Abstract
The RecA protein of Deinococcus radiodurans (RecADr) is essential for the extreme radiation resistance of this organism. The RecADr protein has been cloned and expressed in Escherichia coli and purified from this host. In some respects, the RecADr protein and the E. coli RecA (RecAEc) proteins are close functional homologues. RecADr forms filaments on single-stranded DNA (ssDNA) that are similar to those formed by the RecAEc. The RecADr protein hydrolyzes ATP and dATP and promotes DNA strand exchange reactions. DNA strand exchange is greatly facilitated by the E. coli SSB protein. As is the case with the E. coli RecA protein, the use of dATP as a cofactor permits more facile displacement of bound SSB protein from ssDNA. However, there are important differences as well. The RecADr protein promotes ATP- and dATP-dependent reactions with distinctly different pH profiles. Although dATP is hydrolyzed at approximately the same rate at pHs 7.5 and 8.1, dATP supports an efficient DNA strand exchange only at pH 8.1. At both pHs, ATP supports efficient DNA strand exchange through heterologous insertions but dATP does not. Thus, dATP enhances the binding of RecADr protein to ssDNA and the displacement of ssDNA binding protein, but the hydrolysis of dATP is poorly coupled to DNA strand exchange. The RecADr protein thus may offer new insights into the role of ATP hydrolysis in the DNA strand exchange reactions promoted by the bacterial RecA proteins. In addition, the RecADr protein binds much better to duplex DNA than the RecAEc protein, binding preferentially to double-stranded DNA (dsDNA) even when ssDNA is present in the solutions. This may be of significance in the pathways for dsDNA break repair in Deinococcus.
Bacteria belonging to the family Deinococcaceae are the most radiation-resistant organisms known (2, 19, 40). Despite ubiquitous distribution and ancient derivation, only seven species of this family have been described (35). Of these species, Deinococcus radiodurans is the only one for which systems of genetic manipulation have been developed, and these have been used in the molecular analysis of its DNA repair pathways (15, 16) and in its development for bioremediation of radioactive waste environments (5, 14, 27). Adding to the growing resource of genetic technologies available for D. radiodurans is the recent whole-scale sequencing, annotation, and analysis of its genome (28, 35, 36, 56). D. radiodurans is the first representative with a completely sequenced genome from a bacterial branch of extremophiles—the Thermus/Deinococcus group. Phylogenetic tree analysis, combined with the identification of several synapomorphies (shared derived characters) between Thermus and Deinococcus, support that it is a very ancient branch localized in the vicinity of the bacterial tree root (35).
D. radiodurans is a polyextremophile (48), showing remarkable resistance to a range of severe damage caused by ionizing radiation, desiccation, UV radiation, oxidizing agents, or electrophilic mutagens (40). This bacterium is most famous for its resistance to ionizing radiation. It survives acute exposures to gamma radiation that exceed 1,700,000 rads without lethality or induced mutation (18) and is capable of vigorous growth in the presence of chronic irradiation (6,000 rads per h) (27, 55). D. radiodurans can survive 100 to 200 irradiation-induced DNA double-stranded breaks (DSBs) per haploid genome, yielding >1,000 DSB fragments in a typical polyploid cell (18, 28). Although its myriad resistance phenotypes stem from efficient DNA repair processes, the mechanisms underlying this repair remain poorly understood (35). Its extreme DNA damage resistance phenotype appears to be very complex, determined collectively by features revealed by analysis of its genome (35, 56) as well as by many more subtle structural peculiarities of proteins and DNA that are not readily inferred from the sequences. Remarkably, the number of DNA repair enzymes identified in D. radiodurans by genomic annotation (35, 56) is less than reported for Escherichia coli. Further, a recent analysis of predicted expression levels [E(g)] (based on codon usage) of the main repair proteins of D. radiodurans and E. coli showed that neither set of repair genes is likely to be expressed at high levels (22). There is one exception. RecA protein is predicted to be highly expressed in both organisms; the level of E. coli RecA (RecAEc) expression was predicted to be high [E(g) = 1.48], and D. radiodurans RecA (RecADr) expression was predicted to be dramatically high [E(g) = 2.04]. Given these general similarities in expression of repair functions, another explanation for the radioresistance of D. radiodurans has been sought. For example, unlike E. coli, the repair proteins of D. radiodurans may operate in the context of highly organized clusters of aligned multiple identical chromosomes (17, 35). This chromosomal structure would simplify the search for homology during repair and facilitate the function of what appears to be a conventional set of repair enzymes in D. radiodurans.
RecA or RecA-like proteins have been found throughout the phylogenetic tree, including archaea, bacteria, yeast, plants, and mammals; they play a critical role in biological processes that require homologous DNA pairing and recombination (4, 10, 49). D. radiodurans is no exception, and its recA gene appears very similar to those found in Thermus spp. and other gram-positive bacteria. For example, the RecAs of D. radiodurans and Thermus aquaticus have 69% amino acid sequence identity. In comparison, the RecAs of E. coli and D. radiodurans exhibit a 53% sequence identity. A functional RecADr protein is central to the expression of the resistance phenotypes of D. radiodurans (41). For example, disruption of recADr greatly diminishes the bacterium's ability to recover from acute DNA damage (6, 18, 20, 41) and prevents its growth in the presence of chronic radiation (55). A direct comparison of the D. radiodurans and E. coli RecA proteins could prove enlightening. The biological function of the RecADr protein has been tightly linked to efficient repair of DSBs. The primary function of the RecAEc protein appears to lie in the repair of a broad range of stalled replication fork structures (9, 11, 12, 25).
RecADr is normally expressed constitutively at low levels in D. radiodurans (M. Lipton, Pacific Northwest National Laboratory, Richmond, Wash., personal communication) and is only transiently expressed at high levels following extreme DNA damage (6). Thus, it has been difficult to purify significant quantities of this recombinase directly from D. radiodurans, and several previous attempts to purify cloned RecADr in E. coli were unsuccessful (6, 20, 40).
These difficulties have been overcome, and we report here the cloning and expression of RecADr protein in two different E. coli expression systems. One involves the construction of a fusion protein to aid in purification, and the other is a direct expression of the native protein. Both proteins have been purified to near-homogeneity, and their in vitro activities have been examined at some length. In general, the RecADr protein forms filaments on DNA and possesses activities that resemble those of the RecAEc protein. However, certain molecular functions, particularly binding to duplex DNA, the hydrolysis of nucleoside triphosphate (NTP) cofactors, and the response of DNA strand exchange to NTP hydrolysis, provide a source of some sharp contrasts between the E. coli and D. radiodurans RecA proteins.
MATERIALS AND METHODS
Enzymes and reagents.
Restriction endonucleases, T4 DNA ligase, the IMPACT-T7 cloning system, chitin beads, and E. coli RecA protein were purchased from New England Biolabs. E. coli single-stranded DNA (ssDNA) binding protein (SSB) was purified as described previously (31) and stored frozen at −70°C in a buffer containing 20 mM Tris-HCl (40% cation, pH 8.4), 0.15 M NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, and 50% glycerol. The concentration of SSB was determined by absorbance at 280 nm using an extinction coefficient of ɛ280 = 1.5 A280 mg−1 ml−1 (32). Plasmid DNA purification and gel extraction kits were purchased from Qiagen. [γ-S]ATP, Tris buffer, creatine phosphokinase, and phosphocreatine were purchased from Boehringer Mannheim. Restriction endonucleases were purchased from New England Biolabs. Ceramic hydroxyapatite type II was purchased from Bio-Rad. Phosphoenolpyruvate, pyruvate kinase, lactic dehydrogenase, NADH, ATP, and proteinase K were purchased from Sigma.
DNA.
Circular duplex and single-stranded DNA (ssDNA) from M13mp8 and its derivatives (M13mp8.52) were prepared using methods described previously (51). Circular ssDNA, supercoiled double-stranded DNA (dsDNA), and nicked circular φX174 DNA were purchased from New England Biolabs. The concentrations of ssDNA and dsDNA were determined by absorbance, using A260 = 1 as equivalent to 36 and 50 μg ml−1, respectively. Linear duplex φX174 DNA was prepared by digestion of supercoiled DNA with PstI endonuclease. Linear ssDNA was prepared by annealing a synthetic oligonucleotide to the circular single-stranded φX174 DNA at a site encompassing the unique PstI recognition sequence, with digestion with PstI endonuclease. M13mp8 dsDNA was linearized by digestion with EcoRI. M13mp8.52 dsDNA was linearized by cleavage with the restriction endonuclease MscI. The linear ssDNA was purified using the Qiaquick gel extraction kit (Qiagen).
Cloning of D. radiodurans recA gene.
The recA gene from D. radiodurans used in this study was cloned in two different ways.
