Abstract
The bchP gene product of Rhodobacter sphaeroides is responsible for the reduction of the isoprenoid moiety of bacteriochlorophyll (Bchl) from geranylgeraniol (GG) to phytol; here, we show that this enzyme also catalyzes the reduction of the isoprenoid moiety of bacteriopheophytin (Bphe). In contrast, we demonstrate that a newly identified homolog of this gene in Rhodospirillum rubrum encodes an enzyme, GG-Bphe reductase, capable of reducing the isoprenoid moiety of Bphe only. We propose that Rhodospirillum rubrum is a naturally occurring bchP mutant and that an insertion mutation may have been the initial cause of a partial loss of function. Normal BchP function can be restored to Rhodospirillum rubrum, creating a new transconjugant strain possessing Bchl esterified with phytol. We speculate on the requirement of Rhodospirillum rubrum for phytylated Bphe and on a potential link between the absence of LH2 and of phytylated Bchl from the wild-type bacterium. The identification of a second role for the fully functional BchP in catalyzing the synthesis of phytylated Bphe strongly suggests that homologs of this enzyme may be similarly responsible for the synthesis of phytylated pheophytin in organisms possessing photosystem 2. In addition to bchP, other members of a photosynthesis gene cluster were identified in Rhodospirillum rubrum, including a bchG gene, demonstrated to encode a functional Bchl synthetase by complementation of a Rhodobacter sphaeroides mutant.
Purple photosynthetic bacteria contain two types of antenna complex, responsible for the efficient transfer of light energy to the reaction center (RC), the site of charge separation: light-harvesting complex 1 (LH1) surrounds the RC (35, 36), whereas light-harvesting complex 2 (LH2) is on the periphery of the photosynthetic unit and donates excitation energy to LH1 (12). Both complexes comprise multiple copies of two integral membrane polypeptides, termed α and β, to which bacteriochlorophyll (Bchl) and carotenoid are bound (8). However, not all purple photosynthetic bacteria have the same complement of antenna complexes; for example, Rhodospirillum rubrum lacks LH2.
The LH1 of Rhodospirillum rubrum is the best characterized of any such complex, with a projection map at 8.5 Å resolution generated from electron microscopy studies (19). This revealed a ring structure composed of 16 αβ heterodimers, the central hole of which is now known to be occupied by the RC (35). The Bchla with which these complexes are assembled is esterified with the C20 isoprenoid alcohol geranylgeraniol (GG) (20). In this respect, Rhodospirillum rubrum again differs from other purple bacteria, including the well-characterized Rhodobacter species Rhodobacter sphaeroides and Rhodobacter capsulatus. In these species at least, the major alcohol component of the Bchl pigment is phytol (6, 7), which is the commonest esterifying alcohol of chlorophylls and Bchls (32). Like GG, phytol is a C20 isoprenoid alcohol, but phytol differs in being more highly saturated. It appears that a GG moiety is a biosynthetic precursor of a phytol moiety in both of these Rhodobacter species and that the product of a single gene is responsible for the three necessary reduction steps (3, 6). This gene, bchP, encodes the enzyme GG-Bchl reductase and resides among other photosynthesis-related genes in a photosynthesis gene cluster (PGC) (4, 25).
Our previous work has indicated that although both LH1 and LH2 assemble with GG-esterified Bchl in place of the usual phytylated pigment in a bchP mutant of Rhodobacter sphaeroides, LH2 is severely affected by this change, while LH1 is not (3). It is possible that the absence of LH2 and phytylated Bchl (BchlaP) from wild-type Rhodospirillum rubrum may be linked. Homologs of bchP have been found in cyanobacteria and plants, in which the gene product has been identified as the catalyst for the reduction of the isoprenoid moiety of chlorophyll (Chl) (2, 21).
