultured macrophages (10), indicating that magnesium limitation is not required for expression of SsrAB-activated genes. In addition, these media contained 1 mM phosphate, indicating that phosphate limitation is not required for SsrAB activation.
FIG. 1.
Expression of sspH2::f-luc in the presence of 8 or 200 μM MgCl2 at pH 4.5 or 7. (A) Overnight LB medium cultures of bacteria were washed and then diluted to an optical density at 600 nm (OD600) of 0.026 in 70-ml portions of N minimal media containing either 8 or 200 μM MgCl2 at a starting pH of 4.5 or 7. Samples were taken after 1.5 h and at 1-h intervals between 3 and 28 h, and luciferase activity was determined by using the sspH2::f-luc reporter values (30 s) and the constitutive r-luc reporter values (10 s). The values shown were determined by dividing the f-luc value by the r-luc value at each time point for the wild type (▪) and the ssrA mutant (•). Each data point is the value for a single sample taken from a culture and is representative of three experiments. The adjusted optical densities at 600 nm for the wild type (□) and the ssrA mutant (○) were determined by diluting bacterial cultures so that the densities were within the linear range of measurement for a Spectronic Genesys 5 spectrometer and then correcting for the dilution factor. pH was monitored with colorpHast pH strips (Fisher Scientific), which were accurate to within approximately 0.5 pH unit for the wild-type strain (squares) and the ssrA strain (circles). (B) Induction of sspH2::f-luc, as calculated by dividing the wild-type f-luc/r-luc value by the ssrA f-luc/r-luc value at each time point. In several cases the f-luc value was below the limit of detection (800 relative light units [RLU]) for the ssrA mutant, and the induction shown is the minimum induction obtained by dividing the wild-type f-luc/r-luc value by the limit of detection for the ssrA mutant, which was 800/r-luc. This was the case for pH 4.5 medium containing 8 μM MgCl2 at 15 to 28 h (except for the 18-h time point), for pH 4.5 medium containing 200 μM MgCl2 at 10 to 28 h, and for pH 7 medium containing 200 μM MgCl2 at 15 to 28 h. wt, wild type.
A low Ca2+ concentration induces expression of type III secretion genes in Yersinia (14), and Deiwick et al. suggested that a high calcium concentration represses SsrAB-dependent gene expression. Because the media described above did not contain added calcium, we attempted to repress expression of sspH2::f-luc by adding Ca2+ to the media. Addition of 2 mM CaCl2 and addition of 0.5 mM CaCl2 to the media (pH 6.5 buffered media containing 200 μM MgCl2 [see Fig. 3]) did not repress sspH2::f-luc expression (f-luc/r-luc ratios at 16 h, 3.9 and 5.4, respectively) or regulation by ssrA (52- and 73-fold induction at 16 h, respectively). Thus, we detected levels of SsrAB-dependent gene expression comparable to the levels found in intracellular bacteria in media in which magnesium, calcium, and phosphate were not limiting.
FIG. 3.
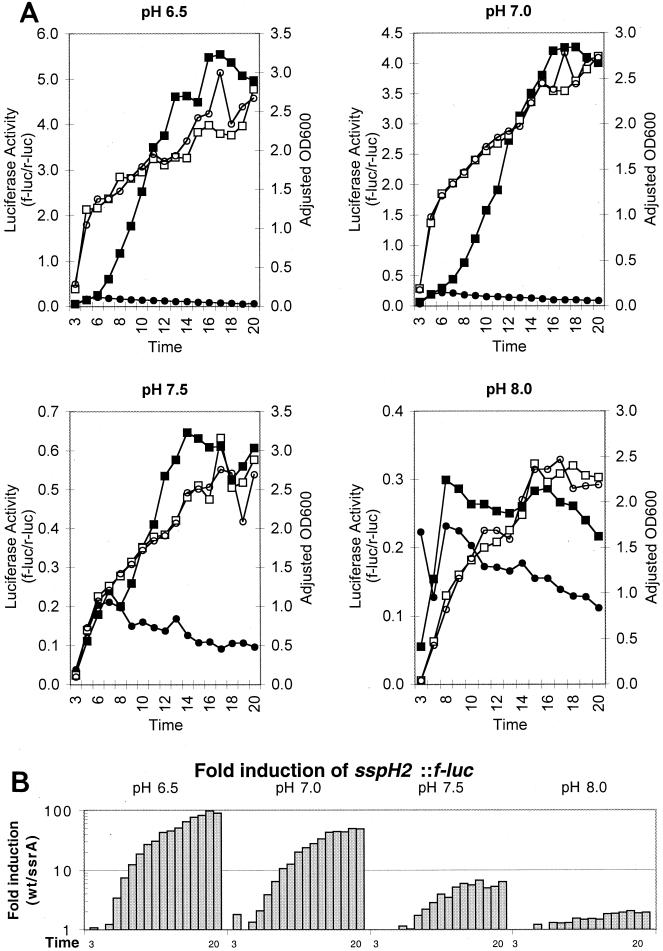
Effect of pH on sspH2::f-luc expression. Overnight cultures of bacteria were washed and then diluted to an optical density at 600 nm (OD600) of 0.026 in 40-ml portions of N minimal media containing 200 μM MgCl2 supplemented with 100 mM HEPES and 100 mM bis-Tris at pH 6.5, 7.0, 7.5, or 8. Samples were taken after 3 h and every hour between 5 and 20 h after inoculation, and luciferase activity was determined for the wild type (wt) (▪) and ssrA mutants (•) as described in the legend to Fig, 1. The adjusted optical densities at 600 nm were determined by dilution in saline for the wild type (□) and the ssrA mutant (○). (B) Induction of sspH2::f-luc was determined as described in the legend to Fig. 1. All values were above the limit of detection.