The first utilized the IMPACT-T7 expression system as described previously (20) and subcloned as pJDC101 (6). To clone the recA gene into the IMPACT-T7 vector, two PCR forward and reverse primers were designed with an NdeI site (underlined) flanking at the N-terminal side (5′ TTTTTTCATATGAGCAAGGACGCCACCAAAG 3′) and a SmaI site (underlined) at the C-terminal side (5′ TTTTTTCCCGGGCGCTTCGGCGGCTTCGGGC 3′). DNA was amplified by PCR using an Advantage-GC Genomic PCR kit (Clontech) as described by the manufacturer. The PCR product was agarose gel purified, and the DNA was digested with the NdeI and SmaI restriction enzymes. The cleaved DNA was ligated in frame to the pTYB2 plasmid vector of the IMPACT-T7 system, which was also restricted with NdeI/SmaI. The result is a cloned gene in which the recADr gene is fused to an intein- and chitin-binding domain. The recombinant plasmid DNA was transformed into competent cells of the E. coli strain BLR(DE3) (Novagen) and plated onto Luria-Bertani (LB) agar plates containing ampicillin (100 μg/ml). The colonies were picked and grown in LB liquid media, and plasmid DNA was prepared from different bacterial colonies. Plasmids were checked for the correct insert DNA by restriction analysis and DNA sequencing.
The second method involved direct cloning of the recADr gene into the pET21A vector (Novagen). The recADr gene was PCR amplified from genomic DNA extracted from D. radiodurans bacterial cells (ATCC 13939), using the following primers: 5′CCAATAGCATATGAGCAAGGACGCCA and 5′CGGGATCCCAAGAGGAGGTTTAC. The 1,123-bp PCR product was gel purified and then digested overnight with 30 U each of NdeI and BamHI. It was ligated to pET21A that had been digested with the same enzymes and transformed into the nonexpression host DH5α. Several clones were sequenced. The first three had point mutations, but the fourth had the complete wild-type recADr gene and was selected as pEAW158. The pEAW158 construct was transformed into the host STL2669/pT7pol26 (a gift from Susan Lovett, Brandeis University) for expression. This strain lacks the recAEc gene and carries a plasmid expressing the bacteriophage T7 RNA polymerase under lac control.
Purification of RecADr protein.
The recombinant plasmid containing D. radiodurans recA gene fused to the intein-, chitin-binding domain (pAS18) in E. coli was inoculated into LB medium containing 100 μg of ampicillin/ml at 37°C. When the culture reached an optical density at 600 nm of 0.6, it was induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration) for 4 h. The cells were harvested, resuspended in buffer A (20 mM Tris Cl [pH 7.5]-1 mM EDTA-1.5 M KCl-0.1% Tween 20) containing pepstatin and leupeptin protease inhibitors (final concentration, 1 μg/ml each), and lysed by passing the cells three times through a French press cell (American Instrument Co.) at 900 lb/in2. The lysate was incubated at 4°C for 1 h with slow shaking and centrifuged at 27,000 × g for 20 min, and the clarified crude cell lysate was loaded onto a chitin agarose column. The chitin column was then rinsed with 10 volumes of buffer A and then quickly flushed with buffer B (20 mM Tris Cl [pH 7.5]-0.5 M NaCl-1 mM EDTA) containing 50 mM dithiothreitol (DTT). The column was then left at 4°C for about 16 h. This step produces efficient self-cleavage of the RecA protein from the intein portion of the vector. The latter portion remains attached to the chitin agarose column. Then the protein was eluted from the column with 5 volumes of buffer B. The protein was further concentrated using an ultrafiltration cell concentrator (Amicon) and dialyzed against buffer C (20 mM phosphate buffer [pH 7.5]-0.1 mM EDTA). This protein was loaded on a hydroxyapatite column and rinsed with 10 volumes of buffer C. The protein was then eluted with a phosphate buffer step gradient (50 to 400 mM). The individual protein fractions were checked for nuclease contamination by incubating them with φX174 RFI DNA, linear dsDNA, and circular ssDNA. In each case, the amount of RecADr used was twice that used in typical experiments, the incubations were carried out under standard reaction conditions for 2 h, and the reactions were analyzed by gel electrophoresis. The fractions without any detectable nuclease contamination were pooled and dialyzed extensively against buffer D (20 mM Tris Cl [pH 7.5]-0.1 mM EDTA-1 mM DTT-10% glycerol). As described in Results, the resulting protein has one extra glycine residue attached to the C terminus, and we refer to this protein as RecA*Dr.
The native RecADr protein was purified using a different procedure. The STL2669/pT7pol26 cells containing pEAW158 were grown in LB broth containing 100 μg of ampicillin/ml and 40 μg of kanamycin/ml at 37°C to an optical density at 600 nm of just greater than 0.5. They were then induced with 0.4 mM IPTG and grown for three more hours at 37°C before harvesting. The cells (50 g) from a 6-liter culture were resuspended in a buffer (200 ml) containing 25% sucrose and 250 mM Tris-HCl (80% cation, pH 7.5) and lysed by adding lysozyme (final concentration, 1.4 mg/ml). After 1 h of stirring at 4°C, EDTA (10 mM final concentration) was added and the cells were sonicated. The insoluble material was removed by centrifugation at 31,000 × g for 1 h at 4°C. The DNA and proteins were precipitated by adding 5% Polymin P (final concentration, 0.5% vol/vol) slowly to the supernatant. The lysate was stirred for 30 min at 4°C and centrifuged at 9,000 rpm for 15 min. The pellet was washed twice with R buffer (20 mM Tris-HCl, 80% cation [pH 7.5], 10% glycerol, 1 mM DTT) plus 50 mM ammonium sulfate, by resuspending it with stirring for 15 min at 4°C followed by centrifugation at 9,000 rpm for 20 min each time. Using the same procedure, the final pellet from the above wash steps was washed with R buffer plus 200 mM ammonium sulfate to elute the RecADr protein, followed by centrifugation at 14,000 rpm for 30 min. Ammonium sulfate (0.2002 g/ml) was added to the supernatant, and the precipitated proteins were removed by centrifugation. The pellet was discarded. Additional ammonium sulfate (0.145 g/ml of supernatant) was then added to precipitate the RecADr protein, followed by centrifuged at 14,000 rpm for 30 min. The pellet was washed twice with R buffer plus 0.377 g of ammonium sulfate/ml to remove as much Polymin P as possible and then dissolved in R buffer plus 200 mM ammonium sulfate and dialyzed against R buffer containing 50 mM KCl. The dialyzed solution was loaded at a flow rate of 150 ml/h onto a 60-ml DEAE-Sepharose column, equilibrated with R buffer plus 50 mM KCl. The column was washed with R buffer plus 50 mM KCl. Fractions containing D. radiodurans RecA protein were combined and dialyzed against P (350 mM) buffer containing 350 mM potassium phosphate (pH 7.5), 10% glycerol, 0.1 mM EDTA, and 1 mM DTT, and then loaded onto a 70-ml Bio-Gel hydroxyapatite column, equilibrated with the same P (350 mM) buffer. The RecA protein was eluted from the column with 2 volumes of P (500 mM phosphate) buffer. The individual protein fractions were checked for nuclease contamination by incubating fractions with φX174 RFI DNA, linear dsDNA, and circular ssDNA as described above. Fractions containing purified RecA protein were pooled and dialyzed against storage buffer (20 mM Tris-HCl, 80% cation [pH 7.5], 10% glycerol, 1 mM DTT), aliquoted, and stored at −80°C. Typical yields of the native protein were 0.3 to 0.5 mg per g of cells.
The extinction coefficient for RecADr protein was determined by a published procedure (30), modified as described previously (37). The results of three determinations were averaged to give an extinction coefficient of RecADr protein as ɛ280 = 0.372 A280 mg ml−1. The RecADr protein contains no tryptophan residues. N-terminal sequence analysis was carried out at the Protein and Nucleic Acid Chemistry Laboratories, Department of Molecular Biology and Pharmacology, Washington University School of Medicine, St. Louis, Mo., and was carried out with protein samples that were not identified to the sequencing laboratory personnel. Matrix-assisted laser desorption ionization (MALDI) mass spectrometry was carried out in the Mass Spectrometry Bioanalytical Facility at the University of Wisconsin Biotechnology Center.
DNA strand exchange reactions.