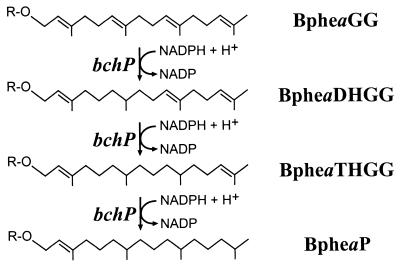
In addition to Bchl, the RC of purple bacteria contains two bacteriopheophytin (Bphe) molecules, one of which is the first clearly resolved acceptor of electrons, following electron transfer from the Bchl dimer (see reference 13 for a review). This situation is mirrored in higher photosynthetic organisms, in which pheophytin serves an identical role within photosystem 2 (PS2) (30). Although the Bchl of Rhodospirillum rubrum is esterified with GG and not with phytol, it has previously been reported that the Bphe of this organism is esterified with phytol, as it is in Rhodobacter sphaeroides (11). This discrepancy implied that an enzyme capable of reducing the isoprenoid moiety of BpheaGG but not that of BchlaGG existed in Rhodospirillum rubrum (Fig. 1); such an enzyme might be expected to show some homology with GG-Bchl reductase.
FIG. 1.
Proposed pathway for reduction of BpheaGG in Rhodospirillum rubrum. The sequence of the hydrogenation reactions is based on that which occurs during Chl biosynthesis in greening plants (33). Reduction proceeds via dihydro-GG-esterified (BpheaDHGG) and tetrahydro-GG-esterified (BpheaTHGG) intermediates to the final product, phytylated Bphe (BpheaP). R represents the tetrapyrrole-derived moiety of Bphe.
We searched for and found a Rhodospirillum rubrum homolog of bchP and demonstrated that the gene product does have the ability to reduce the isoprenoid moiety of Bphe but not that of Bchl. In contrast, it was demonstrated that the Rhodobacter sphaeroides bchP gene product catalyzes the reduction of both pigment types. We propose that the homologs of bchP in PS2-containing organisms are similarly responsible for the reduction of the isoprenoid moieties of both Chl and pheophytin.
Unequivocal proof of the difference in the activities of the Rhodobacter sphaeroides and Rhodospirillum rubrum bchP gene products was provided by introducing the Rhodobacter sphaeroides bchP gene into Rhodospirillum rubrum, generating a novel transconjugant strain of Rhodospirillum rubrum in which the Bchls are esterified with phytol. We propose that Rhodospirillum rubrum is a naturally occurring bchP mutant and that an insertion mutation may have been the initial cause of a partial loss of function. A gene sited close to bchP in Rhodospirillum rubrum has been demonstrated to encode a functional Bchl synthetase, the enzyme responsible for the addition of the isoprenoid moiety to bacteriochlorophyllide (1, 6, 27).
MATERIALS AND METHODS
Isolation of a bchP homolog and other members of a PGC from Rhodospirillum rubrum.
Degenerate oligonucleotides, designed according to the regions of derived amino acid similarity between the Rhodobacter capsulatus bchP gene and an expressed sequence tag from Arabidopsis thaliana (2), were used to PCR amplify a 278-bp fragment from Rhodospirillum rubrum genomic DNA. This was cloned and sequenced, and the deduced amino acid sequence was found to display significant similarity to each of the original sequences and to other GG-(B)chl reductase genes.
The PCR fragment was radiolabeled and used to probe a partial genomic library of Rhodospirillum rubrum constructed in the pBluescript II SK+ phagemid (Stratagene), resulting in the isolation of overlapping KpnI and NotI-EcoRI fragments spanning 9.3 kb of the genome. DNA was sequenced using an ABI 373A sequencer. Sequence analysis was performed mainly with Lasergene software (DNAStar), with additional use of the European Molecular Biology Open Software Suite (EMBOSS) at the Sanger Centre, Cambridge, United Kingdom. For codon usage analysis, a codon usage table was created using 43 miscellaneous Rhodospirillum rubrum gene sequences.
Growth of Rhodobacter sphaeroides and Rhodospirillum rubrum.
Rhodobacter sphaeroides and Rhodospirillum rubrum strains were grown in M22+ medium (16). Rhodobacter sphaeroides was grown semiaerobically in the dark at 34°C; Rhodospirillum rubrum was grown photosynthetically at low light intensity (3 W/m2) at 20°C. Antibiotic concentrations were 20 μg/ml for neomycin, 5 μg/ml for streptomycin, and 1 μg/ml for tetracycline. To overcome problems of phototoxicity, tetracycline was substituted with 250 ng of doxycycline per ml in photosynthetically grown cultures.
Spectroscopic analysis of intracytoplasmic membrane.
Cells were disrupted, and cell-free intracytoplasmic membrane fractions were isolated as described previously (26). Absorbance spectra were recorded on a Beckman DU640 spectrophotometer.
Mutagenesis of bchP in Rhodobacter sphaeroides mutants DBCΩ and DPF2.