PhoPQ is not required for activation of SsrAB.
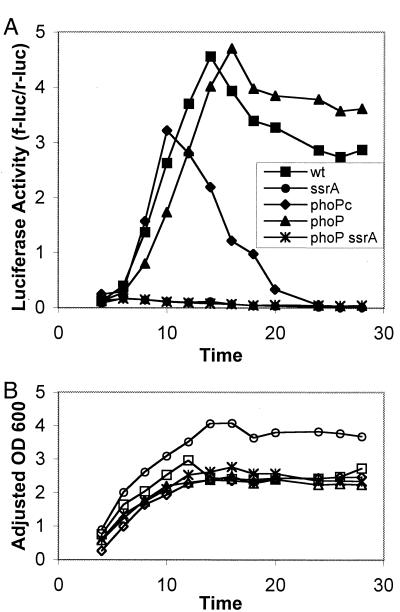
Deiwick et al. previously examined the effect of a phoP mutation on SPI-2 gene expression and concluded that phoPQ was required for SsrAB activation (3). Since PhoQ is activated in response to magnesium limitation (4), this finding supported the hypothesis that magnesium limitation stimulates SsrAB activation. These authors observed a decrease in SsrAB-dependent gene expression in phoP mutants grown in media in which magnesium was limiting (8 μM magnesium). Under these conditions, however, the growth of phoP mutants was severely limited compared to the growth of wild-type bacteria; thus, it was not clear if the decreased SsrAB-dependent gene expression in phoP mutants was due to direct regulation by PhoPQ or indirect effects on bacterial physiology induced by growth arrest. In order to more thoroughly examine the effect of PhoPQ on SsrAB-dependent gene expression, sspH2::f-luc expression in media containing 200 μM MgCl2 was examined.
If activation of PhoPQ contributes to SsrAB-dependent gene expression, enhanced expression of sspH2::f-luc would be expected in strains carrying the pho-24 (12) mutation, which increases net PhoP phosphorylation by PhoQ. However, the sspH2::f-luc expression in pho-24 or phoP102::Tn10d-Cm (11) mutants was similar to the expression observed in wild-type bacteria (Fig. 2), indicating that PhoPQ activation alone does not enhance expression of SsrAB-regulated genes. The relatively rapid decrease in sspH2::f-luc activity in pho-24 mutants after 16 h of growth was probably due to alterations in the physiological state of the bacteria resulting from constitutive activation of PhoP, which may have resulted in nonspecific alterations in transcription, mRNA stability, translation, or protein stability. The lack of specificity of this premature decline in activity was demonstrated by expression of the constitutively expressed r-luc reporter, whose activity was found to decrease dramatically after 20 h compared to the activity in wild-type or phoP null bacteria (data not shown).
FIG. 2.
Expression of sspH2::f-luc in phoP mutants. Wild-type bacteria (wt), ssrA mutants, pho-24 mutants, phoP mutants, and phoP ssrA mutants were grown in buffered pH 6.5 media as described in the legend to Fig. 3. Samples were taken approximately every 2 h from 2 to 28 h, and luciferase activity (A) and adjusted optical density at 600 nm (OD 600) (B) were determined as described in the legend to Fig. 1. All values were above the limit of detection except the values for the pho-24 mutant at 26 and 28 h.
An alternative means of determining the effect of phoP on SsrAB activation is to examine the ability of phoP mutants to activate SsrAB within the phagosomes of cultured macrophage cells. Because phoP mutants exhibit a dramatic growth defect compared to wild-type bacteria in the intracellular environment, the strains cannot be directly compared. On the other hand, we have found that both phoP and phoP ssrA mutants have similar growth and survival characteristics during a 6-h infection (1 h of infection plus 5 h of gentamicin treatment) in RAW264.7, a macrophage-like cell line (data not shown). Luciferase activity recovered from these bacteria revealed that despite an approximately 17-fold replication defect compared to wild-type bacteria (data not shown), the phoP mutant induced sspH2::f-luc expression 13-fold in an SsrA-dependent fashion (f-luc/r-luc ratios for phoP, 0.66 ± 0.04; f-luc/r-luc ratios for phoP ssrA, 0.05 ± 0.01). The results of this experiment suggest that SsrAB can activate gene expression in phoP mutants in vivo.