RecA-dependent DNA strand exchange reactions were carried out as described previously (3, 13), between circular ssDNA and the linear duplex DNA (derived from either φX174 or M13mp8). Unless otherwise stated, all reactions were carried out at 37°C in solutions containing 25 mM Tris-acetate (80% cation, pH 7.5), 1 mM DTT, 5% glycerol, 3 mM potassium glutamate, 10 mM (or the indicated concentration) magnesium acetate, and an ATP-regenerating system (10 U/ml of pyruvate kinase, 3.3 mM phosphoenolpyruvate, or 10 U of creatine kinase/ml, 12 mM phosphocreatine). DNA, SSB, ATP (or dATP), and RecA protein concentrations are indicated for each experiment. A preincubation of ssDNA with RecADr protein at 37°C for 5 min was followed by addition of ATP and SSB. After an additional 5-min incubation, linear duplex DNA was added to start the DNA strand exchange reactions. The order of addition is described in figure legends for each experiment in which it was changed. Aliquots (20 μl unless otherwise indicated) of strand exchange reactions described above were removed at each time point, and the reactions were stopped by addition of 5 μl of gel loading buffer (0.125% bromophenol blue, 25 mM EDTA, 25% glycerol, 5% sodium dodecyl sulfate [SDS]). These aliquots were stored on ice until after the last time point was taken. Samples were electrophoresed in an 0.8% agarose gel with TAE buffer (40 mM Tris-acetate [pH 8.0], 1 mM EDTA), stained with GelStar nucleic acid gel stain (FMC BioProducts) or ethidium bromide, and photographed with UV light using a gel document camera. The DNA bands were quantified with ImageQuant software (version 4.2). In order to correct for variability in sample loading onto the agarose gel, the band corresponding to full-length circular hybrid duplex product was quantified as the fraction of the total fluorescing DNA in a given gel lane, excluding only the band corresponding to the ssDNA.
ATPase assay.
ATP hydrolysis was monitored as previously described (3, 21, 42). All reactions were done at 37°C. ATP (or dATP) hydrolysis was measured by a coupled spectrophotometric assay. Absorbance measurements were obtained on a Cary 300 spectrophotometer equipped with two six-position, thermojacketed cuvette holders attached to a constant-temperature water circulator. Reaction mixtures contained 25 mM Tris-acetate (80% cation, pH 7.5), 1 mM DTT, 5% glycerol, 3 mM potassium glutamate, and 10 mM magnesium acetate. Concentrations of RecADr protein, DNA, SSB, and ATP (or dATP) are indicated in figure legends. Where the pH was varied, the buffer was a 25 mM concentration of either Tris-acetate or morpholineethanesulfonic acid-NaOH. An ATP regenerating system (3 mM phosphoenolpyruvate, 10 U of pyruvate kinase ml−1, and 3 mM potassium glutamate) and a coupling system (2 mM NADH and 10 U of lactate dehydrogenase ml−1) were also included. When dATP was used, the ATP regenerating and coupling enzymes were increased to 25 U of pyruvate kinase ml−1 and 60 U of lactate dehydrogenase ml−1. Reactions were initiated by the addition of ATP (or a mixture of ATP and SSB) after all other components were incubated at 37°C for 10 min. Changes in the order of addition are indicated in figure legends. Regeneration of ATP from ADP and phosphoenolpyruvate is coupled to the conversion of NADH to NAD+ (in the reaction catalyzed by lactate dehydrogenase), which can be monitored by a decrease in absorbance at 380 nm. Although the absorbance maximum for NADH occurs at 340 nm, absorbances were measured at 380 nm to remain within the linear absorbance range of the spectrophotometer for an extended length of time as required by these experiments. Rates of ATP hydrolysis (μM/min) were calculated from ΔA380 min−1 obtained at steady state, using an extinction coefficient of 1,210 M−1 cm−1 at 380 nm for NADH. In most cases, absorbances were continuously measured over a period of 2 h.
Electron-microscopic studies.
The formation of RecA filament onto circular ssDNA, linear ssDNA, and dsDNA was visualized by electron microscopy. Carbon films mounted on electron microscopy grids were activated with denatured bovine serum albumin (BSA) in the following way. A solution containing 0.1% BSA and 0.5% N-lauroyl sarcosine (sodium salt) was placed in a boiling water bath for 10 min. After cooling to 25°C, carbon grids were floated on drops of the denatured BSA for 5 min and then on drops of water followed by immersion in water (three times for 10 s). Grids were finally dried under a heat lamp and stored in a desiccator.
Reactions between RecADr and circular ssDNA or linear dsDNA (φX174) consisted of RecA*Dr (2 uM), HEPES (25 mM, pH 7.5), ssDNA or dsDNA (6 uM), creatine phosphokinase (10 U/ml), phosphocreatine (12 mM). After incubation for 10 min at 37°C, SSB and dATP were added (2 μM and 3.3 mM, respectively) followed by a further incubation for 1 min at 37°C. This was then immediately followed by the addition of γ-S-ATP (final concentration, 1 mM).
The above-described samples were immediately diluted 10-fold with 200 mM ammonium acetate plus 10% glycerol, adsorbed to denatured BSA-activated carbon grids for 3 min, washed with 200 mM ammonium acetate plus 10% glycerol, and stained with uranyl acetate (5%) plus 10% glycerol. Grids were then immersed in water for 10 s and dried. Grids were finally rotary shadowed with platinum at an angle of about 7 degrees.
RESULTS
Experimental rationale.
The goal of these experiments was straightforward. We wished to characterize the RecADr protein as part of a larger effort to discover the molecular basis of the extreme radioresistance of the bacterium D. radiodurans. These initial efforts focused on protein purification and an examination of classical RecA activities.
RecA gene cloning strategy.
Because very low-level expression of the RecADr gene in the absence of inducer (e.g., IPTG) was reported to be lethal to E. coli cells (20), we tried several different expression systems. We were eventually able to successfully express the RecADr protein in two ways, as a fusion protein with an intein at the C-terminal region (using the IMPACT-T7 system in E. coli) and as the native protein (in the pET21A vector). The pTYB2 plasmid vector is designed to create a fusion between the gene of interest (in this case D. radiodurans RecA), 55-kDa intein, and the DNA encoding a small 5-kDa chitin binding domain at the C terminus of the intein for efficient column affinity purification of the three-part fusion protein. The N terminus of intein undergoes a self-cleavage at low temperatures in the presence of thiols, such as DTT, β-mercaptoethanol, or cysteine (7, 8). The recADr gene flanking NdeI at the N terminus and SmaI at the C terminus was cloned into pTYB2 vector as described in Materials and Methods. In this case, the removal of the intein leaves one extra glycine residue at the C terminus of the RecADr protein. Although this protein promoted many RecA activities, as described below, the one extra glycine residue did appear to have modest effects on the function of the RecADr protein, as described in Results, and we refer to this protein as the RecA*Dr protein.
In cloning the native protein, we found an unusual number of point mutations among the isolated clones. These mutations included F229L, K245E, and A348V. Based on the previous reports of RecADr toxicity, it is possible that these changes inactivated the protein, but we did not follow up on them. We did isolate one clone that lacked sequence alterations and used this one for expression and purification (plasmid pEAW158). We do not know if a mutation elsewhere in the plasmid or in the host strain genome permitted the survival of this clone. However, it has remained stable (subsequent sequencing—twice over a period of more than a year—has revealed no new sequence changes) and expresses the RecADr protein at high levels after induction.
Purification of recombinant RecADr protein from E. coli.
In order to ascertain the biochemical properties of the D. radiodurans RecA protein, it was necessary to purify this protein to homogeneity. After expression and purification on a chitin column, the RecA-intein fusion was cleaved by DTT under conditions described by New England Biolabs. However, residual nucleases were still present in the preparation after these procedures were completed. Therefore, several additional purification steps were explored, with success achieved using a hydroxyapatite column, which efficiently removed the nuclease contamination. The native protein was purified with a combination of Polymin P precipitation and chromatography on DEAE-Sepharose and hydroxyapatite, a general strategy that we have employed successfully for a number of bacterial RecA proteins and RecA mutants.
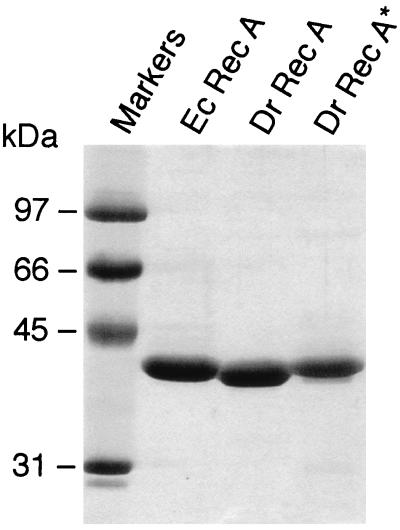
The purity of D. radiodurans RecA protein was checked by SDS-polyacrylamide gel electrophoresis after Coomassie blue staining (Fig. 1). D. radiodurans RecA* migrates somewhat more slowly than native RecADr, even though the two differ by only one glycine residue. Interestingly, the native RecADr protein migrated slightly faster than the E. coli RecA protein. Although RecADr is nine amino acids longer (20), the calculated Mr for native RecADr protein is 38,013 (361 amino acid residues excluding the initiating methionine), only slightly higher than the 37,842 calculated for the RecAEc protein (352 residues without the initiating Met). Direct N-terminal sequencing of the native RecADr protein purified from E. coli confirmed that the N-terminal Met residue was absent in the protein purified from E. coli, but otherwise it perfectly conformed to the expected RecADr sequence (SKDATKE). In addition, a sample of the native RecADr protein was subjected to MALDI mass spectrometry. This yielded a measured mass of 37,987 and 37,970 Da in two trials, 26 and 43 less than the calculated mass, respectively. These values are well within the error of the method (0.2%). For comparison, a sample of pure RecAEc protein was subjected to MALDI mass spectrometry in the same series of trials, yielding a mass of 37,802 Da (40 less than the calculated mass). The RecA*Dr protein was subjected to Western blotting along with the RecAEc protein, and both proteins reacted strongly with antibodies to the RecAEc protein (data not shown). We estimate that the RecADr protein preparations used in this study were more than 95% homogeneous, and all were free of detectable endo- and exonucleases.