Plasmid pSCN6-G:T6G5, which carries the Tn5-mutated Rhodobacter sphaeroides bchP gene (3), was transferred into the genomes of strains DBCΩ (18) and DPF2 (15) by conjugation and homologous recombination (9, 34), generating mutants DBCΩ:T6G5 and DPF2:T6G5, respectively. DBCΩ carries a streptomycin resistance gene in place of the pucBA genes, and so is LH2−; DPF2 has LH2 only. Dot blots showing the absence of the parental vector, pSUP202 (34), confirmed that homologous recombination had occurred.
Complementation of Rhodobacter sphaeroides Bchl biosynthesis mutants.
The oligonucleotides 5′-CATCTAGAGGAGACGACCATATGCAACGGACCGCAGTTCTGC-3′ and 5′-CTGCAAGCTTGGATCCTCATGGGGCCGCCCCGATC-3′ were used to PCR amplify the Rhodospirillum rubrum bchG gene and introduce XbaI and HindIII sites for cloning into pRKSK1 (14), which provides gene expression under the transcriptional control of the Rhodobacter sphaeroides puc operon promoter, yielding plasmid pSK1RRbchG. Similarly, the oligonucleotides 5′-GGTCTAGAGGAGACGACCATATGGACGCCGTCGTCGCCAC-3′ and 5′-CGCGAAGCTTGGATCCTCAAGACTTGGCGATGCCGA-3′ permitted cloning of the Rhodospirillum rubrum bchP homolog into pRKSK1 to give plasmid pSK1RRbchP. Additionally, the first bchP oligonucleotide was used with the oligonucleotide 5′-TCGCTGCAGCACCGAGCTGGTCGGGGCGCGGATGATTTCGTGA-3′, and the second with 5′-CCCCTGCAGTATGACCCGGCGCGCTGCGA-3′, to separately amplify the 5′ and 3′ ends of Rhodospirillum rubrum bchP and to allow the fusion of the two sections via a PstI site encoded within the new oligonucleotides, creating modified gene bchPΔx. This was cloned into pRKSK1, yielding plasmid pSK1RRbchPΔx.
Another pair of pRKSK1 derivatives was made, each with an insert consisting partly of Rhodospirillum rubrum bchP and partly of Rhodobacter sphaeroides bchP. The Rhodospirillum rubrum fragments were as in plasmid pSK1RRbchPΔx. Equivalent fragments of Rhodobacter sphaeroides bchP were amplified by PCR using existing oligonucleotides designed for amplification of the entire coding region (3) in conjunction with two new oligonucleotides, 5′-CATCTGCAGCACCGAGCTGGTCGGCGGCT-3′ and 5′-CACATGCATTACGACCCTACCCGCTGCGAC-3′. The former of these encodes a PstI site; the latter, an NsiI site, which gives PstI-compatible ends. The 5′ end of Rhodobacter sphaeroides bchP was fused to the 3′ end of Rhodospirillum rubrum bchP, generating plasmid pSK1bchPS/PR. Similarly, the 5′ end of Rhodospirillum rubrum bchP and the 3′ end of Rhodobacter sphaeroides bchP yielded pSK1bchPR/PS. As a positive control, the two Rhodobacter sphaeroides bchP fragments were also fused in plasmid pSK1bchPS/PS. Each pRKSK1 derivative was transformed into Escherichia coli strain S17-1 (34) for conjugative transfer into Rhodobacter sphaeroides.
HPLC of pigments.
The Bchls of Rhodobacter sphaeroides and Rhodospirillum rubrum strains were extracted and analyzed as described previously (3), which also permitted identification of Bphe pigments. Elution times were approximately 22.6 min for BpheaGG and 27.4 min for phytylated Bphea (BpheaP). High-pressure liquid chromatography (HPLC) standards of these two pigments were prepared from BchlaGG and BchlaP extracted from Rhodobacter sphaeroides strains DPF2:T6G5 and DPF2, respectively, by the method of Perkins and Roberts (28). These strains were used because they lack RCs and are thus devoid of native Bphe.
Heterologous expression of Rhodobacter sphaeroides bchP in Rhodospirillum rubrum.