While the results of the in vitro and in vivo experiments described above do not completely eliminate the possibility that PhoPQ has some effect on SsrAB-dependent gene expression, they provide no evidence that supports this hypothesis.
Alkaline pH inhibits SsrAB activation.
The buffering capacities of the media used in the experiments whose results are shown in Fig. 1 were insufficient to prevent acidification during bacterial growth (Fig. 1). During maximal induction in media with an initial pH of 7.0, the measured pH was between 6 and 6.5 (Fig. 1). Thus, acidification to pH 4.5 was not required for high levels of SsrAB activation.
In order to more accurately determine the effect of pH on SsrAB activation, buffered media were used to prevent acidification during bacterial growth. When cultures were grown in the presence of 20 mM bis-Tris and 10 mM HEPES in addition to the 100 mM Tris present in N minimal media, acidification of the media was observed in media having initial pHs between 6.5 and 8.0. Using the Henderson Hasselback equation, we calculated that growth of Salmonella serovar Typhimurium under these conditions resulted in more than 28 mM H+, which overwhelmed the three buffers. We calculated that in order to prevent medium acidification, 100 mM Tris, 100 mM bis-Tris, and 100 mM HEPES should be included in the media. Under these buffering conditions only minor pH changes were observed for at least 20 h during growth of Salmonella serovar Typhimurium.
Using these buffered media, we analyzed the effect of pH on SsrAB activation. Salmonella serovar Typhimurium expressing sspH2::f-luc was grown in buffered media having initial pHs of 6.5, 7.0, 7.5, and 8.0. The final pHs of these media after growth of either wild-type or ssrA mutant bacteria for 20 h were determined to be 6.24, 6.90, 7.45, and 7.91, respectively, by centrifuging the media and measuring the pHs with a pH meter. Firefly luciferase activity was determined hourly for 20 h for wild-type bacteria or for a strain carrying an ssrA mutation. sspH2::f-luc was highly induced at pH 6.5 and 7.0 (Fig. 3). The maximal level of induction in wild-type bacteria compared with the level of induction in ssrA mutants occurred at 19 h, when wild-type bacteria exhibited 97- and 49-fold-greater activity than an ssrA mutant in pH 6.5 and 7.0 media, respectively (Fig. 3). These levels of induction were similar to the 100- to 130-fold levels of induction observed in media having estimated pHs of 6 and 4 (Fig. 1). In media buffered at more basic pHs, expression of sspH2::f-luc was dramatically decreased. However, wild-type bacteria did exhibit higher levels of expression than ssrA mutants; sevenfold induction and twofold induction dependent on SsrAB were observed in the pH 7.5 and 8.0 media, respectively (Fig. 3). Similar results were obtained with an ssaH::f-luc reporter at 16 h, and f-luc/r-luc activity decreased with increasing pH (f-luc/r-luc ratio at pH 6.5, 15.1; f-luc/r-luc ratio at pH 7.0, 10.8; and f-luc/r-luc ratio at pH 7.5, 6.3). These results may have relevance for salmonellae ingested by animal hosts as the lumen of the intestine is alkaline. Subsequently, internalization by macrophages in acidified spacious phagosomes would induce expression of the SPI-2 TTSS.
In conclusion, we examined the effects of various environmental and genetic conditions on sspH2::f-luc expression. The SsrAB-dependent expression of sspH2 in minimal media is similar to that of bacteria recovered from infected macrophages, while SsrAB-dependent expression is not observed in bacteria grown in Luria-Bertani (LB) medium (10). Magnesium limitation, acidic pH, and PhoPQ are not required for expression of the SsrAB regulon in minimal media. In contrast, an alkaline pH represses full activation of sspH2 by SsrAB. However, it is unlikely that the alkaline pH is the only component of LB medium which represses SsrAB-dependent gene expression, since LB medium buffered at an acidic pH (pH 6.0) and low osmolarity (no added NaCl) does not stimulate sspH2 expression (data not shown). Therefore, factors other than alkaline pH may control the repression of SPI-2 gene expression in LB medium.
Acknowledgments
This work was supported by the Poncin Scholarship Fund and a Paul Allen Fellowship (to E.M.) and by grant RO1 AI48683 from the National Institutes of Health (to S.M.).
REFERENCES
- 1.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 3.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 4.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 6.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas, R. L., and C. A. Lee. 2000. Unravelling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36:1024-1033. [DOI] [PubMed] [Google Scholar]
- 9.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Bäumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 11.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, S. I., and J. J. Mekalanos. 1990. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol. 172:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rakeman, J. L., and S. I. Miller. 1999. Salmonella typhimurium recognition of intestinal environments. Trends Microbiol. 7:221-223. [DOI] [PubMed] [Google Scholar]
- 14.Straley, S. C., G. V. Plano, E. Skrzypek, P. L. Haddix, and K. A. Fields. 1993. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol. Microbiol. 8:1005-1010. [DOI] [PubMed] [Google Scholar]
- 15.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]