FIG. 1.
SDS-polyacrylamide gel electrophoresis of purified RecA proteins. Lane 1 contains molecular size markers, as indicated. The RecADr and RecA*Dr proteins are compared, with the RecAEc protein included as a reference.
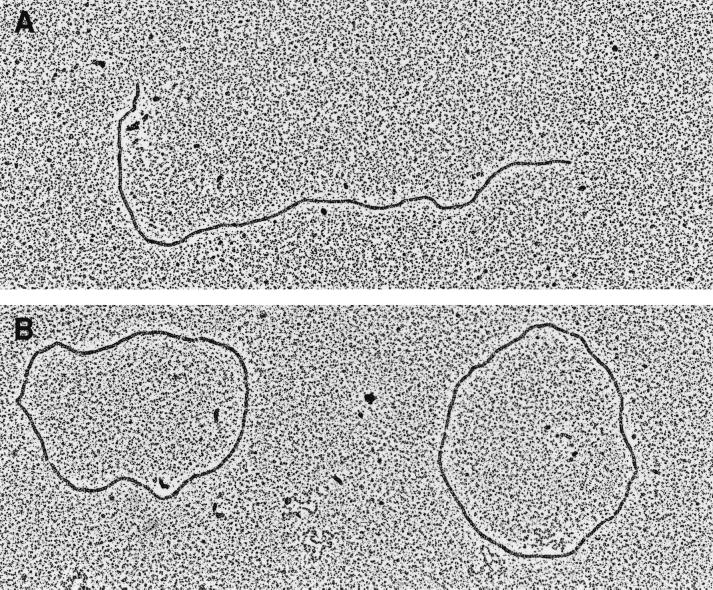
Electron microscopy. RecA*Dr protein was bound to circular ssDNA or linear dsDNA in the presence of γ-S-ATP, and the mixtures were spread and examined. The RecA*Dr protein formed filaments on both DNAs that were essentially indistinguishable from the filaments formed by the RecAEc protein (Fig. 2). The filaments had the characteristic striated appearance of normal RecA filaments, and the DNA was extended. When RecADr was incubated with linear dsDNA, as described in Materials and Methods, essentially all of the DNA was completely filamented by RecA*Dr (Fig. 2A). These experiments also suggested to us that the RecADr protein bound to dsDNA more readily than the RecAEc protein. Similar experiments with RecA*Dr protein bound to circular ssDNA are shown in Fig. 2B. A large proportion of the DNA molecules were completely filamented in these trials. A minor proportion of the filamented DNA had variable regions that are bound with SSB. Additionally, there was a minor proportion of ssDNA circles that were not filamented but rather bound with SSB (examples of the low-contrast and highly condensed, SSB-bound DNA circles can also be seen in Fig. 2B). There appeared to be a subtle increase in the clarity of the striations in the dsDNA complexes, relative to those on ssDNA, which was reproduced in several experiments.
FIG. 2.
Electron microscopy of the RecA*Dr protein bound to DNA derived from bacteriophage φX174. (A) RecADr filamented linear dsDNA. (B) RecADr filamented circular ssDNA. This view was chosen because it includes in the background, at lower contrast, several collapsed DNA circles bound with SSB.
ATPase activity.
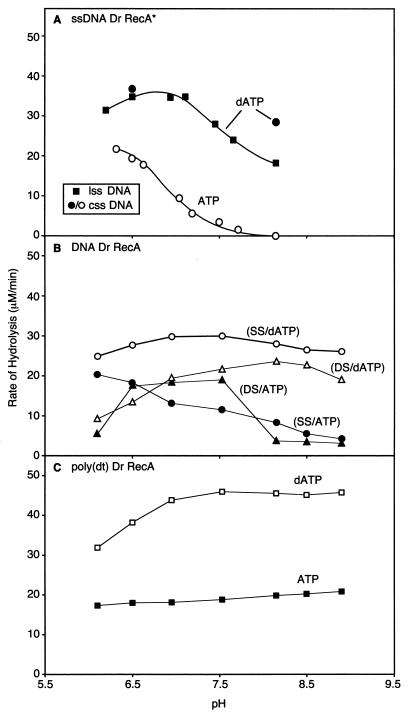
ATP hydrolysis was monitored with a coupled spectrophotometric assay (29, 42). As seen in Fig. 3, the RecA*Dr protein will catalyze the hydrolysis of either ATP or dATP. Using linear ssDNA as a cofactor, the dATP hydrolysis exhibits an apparent pH optimum between pHs 6.3 and 7.2. The decline above pH 7.2 is more gradual than would be expected if it reflected the simple ionization of an amino acid residue on the protein, and we therefore suspected that the decline might reflect a complex effect of pH on binding of RecADr to the linear DNA. The assembly and disassembly of the RecAEc protein is unidirectional (10), and there are multiple steps that could be affected by pH. While not exploring all of these parameters in detail, we carried out a few experiments with circular ssDNA. Here, nucleation of filament formation should result in a completely coated DNA (assuming the nucleation and extension of filament formation are similar to those of the RecAEc protein). At high pH, we observed that the rate of dATP hydrolysis increased when the circular DNA was used, suggesting that the inherent rate of dATPase was constant or nearly so over a broader range of high pHs.
FIG. 3.
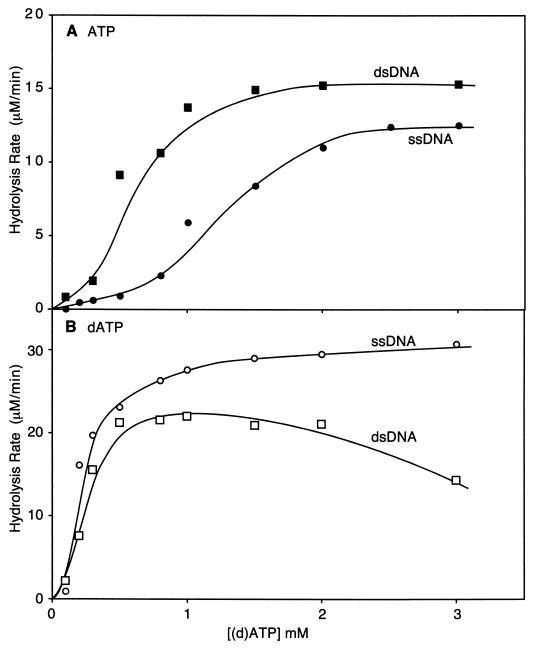
Effects of pH on the hydrolysis of ATP and dATP by the RecADr protein. Circular single-stranded (css or SS), linear single-stranded (lss), and linear double-stranded DNA from φX174 linearized with PstI restriction enzyme (DS) were used as DNA cofactors. (A) (d)ATP hydrolysis reactions of the RecA*Dr protein. Closed and open symbols represent dATP and ATP hydrolysis, respectively. Circles and squares represent reactions with circular and linear ssDNA, respectively. Reactions were carried out as described in Materials and Methods, and reaction mixtures contained 5 μM ssDNA, 3 μM RecA*Dr protein, 0.5 μM E. coli SSB protein, and a 3 mM concentration of either ATP or dATP. (B) Effect of pH on the DNA-dependent (d)ATP hydrolytic activities of the native RecADr protein. Reactions were carried out as described in Materials and Methods, and reaction mixtures contained 2 mM ATP (or dATP), 2 μM RecADr, 0.5 μM E. coli SSB protein, and 5 μM ssDNA (or 10 μM dsDNA). The E. coli SSB protein was omitted in the dsDNA-dependent reactions. (C) Reactions were the same as the ssDNA reactions in panel B, but poly(dT) was substituted for the ssDNA.
In contrast, very little hydrolysis of ATP was observed under the conditions normally used to monitor this activity with the RecAEc protein (pH 7.5). This result was obtained consistently in four trials. This property was reminiscent of those observed with the RecA protein from Bacillus subtilis, which also promoted the hydrolysis of dATP but not ATP (33). More recently it has been shown that modest adjustments of the reaction conditions would unmask ATPase activity in the B. subtilis RecA protein (52), and we decided to further investigate the use of ATP by the RecADr protein. A pH-rate profile showed that this RecA protein does hydrolyze ATP, but only at pHs below 7.5 (Fig. 3A).