Plasmid pSK1bchP (3) is a pRKSK1-based expression plasmid for Rhodobacter sphaeroides bchP. Saegesser et al. (31) have demonstrated the suitability of RK2-based vectors, of which pRKSK1 (14) is an example, for the expression of genes in Rhodospirillum rubrum. Here, both pSK1bchP and pRKSK1 were introduced into Rhodospirillum rubrum wild-type strain S1 by conjugation, as for Rhodobacter sphaeroides.
Nucleotide sequence accession number.
The Rhodospirillum rubrum DNA sequence encompassing bchP has been assigned EMBL accession number AJ310779.
RESULTS
Identification of a bchP homolog and other members of a PGC within the genome of Rhodospirillum rubrum.
A 278-bp stretch of DNA was isolated from Rhodospirillum rubrum genomic DNA by heterologous PCR. This bore strong homology in its deduced amino acid sequence to GG-(b)chl reductases from photosynthetic bacteria and plants. Subsequently, approximately 9 kb of flanking DNA was sequenced; putative open reading frames (ORFs), conforming to a compiled codon usage table for Rhodospirillum rubrum genes, were identified through homology with genes present in the PGC of Rhodobacter sphaeroides (25).
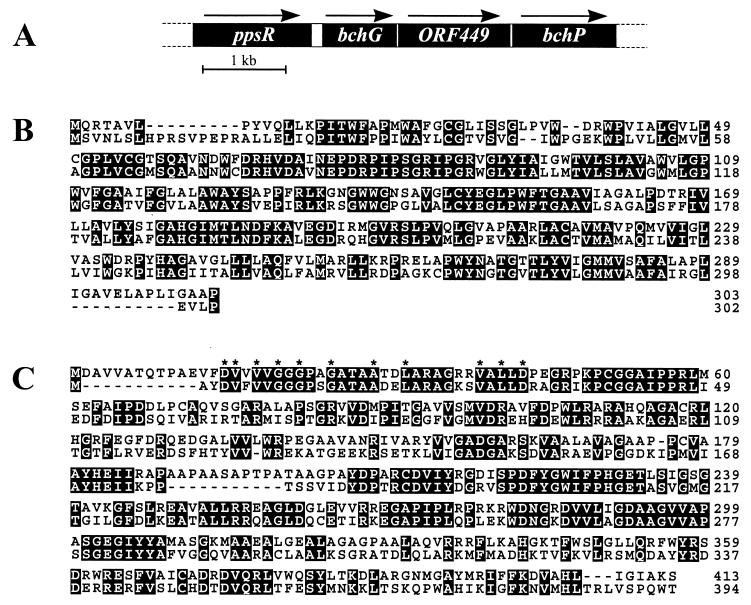
In addition to the bchP homolog, there are equivalents of ppsR, which is involved in the repression of pigment biosynthesis; bchG, which encodes GG-Bchl synthetase; and ORF427, which may encode an assembly factor for photosynthetic complexes. The Rhodospirillum rubrum homolog of ORF427 is ORF449 (designated ORF1 in the sequence accession) (Fig. 2A). The sequence identity between the predicted bchG gene products of Rhodospirillum rubrum and Rhodobacter sphaeroides is 58.8% (Fig. 2B). In both Rhodobacter sphaeroides and Rhodobacter capsulatus, bchG, ORF427 (ORF428 in Rhodobacter capsulatus), and bchP are members of a more extensive operon (4); upstream of bchG, this operon includes two further Bchl biosynthesis genes, bchE and bchJ, while immediately downstream of bchP lies an isopentenyl diphosphate isomerase gene (25).
FIG. 2.
(A) Physical map of part of a Rhodospirillum rubrum PGC. Arrows indicate the direction of transcription of the labeled genes. (B) Deduced amino acid sequence alignment between Rhodospirillum rubrum bchG (top) and Rhodobacter sphaeroides bchG (bottom). (C) Deduced amino acid sequence alignment between Rhodospirillum rubrum bchP (top) and Rhodobacter sphaeroides bchP (bottom). Asterisks above the alignment mark the positions of significant amino acids within a putative ADP-binding βαβ fold (37).
In each of these species, ppsR (ORF469 in Rhodobacter capsulatus) is sited upstream of bchE. Downstream of bchP in Rhodospirillum rubrum we have identified a partial ORF, putatively encoding a component of the sensory apparatus of this organism. The product of this ORF shows strong homology to Ptr, a component of the photosensory apparatus of Rhodospirillum centenum, but only in a putative cytoplasmic output domain (17); the likely periplasmic sensory domain is more homologous to those of chemoreceptors, such as Rhodobacter capsulatus McpA (24). Between bchP and this partial ORF lies a complete ORF of unknown function, transcribed in the opposite direction.