A more extended set of experiments was carried out with the native RecADr protein (Fig. 3). The results with circular ssDNA and ATP were quite similar to those obtained with RecA*Dr, although there was somewhat more ATP hydrolysis detected at the higher pHs (Fig. 3B). For dsDNA, a pH optimum for ATP hydrolysis was observed between pHs 6.5 and 7.5, with a severe drop-off in rates above pH 7.5. Unlike RecAEc protein, the RecADr protein hydrolyzes ATP more rapidly on dsDNA than on ssDNA at pH 7.5. This provided another clue that the RecADr protein possessed an enhanced capacity to bind dsDNA relative to the RecAEc protein. For dATP hydrolysis, rates were generally a little faster, with a broader optimum between pHs 7 and 9 (Fig. 3B). The rates with ssDNA were obtained in the presence of the E. coli SSB protein. We also examined the pH rate profile of hydrolysis using poly(dT) as the DNA and without SSB. These rates were substantially higher for both ATP and dATP and exhibited a broader pH optimum (Fig. 3C). This could reflect some special property of poly(dT) as a cofactor, or an inhibitory effect of the SSB protein, and is explored further below.
The time course of NTP hydrolysis with a ssDNA cofactor was generally linear with time under these conditions (with excess RecADr protein), beginning a minute or so after the reaction was initiated (data not shown). With dsDNA as a cofactor, a lag was observed before the establishment of a steady state under most conditions (data not shown). For ATP, steady-state hydrolysis was observed with a lag of less than 3 min at pH 6.5. The lag increased to just over 13 min at pH 7.5. At pH 8.5 the lag disappeared but the rate slowed markedly. For dATP, the reaction at pH 6.5 was relatively slow but exhibited no evident lag. A slight lag and a higher rate appeared at pH 7.5, and a longer lag and a higher ultimate rate was seen at pH 8.5. In general, the RecADr protein appeared to bind to dsDNA faster than the E. coli RecA protein (45, 46) under many conditions, especially at intermediate pHs for ATP and higher pHs for dATP.
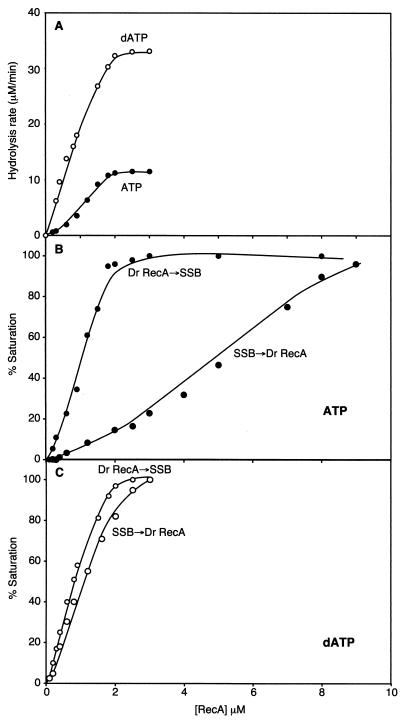
To learn more about the competition between E. coli SSB and RecADr protein, we carried out RecADr protein titrations using 6 μM circular ssDNA (Fig. 4), employing the native RecADr protein and the ATPase assay as an indirect measure of binding of RecADr protein to DNA. When the standard order of addition was observed [RecADr protein and DNA incubated together first, with SSB and (d)ATP added to start the reaction], the rates of hydrolysis for both ATP and dATP leveled off abruptly at an approximately 2 μM concentration of RecADr protein, suggesting a binding stoichiometry of one RecADr monomer per three nucleotides of ssDNA (Fig. 4A). This binding stoichiometry is the same as that observed for the RecAEc protein, and the transition in the binding curve is quite sharp. This suggests that the higher rates observed on poly(dT) (Fig. 3) reflect some special stimulatory property of the poly(dT) and do not indicate that the rate of hydrolysis on other DNA substrates is limited by competition with SSB.
FIG. 4.
Effect of native RecADr protein concentration on the rate of (d)ATP hydrolysis. (A) Reactions were carried out as described in Materials and Methods, and reaction mixtures contained 6 μM circular single-stranded φX174 DNA, 0.6 μM E. coli SSB, 2 mM ATP (or dATP), and the indicated concentration of native RecADr protein. Reactions carried out with the two different nucleotide cofactors are identified in the figure. (B) Order of addition effects in the presence of ATP. Reactions were carried out as for panel A, with 2 mM ATP. RecA protein was added to ssDNA, which was followed by addition of ATP and SSB (RecADr→SSB reactions), or SSB protein was added to ssDNA, which was followed by addition of ATP and RecA protein (SSB→RecADr reactions). (C) Reactions were carried out as for panel B, but with dATP replacing ATP.
The competition with SSB follows patterns reminiscent of the results reported for the RecAEc protein (39). When ATP is used as a cofactor, the addition of E. coli SSB prior to the RecADr protein has a strong inhibitory effect on the apparent binding of the RecADr protein to the ssDNA (Fig. 4B). This is similar to the effects seen with the RecAEc protein, in which SSB addition prior to RecA produces an inhibition of filament nucleation. However, if RecA is bound first, SSB facilitates complete RecA filament formation by eliminating the secondary structure before being displaced by RecA filament extension. We note that even when RecADr was added first, rates of ATP hydrolysis declined somewhat over time when the RecADr protein was present at subsaturating concentrations, suggesting that SSB could gradually displace the RecADr protein, as described below. The rates of ATP hydrolysis reported in Fig. 3B and 4 reflect the initial rate observed during the first 10 min of the reaction.
With dATP as a cofactor, the order of addition makes little difference, and both the rates and the transition point in the titration are similar (Fig. 4C). This suggests that the RecADr protein competes very well for ssDNA binding with E. coli SSB in the presence of dATP.
In similar titrations with the RecA*Dr protein in the presence of dATP, we did note some apparent inhibition of ssDNA binding by E. coli SSB when the two proteins were added at the same time. This was reflected in a titration curve that did not level off until twice as much RecA*Dr was present than should be needed to saturate the DNA (data not shown). Thus, the extra Gly residue in RecA*Dr may compromise somewhat the capacity of this mutant protein to compete with E. coli SSB.
Titrating with the nucleotide cofactor allowed an estimation of the S0.5 for ATP and dATP at pH 7.5 (Fig. 5). The S0.5 (point for half saturation) for ATP appears to be quite high (Fig. 5A). When RecADr protein is bound to ssDNA, the S0.5 is approximately 1.2 mM. On dsDNA, ATP is not only hydrolyzed faster, it exhibits a lower S0.5 of approximately 600 μM. The observed S0.5 values for dATP were lower, about 250 μM for both ssDNA and dsDNA. The S0.5 observed for the hydrolysis of dATP by RecA*Dr bound to ssDNA was nearly 1 mM, again suggesting that this form of the protein is somewhat compromised in some functions (data not shown).
FIG. 5.
Effect of (d)ATP concentration on RecADr-mediated and DNA-dependent (d)ATP hydrolysis. Reactions were carried out as described in Materials and Methods, and reaction mixtures contained 5 μM circular φX174 ssDNA (or 10 μM φX174 linear dsDNA), 2 μM native RecADr protein, 0.5 μM E. coli SSB, and the indicated concentration of ATP (A) or dATP (B).
DNA strand exchange.
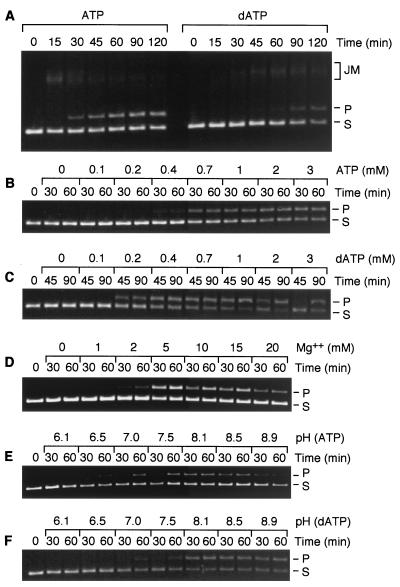
The RecADr protein will promote DNA strand exchange with either ATP or dATP (Fig. 6). However, the two nucleotide cofactors have different effects on the reactions. At pH 7.5, dATP is hydrolyzed two to three times faster than ATP (Fig. 3 and 4). Nevertheless, the DNA strand exchange promoted in the presence of ATP is much more efficient, reaching completion in a quarter or less of the time needed for the dATP-dependent reaction. Direct measurements of the ATP and dATPase activities during DNA strand exchange indicated that the slow reaction in the presence of dATP was not caused by a large decline in dATP hydrolysis (Table 1). For ATP, rates of hydrolysis actually increased somewhat during DNA strand exchange relative to the ssDNA-dependent ATPase rates. Essentially identical DNA strand exchange reactions were observed under these conditions for the RecA*Dr protein (data not shown). Thus, the apparently weaker competition of this RecADr variant with E. coli SSB and the higher S0.5 for dATP do not appear to affect the DNA strand exchange activities of that protein.
FIG. 6.