The predicted gene product of Rhodospirillum rubrum bchP has 52.5% sequence identity with Rhodobacter sphaeroides BchP (Fig. 2C) and shows strong homology throughout virtually its entire length to this and other known GG-(b)chl reductase enzymes. In common with these enzymes, close to the N terminus it aligns with an amino acid fingerprint predicting the occurrence of an ADP-binding βαβ fold, which may bind the ADP moiety of a flavin adenine dinucleotide, NADH, or NADPH cofactor (37). Rhodospirillum rubrum BchP has identical amino acids with Rhodobacter sphaeroides BchP at all significant positions within this fingerprint, suggesting that its cofactor requirement is likely to be identical.
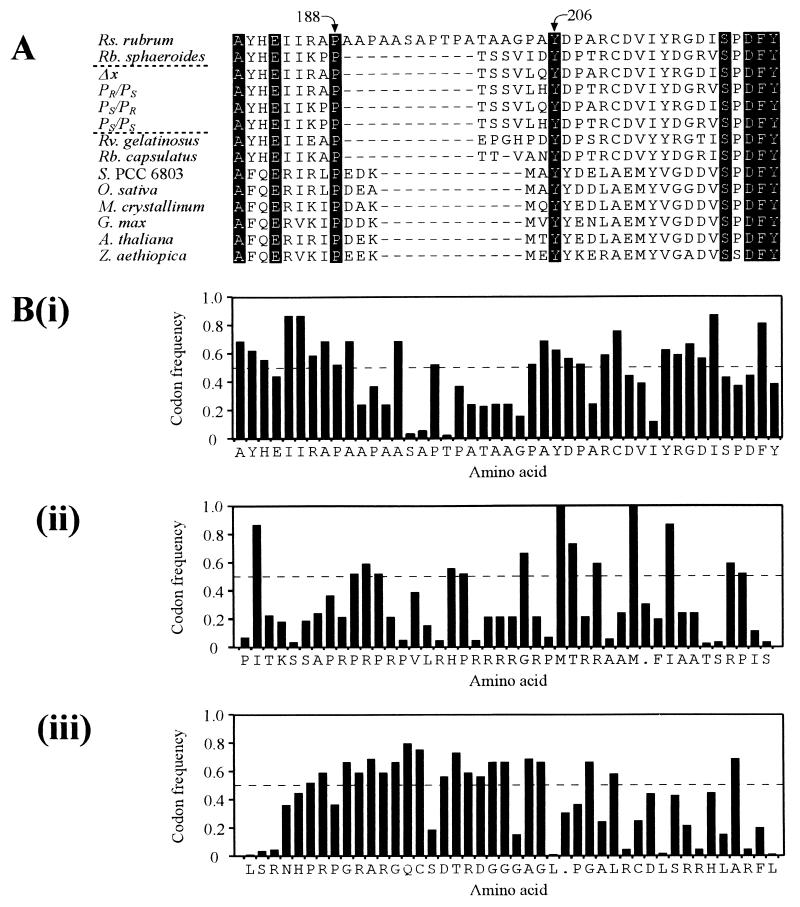
One difference is, however, immediately apparent upon comparison with the GG-(b)chl reductase sequences. Near the center of each there are two highly conserved residues, a proline and a tyrosine (P188 and Y206 of the Rhodospirillum rubrum sequence). There are only five to six amino acids between these residues in each of the sequences, with the single exception being that from Rhodospirillum rubrum, in which 17 amino acids are present (Fig. 3A). The DNA encoding these amino acids displays atypical codon usage for Rhodospirillum rubrum genes, but in another reading frame, typical codon usage is found (Fig. 3B).
FIG. 3.