DNA strand exchange promoted by the RecADr protein. Reactions were carried out as described in Materials and Methods, and reaction mixtures contained 6 μM φX174 circular ssDNA, 12 μM φX174 linear dsDNA, 2 μM native RecADr protein, and 0.6 μM E. coli SSB. Reactions also contained 2 mM ATP (or dATP) unless otherwise indicated. Substrate DNA (φX174 linear dsDNA), product (nicked circular dsDNA), and reaction intermediates (joint molecules) are denoted as S, P, and JM, respectively. (A) Comparison of ATP and dATP reactions at pH 7.5. (B) Effects of ATP concentration. Reactions were carried out at pH 8.1. (C) Effects of dATP concentration. Reactions were carried out at pH 8.1. (D) Effect of Mg2+ concentration on DNA strand exchange. Reactions were carried out at pH 7.5. The standard reaction mixture described above and in Materials and Methods was altered by including only the indicated concentration of Mg(acetate)2. (E) Effects of pH on the ATP-dependent DNA strand exchange reaction. The standard reaction was altered by including buffers of the indicated pH. (F) Effects of pH on the dATP-dependent DNA strand exchange reaction. The standard reaction was altered by including buffers of the indicated pHs.
TABLE 1.
Rates of ATP (or dATP) hydrolysisa
| Activity and pHs | Rate of hydrolysis (μM/min)
|
||
|---|---|---|---|
| dsDNA-dependent | During strand exchange | ssDNA-dependent | |
| ATP hydrolysis | |||
| pH 7.5 | 15.5 | 15.7 | 11.5 |
| pH 8.1 | 3.6 | 10.2 | 8.3 |
| dATP hydrolysis | |||
| pH 7.5 | 16.9 | 21.5 | 30 |
| pH 8.1 | 20.3 | 26.9 | 28 |
Reaction conditions: 5 μM φX circular ssDNA, 10 μM φX linear dsDNA, 2 μM RecADr, 0.5 μM E. coli. SSB, and 2 mM ATP (or dATP).
Further characterization focused entirely on the native RecADr protein. The effects of pH, magnesium ion concentration, and (d)ATP concentration are also explored in Fig. 6. The effects of pH on strand exchange do not perfectly parallel the effects of pH on DNA-dependent ATP or dATP hydrolysis. With ATP, a good strand exchange reaction is seen between pHs 7 and 8.5. Even though ATP hydrolysis on ssDNA and dsDNA declines above pH 7.5 (Fig. 3), the rates during strand exchange are higher (Table 1). The hydrolysis of dATP is optimal or nearly so between pHs 7 and 9 (Fig. 3 and Table 1), but efficient DNA strand exchange is seen only at the higher pHs (8 and above). The optimal concentration of Mg2+ ions for the ATP-dependent DNA strand exchange reaction at pH 7.5 is between 5 and 15 mM (Fig. 6D). ATP supports DNA strand exchange at concentrations above 700 μM, while dATP is effective at concentrations above 200 μM. The (d)ATP titration of the DNA strand exchange reaction was done at pH 8.1, where both ATP and dATP promote a reaction. The requirements for these nucleotides are consistent with the higher S0.5 value observed for ATP in Fig. 5. At pH 7.5, where the ATP-mediated strand exchange reaction is much faster than the dATP reaction (Fig. 6A), the use of a mixture of ATP and dATP (1 mM each) slowed the strand exchange reaction by a factor of 2 relative to a reaction with 2 mM ATP (data not shown).
The effects of the E. coli SSB were explored at pH 8.1 (data not shown). With either nucleotide cofactor, SSB was extremely important to the yield of a strand exchange reaction. With ATP, essentially no reaction occurred with ATP unless SSB was also present. With dATP, some reaction was seen without SSB, but it was minimal. As with RecAEc protein, the order of addition was important. Addition of SSB before the RecADr protein nearly abolished the strand exchange reaction with ATP and slowed the reaction with dATP.
We also explored the capacity of the RecADr protein to promote DNA strand exchange through a heterologous insertion of 52 bp in the duplex DNA substrate. In these reactions, the φX174 DNAs used in most of the experiments were replaced by M13mp8 circular ssDNA and M13mp8.52 linear duplex DNA. The latter DNA contains a 52-bp heterologous insert in M13mp8 (23, 51) and is linearized so as to place the insertion near the center of the linear duplex DNA substrate. In the presence of ATP, the insertion slowed the strand exchange slightly, but an efficient reaction was observed nevertheless at both pHs 7.5 and 8.1 (data not shown). In contrast, dATP supported the formation of paired joint molecules at both pHs, but very little complete strand exchange product was seen. This suggests that the 52-bp insertion is a much more substantial barrier to reaction when dATP is used as the nucleotide cofactor.
Binding to dsDNA.
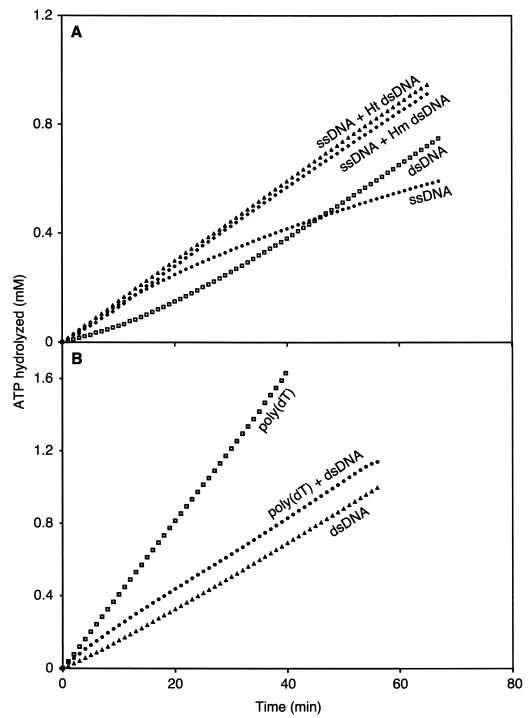
Several results in our initial characterization had suggested that the RecADr protein had an enhanced capacity to bind dsDNA relative to the RecAEc protein. We therefore examined the binding to ssDNA and dsDNA in reactions where both were present in the same test tube, using ATP as a cofactor at pH 7.5. The RecADr protein concentration was set at 1.4 μM, subsaturating relative to the 5 μM ssDNA or 10 μM dsDNA. The rates of ssDNA-dependent ATP hydrolysis (initial kcat = 8.7 min−1) declined slowly with time, probably as a result of SSB-mediated displacement as described above (Fig. 7A). The rates of dsDNA-dependent hydrolysis reached a steady state (kcat = 10.3 min−1) after a 15 min lag. When both dsDNA and ssDNA are included in the reaction mixture, with enough RecADr protein to nearly saturate one but not both, the rates of ATP hydrolysis were those characteristic of binding to dsDNA (Fig. 7A). This was true whether the dsDNA was homologous or heterologous. A lag in ATP hydrolysis was not observed when both DNAs were present.
FIG. 7.
The RecADr protein binds to dsDNA in preference to ssDNA. (A) Reactions were carried out at pH 7.5 and contained 1.4 μM RecADr protein, DNA as indicated (5 μM css φX174 DNA (ssDNA), 10 μM lds φX174 (Hm dsDNA or dsDNA), and/or 10 μM lds M13mp8 DNA (Ht dsDNA), 0.4 μM E. coli SSB, 2 mM ATP, and an ATP regenerating system. (B) Reactions were carried out at pH 8.1, and reaction mixtures contained 1.0 μM RecADr protein, DNA as indicated [4 μM poly(dT) and/or 8 μM lds M13mp8 DNA], no E. coli SSB, 2 mM dATP, and a dATP regenerating system.
To ensure that transfer to dsDNA was not compelled by the presence of the E. coli SSB protein, we carried out additional experiments without SSB and by substituting poly(dT) (which has no secondary structure) for the ssDNA (Fig. 7B). We also used dATP in this experiment. The poly(dT)-dependent dATP hydrolysis was substantially greater than that observed with dsDNA (see Fig. 3). Again, when the DNAs were mixed, the rates of hydrolysis were those characteristic of binding to dsDNA. The experiments of Fig. 7B were carried out at pH 8.1, where there is no evident lag in the direct binding of RecADr protein to dsDNA, as described above. The time course of dATP hydrolysis in the reaction including both DNAs appeared to start out at a higher rate before slowing to the dsDNA-dependent rate after 10 min, suggesting that ssDNA may be bound first and may prime the binding to dsDNA in some way.
DISCUSSION
We report here the purification and initial characterization of the RecA protein from the radioresistant bacterium D. radiodurans. In many respects, the protein has the properties one would expect of a close homologue of the E. coli RecA protein. It forms the striated filaments on DNA that are characteristic of RecA protein and hydrolyzes both ATP and dATP. It also promotes an efficient DNA strand exchange reaction. The contrasts come in the details, and several seem quite significant. The presence of dATP allows the RecADr protein to compete better with E. coli SSB for binding sites on ssDNA, and the S0.5 for dATP hydrolysis is much lower than the S0.5 for ATP hydrolysis. Nevertheless, dATP hydrolysis supports RecADr-mediated DNA strand exchange poorly under many conditions and only minimally supports DNA strand exchange through heterologous insertions in the duplex DNA substrate. ATP, which is hydrolyzed more slowly than dATP under most conditions, supports an efficient DNA strand exchange that readily bypasses heterologous insertions in the DNA substrates. Structural and mechanistic comparisons of these proteins may eventually yield clues to the molecular basis for the coupling between ATP hydrolysis and DNA strand exchange in this system. In addition, the RecADr protein appears to bind dsDNA much faster than does the RecAEc protein. More significantly, the RecADr protein binds preferentially to dsDNA in reactions that also contain ssDNA, under at least one set of reaction conditions. This is true even when the two DNAs are heterologous and cannot undergo DNA strand exchange.
The reactions of bacterial RecA proteins and their eukaryotic homologues are almost always greatly stimulated by ssDNA binding proteins. In many cases, there may be little species-specific protein-protein interaction. The SSBs of many organisms are often interchangeable, with the eukaryotic replication protein A (RPA) stimulating the strand exchange reactions of bacterial RecA proteins and bacterial SSB substituting well for RPA in stimulating the reactions of yeast Rad51 protein (1, 43). For our work on RecADr protein, we do not yet have the cognate D. radiodurans SSB protein and have relied on the E. coli SSB protein. There is a very large stimulation of RecADr-mediated DNA strand exchange by the E. coli SSB. However, we cannot eliminate the possibility that the SSB may alter some of these results, perhaps allowing for better strand exchange under some conditions where it was not observed here. This remains a goal of future research.
The work reported here provides a comparison of two different RecADr proteins, generated by different cloning procedures. The variant we have designated RecA*Dr has an extra glycine at the C terminus. In its capacity to form filaments on DNA and to promote DNA strand exchange, this protein appears to be very similar in its properties to the native RecADr protein isolated from E. coli. The only differences noted in the set of experiments done with this protein were an increased S0.5 for dATP hydrolysis (on ssDNA) and a reduced capacity of this protein to compete with E. coli SSB for ssDNA binding sites in the presence of dATP. The C-terminal domain of RecA protein protrudes from the RecA filament, and there is no evidence that it plays a role in monomer-monomer interactions in the filament (49, 53). However, the last 25 amino acid residues at the C terminus are not resolved in the published structure (53), and this protein segment could in principle fold back to interact with many parts of the RecA protein surface. The effects of this extra glycine at the C terminus may suggest an interaction or function of the C terminus that affects binding of RecA to both DNA and nucleotide cofactors.
Stole and Bryant (52a) have shown that combinations of wild-type or mutant RecAEc proteins and nucleotide cofactors with an S0.5 value of less than about 200 μM are generally unable to establish the active conformation needed for RecA-mediated DNA strand exchange. This does not seem to be the case for the RecADr protein. The S0.5 value for ATP hydrolysis at pH 7.5, when the RecADr protein is bound to an ssDNA, is more than 1 mM. However, a facile DNA strand exchange reaction is seen under these same conditions as long as the ATP concentration is high enough. Interestingly, the S0.5 value for ATP hydrolysis in the presence of dsDNA (about 600 μM) appears to correlate better with the ATP requirements seen in DNA strand exchange (Fig. 5A) than does the S0.5 value for ATP hydrolysis in the presence of ssDNA (about 1.2 mM).
The significance of some of the RecADr observations will be unclear until work is done to examine intracellular conditions in D. radiodurans. The few measurements that have been reported indicate that the normal concentration of dNTPs in an E. coli cell is generally less than 200 μM (44). The size of the nucleotide pools in D. radiodurans is not known. If they are similar in size to those in E. coli (and if the intracellular pH is above 7), then the RecADr protein may have a limited capacity to hydrolyze dATP under normal in vivo conditions. In view of the generally negative effects that dATP hydrolysis has on the DNA strand exchange reactions of the RecADr protein, one might expect that in vivo conditions would favor the hydrolysis of ATP. The relatively low levels of RecADr-mediated ATP hydrolysis at pH 7.5 is similar to the lack of ATP hydrolysis reported for the B. subtilis RecA protein (33), although more recent data indicates that ATP is hydrolyzed by the B. subtilis protein if the conditions are altered somewhat (52).
The RecADr protein promotes complete DNA strand exchange with full-sized DNA substrates derived from φX174 or M13mp8 with good efficiency. We presume that ATP binding and/or hydrolysis play a role in this reaction, since DNA strand exchange does not occur in the absence of NTP or dNTP cofactor. It has been established that the RecA protein and RecA homologues, such as the Rad51 protein, promote DNA pairing and strand exchange with little or no need for the hydrolysis of NTP cofactors (23, 24, 26, 38, 47, 51, 54). In the case of RecA protein, NTP hydrolysis may contribute a motor function that augments the inherent DNA strand exchange reaction, permitting it to move past DNA structural barriers, rendering it unidirectional, and allowing 4-strand exchange reactions to occur (3, 23, 24, 34, 50, 51). Such activities might be useful in a chromosomal repair capacity. This work provides a jumping-off point for a more complete analysis of the recombinational DNA repair processes in D. radiodurans and for further investigations into the role of ATP hydrolysis in RecA-mediated DNA strand exchange.
Acknowledgments
We thank Nirmala D. Sharma for technical assistance in the cloning, expression, and purification of recombinant RecA*Dr protein; Michael N. Flora of the USUHS Biomedical Instrumentation Center for oligonucleotide synthesis and DNA sequencing; and Maria Schnös and David Inman for assistance in electron microscopy.
This work was supported by grants GM32335-18 (from the National Institutes of Health; to M.M.C.), DE-FG02-98ER6283 (from the Microbial Genome Program, U.S. Department of Energy; to M.J.D.), FG02-97ER62492 and FG07-97ER20293 (from the DOE; to M.J.D.), GM39933-09 (from the National Institutes of Health; to M.J.D.), and GM14711-33 (from the National Institutes of Health; to R.B.I.).
Jong-Il Kim and Ajay K. Sharma contributed equally to this work.
REFERENCES
- 1.Alani, E., R. Thresher, J. D. Griffith, and R. D. Kolodner. 1992. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J. Mol. Biol. 227:54-71. [DOI] [PubMed] [Google Scholar]
- 2.Battista, J. R., A. M. Earl, and M. J. Park. 1999. Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol. 7:362-365. [DOI] [PubMed] [Google Scholar]
- 3.Bedale, W. A., and M. Cox. 1996. Evidence for the coupling of ATP hydrolysis to the final (extension) phase of RecA protein-mediated DNA strand exchange. J. Biol. Chem. 271:5725-5732. [DOI] [PubMed] [Google Scholar]
- 4.Bianco, P. R., R. B. Tracy, and S. C. Kowalczykowski. 1998. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 3:560-603. [Online.] http//www.bioscience.org/. [DOI] [PubMed]
- 5.Brim, H., S. C. McFarlan, J. K. Fredrickson, K. W. Minton, M. Zhai, L. P. Wackett, and M. J. Daly. 2000. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat. Biotechnol. 18:85-90. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, J. D., M. J. Daly, and K. W. Minton. 1996. Expression of recA in Deinococcus radiodurans. J. Bacteriol. 178:130-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chong, S., F. B. Mersha, D. G. Comb, M. E. Scott, D. Landry, L. M. Vence, F. B. Perler, J. Benner, R. B. Kucera, C. A. Hirvonen, J. J. Pelletier, H. Paulus, and M. Q. Xu. 1997. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192:271-281. [DOI] [PubMed] [Google Scholar]
- 8.Chong, S., Y. Shao, H. Paulus, J. Benner, F. B. Perler, and M. Q. Xu. 1996. Protein splicing involving the Saccharomyces cerevisiae VMA intein. The steps in the splicing pathway, side reactions leading to protein cleavage, and establishment of an in vitro splicing system. J. Biol. Chem. 271:22159-22168. [DOI] [PubMed] [Google Scholar]
- 9.Cox, M. M. 2001. Historical overview: searching for replication help in all of the rec places. Proc. Natl. Acad. Sci. USA. 98:8173-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, M. M. 1999. Recombinational DNA repair in bacteria and the RecA protein. Prog. Nucleic Acids Res. Mol. Biol. 63:310-366. [DOI] [PubMed] [Google Scholar]
- 11.Cox, M. M. 2001. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 35:53-82. [DOI] [PubMed]
- 12.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 13.Cox, M. M., and I. R. Lehman. 1981. RecA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc. Natl. Acad. Sci. USA 78:3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly, M. J. 2000. Engineering radiation-resistant bacteria for environmental biotechnology. Curr. Opin. Biotechnol. 11:280-285. [DOI] [PubMed] [Google Scholar]
- 15.Daly, M. J., and K. W. Minton. 1996. An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 178:4461-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daly, M. J., and K. W. Minton. 1995. Interchromosomal recombination in the extremely radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 177:5495-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daly, M. J., and K. W. Minton. 1995. Resistance to radiation. Science 270:1318.. [DOI] [PubMed] [Google Scholar]
- 18.Daly, M. J., L. Ouyang, P. Fuchs, and K. W. Minton. 1994. In vivo damage and recA-dependent repair of plasmid and chromosomal DNA in the radiation-resistant bacterium Deinococcus radiodurans. J. Bacteriol. 176:3508-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira, A. C., M. F. Nobre, F. A. Rainey, M. T. Silva, R. Wait, J. Burghardt, A. P. Chung, and M. S. da Costa. 1997. Deinococcus geothermalis sp. nov. and Deinococcus murrayi sp. nov., two extremely radiation-resistant and slightly thermophilic species from hot springs. Int. J. Syst. Bacteriol. 47:939-947. [DOI] [PubMed] [Google Scholar]
- 20.Gutman, P. D., J. D. Carroll, C. I. Masters, and K. W. Minton. 1994. Sequencing, targeted mutagenesis and expression of a recA gene required for the extreme radioresistance of Deinococcus radiodurans. Gene 141:31-37. [DOI] [PubMed] [Google Scholar]
- 21.Iype, L. E., E. A. Wood, R. B. Inman, and M. M. Cox. 1994. RuvA and RuvB proteins facilitate the bypass of heterologous DNA insertions during RecA protein-mediated DNA strand exchange. J. Biol. Chem. 269:24967-24978. [PubMed] [Google Scholar]
- 22.Karlin, S., and J. Mrazek. 2001. Predicted highly expressed and putative alien genes of Deinococcus radiodurans and implications for resistance to ionizing radiation damage. Proc. Natl. Acad. Sci. USA 98:5240-5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J. I., M. M. Cox, and R. B. Inman. 1992. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. I. Bypassing a short heterologous insert in one DNA substrate. J. Biol. Chem. 267:16438-16443. [PubMed] [Google Scholar]
- 24.Kim, J. I., M. M. Cox, and R. B. Inman. 1992. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. II. Four-strand exchanges J. Biol. Chem. 267:16444-16449. [PubMed] [Google Scholar]
- 25.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 26.Kowalczykowski, S. C., and R. A. Krupp. 1995. DNA-strand exchange promoted by RecA protein in the absence of ATP: implications for the mechanism of energy transduction in protein-promoted nucleic acid transactions. Proc. Natl. Acad. Sci. USA 92:3478-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange, C. C., L. P. Wackett, K. W. Minton, and M. J. Daly. 1998. Engineering a recombinant Deinococcus radiodurans for organopollutant degradation in radioactive mixed waste environments. Nat. Biotechnol. 16:929-933. [DOI] [PubMed] [Google Scholar]
- 28.Lin, J. Y., R. Qi, C. Aston, J. P. Jing, T. S. Anantharaman, B. Mishra, O. White, M. J. Daly, K. W. Minton, J. C. Venter, and D. C. Schwartz. 1999. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science 285:1558-1562. [DOI] [PubMed] [Google Scholar]
- 29.Lindsley, J. E., and M. M. Cox. 1990. Assembly and disassembly of RecA protein filaments occurs at opposite filament ends: relationship to DNA strand exchange. J. Biol. Chem. 265:9043-9054. [PubMed] [Google Scholar]
- 30.Lohman, T. M., K. Chao, J. M. Green, S. Sage, and G. T. Runyon. 1989. Large-scale purification and characterization of the Escherichia coli rep gene product. J. Biol. Chem. 264:10139-10147. [PubMed] [Google Scholar]
- 31.Lohman, T. M., J. M. Green, and R. S. Beyer. 1986. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under λ PL control. Biochemistry 25:21-25. [DOI] [PubMed] [Google Scholar]
- 32.Lohman, T. M., and L. B. Overman. 1985. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 260:3594-3603. [PubMed] [Google Scholar]
- 33.Lovett, C. M. J., and J. W. Roberts. 1985. Purification of a RecA protein analogue from Bacillus subtilis. J. Biol. Chem. 260:3305-3313. [PubMed] [Google Scholar]
- 34.MacFarland, K. J., Q. Shan, R. B. Inman, and M. M. Cox. 1997. RecA as a motor protein. Testing models for the role of ATP hydrolysis in DNA strand exchange. J. Biol. Chem. 272:17675-17685. [DOI] [PubMed] [Google Scholar]
- 35.Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton, E. V. Koonin, and M. J. Daly. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makarova, K. S., Y. I. Wolf, O. White, K. Minton, and M. J. Daly. 1999. Short repeats and IS elements in the extremely radiation-resistant bacterium Deinococcus radiodurans and comparison to other bacterial species. Res. Microbiol. 150:711-724. [DOI] [PubMed] [Google Scholar]
- 37.Marrione, P. E., and M. M. Cox. 1995. RuvB protein-mediated ATP hydrolysis: functional asymmetry in the RuvB hexamer. Biochemistry 34:9809-9818. [DOI] [PubMed] [Google Scholar]
- 38.Menetski, J. P., D. G. Bear, and S. C. Kowalczykowski. 1990. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl. Acad. Sci. USA 87:21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menetski, J. P., and S. C. Kowalczykowski. 1989. Enhancement of Escherichia coli RecA protein enzymatic function by dATP. Biochemistry 28:5871-5881. [DOI] [PubMed] [Google Scholar]
- 40.Minton, K. W. 1994. DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol. Microbiol. 13:9-15. [DOI] [PubMed] [Google Scholar]
- 41.Minton, K. W. 1996. Repair of ionizing-radiation damage in the radiation resistant bacterium Deinococcus radiodurans. Mutat. Res. 363:1-7. [DOI] [PubMed] [Google Scholar]
- 42.Morrical, S. W., J. Lee, and M. M. Cox. 1986. Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of RecA protein and single-stranded DNA. Biochemistry 25:1482-1494. [DOI] [PubMed] [Google Scholar]
- 43.Namsaraev, E. A., and P. Berg. 2000. Rad51 uses one mechanism to drive DNA strand exchange in both directions. J. Biol. Chem. 275:3970-3976. [DOI] [PubMed] [Google Scholar]
- 44.Neuhard, J., and P. Nygaard. 1987. Purines and pyrimidines, p. 445-473. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 45.Pugh, B. F., and M. M. Cox. 1988. General mechanism for RecA protein binding to duplex DNA. J. Mol. Biol. 203:479-493. [DOI] [PubMed] [Google Scholar]
- 46.Pugh, B. F., and M. M. Cox. 1987. Stable binding of RecA protein to duplex DNA. Unraveling a paradox. J. Biol. Chem. 262:1326-1336. [PubMed] [Google Scholar]
- 47.Rehrauer, W. M., and S. C. Kowalczykowski. 1993. Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli recA protein attenuates NTP hydrolysis but not joint molecule formation. J. Biol. Chem. 268:1292-1297. [PubMed] [Google Scholar]
- 48.Richmond, R. C., R. Sridhar, and M. J. Daly. 1999. Physicochemical survival pattern for the radiophile Deinococcus radiodurans: a polyextremophile model for life on Mars. SPIE 3755:210-222.
- 49.Roca, A. I., and M. M. Cox. 1997. RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nucleic Acid Res. Mol. Biol. 56:129-223. [DOI] [PubMed] [Google Scholar]
- 50.Shan, Q., and M. M. Cox. 1998. On the mechanism of RecA-mediated repair of double-strand breaks: no role for four-strand DNA pairing intermediates. Mol. Cell 1:309-317. [DOI] [PubMed] [Google Scholar]
- 51.Shan, Q., M. M. Cox, and R. B. Inman. 1996. DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. J. Biol. Chem. 271:5712-5724. [DOI] [PubMed] [Google Scholar]
- 52.Steffen, S. E., and F. R. Bryant. 1999. Reevaluation of the nucleotide cofactor specificity of the RecA protein from Bacillus subtilis. J. Biol. Chem. 274:25990-25994. [DOI] [PubMed] [Google Scholar]
- 52a.Stole, E., and F. R. Bryant. 1995. Spectroscopic demonstration of a linkage between the kinetics of NTP hydrolysis and the conformational state of the RecA-single-stranded DNA complex. J. Biol. Chem. 270:20322-20328. [DOI] [PubMed] [Google Scholar]
- 53.Story, R. M., I. T. Weber, and T. A. Steitz. 1992. The structure of the E. coli RecA protein monomer and polymer. Nature 355:318-325. [DOI] [PubMed] [Google Scholar]
- 54.Sung, P., and S. A. Stratton. 1996. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271:27983-27986. [DOI] [PubMed] [Google Scholar]
- 55.Venkateswaran, A., S. C. McFarlan, D. Ghosal, K. W. Minton, A. Vasilenko, K. Makarova, L. P. Wackett, and M. J. Daly. 2000. Physiologic determinants of radiation resistance in Deinococcus radiodurans. Appl. Environ. Microbiol. 66:2620-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Y. Qin, L. X. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, C. M. Fraser, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]