(A) Partial alignment of the Rhodospirillum rubrum bchP deduced amino acid sequence (Rs. rubrum) with sequences of known and putative GG-(b)chl reductases from the following species (sequence accession numbers in parentheses): Rubrivivax gelatinosus (EMBL AB034704); Rhodobacter sphaeroides (EMBL AJ010302); Rhodobacter capsulatus (Swissprot P26172); Synechocystis sp. strain PCC 6803 (Swissprot Q55087); Oryza sativa (EMBL AP001080); Mesembryanthemum crystallinum (PIR T12299); Glycine max (EMBL AF068686); Arabidopsis thaliana (EMBL Y14044); and Zantedeschia aethiopica (EMBL AF055296). The numbers above the alignment are amino acid positions within Rhodospirillum rubrum BchP. The sequences labeled Δx, PR/PS, PS/PR, and PS/PS represent the predicted products of the modified and hybrid bchP genes present in plasmids pSK1RRbchPΔx, pSK1bchPR/PS, pSK1bchPS/PR, and pSK1bchPS/PS, respectively. (B) Codon usage analysis for the multialigned region of Rhodospirillum rubrum BchP. Each of the three forward reading frames is presented, with frame i being the true reading frame. Codon frequencies add up to 1.0 for each amino acid species. Average codon frequency for the entire bchP gene (0.497) is indicated by a dashed line. It can be seen that the codon frequency drops substantially for the extra amino acids in the correct frame, but that it is more typical in frame iii in this region.
Functional analysis shows that the bchP homolog from Rhodospirillum rubrum encodes a modified BchP protein identified as GG-Bphe reductase.
It was possible that the Rhodospirillum rubrum bchP homolog encoded a nonfunctional product, which would explain the preponderance of GG-esterified pigments in this organism. Complementation of a Rhodobacter sphaeroides bchP mutant was used to test for this possibility. Plasmid pSK1RRbchP, containing Rhodospirillum rubrum bchP, was transferred into Rhodobacter sphaeroides transposon insertion mutant T6G5 (3) by conjugation. This mutant, which accumulates only GG-esterified Bchl, has now also been found to possess GG-esterified Bphe instead of the phytylated Bphe normally found in Rhodobacter sphaeroides (11), indicating that the bchP gene product is also responsible for the synthesis of this pigment in Rhodobacter sphaeroides. In order to provide an increased ratio of Bphe to Bchl as an aid to pigment analysis, the complementations were actually carried out in an LH1-RC-only background in mutant DBCΩ:T6G5, thereby elevating the Bphe-containing RCs as a proportion of the total photosynthetic complexes.
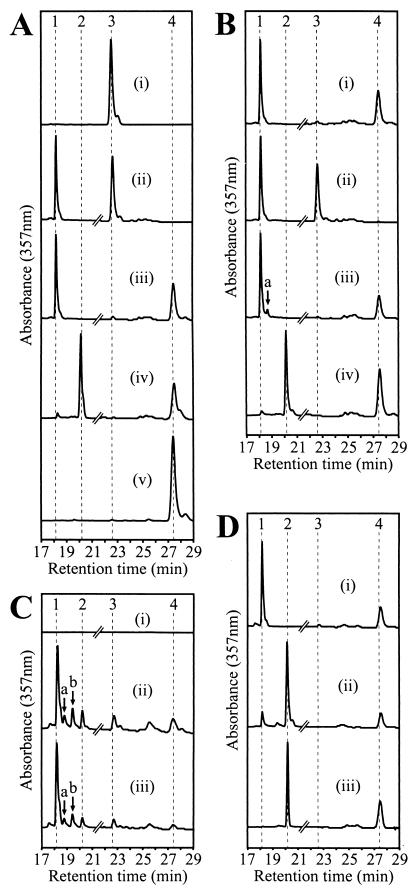
HPLC analysis of a DBCΩ:T6G5[pSK1RRbchP] cell extract (Fig. 4Aiii) failed to reveal any reduction of GG-esterified Bchla to dihydro-GG-esterified, tetrahydro-GG-esterified, or phytylated Bchla; see Fig. 4Ciii for a comparison, in which these pigments are resolved. Significantly, however, the BpheaGG had been replaced with BpheaP as a result of transferring Rhodospirillum rubrum bchP into Rhodobacter sphaeroides, showing that the encoded protein catalyzes the reduction of BpheaGG but not of BchlaGG. This time, comparisons can be made with a BpheaP standard (Fig. 4Av) and with a Rhodospirillum rubrum control which contains BpheaP (Fig. 4Di). In contrast, Rhodobacter sphaeroides BchP performs the conversion of both BchlaGG to BchlaP and BpheaGG to BpheaP; this is shown in Fig. 4Aiv, in which the native bchP gene is used to complement the bchP mutant of Rhodobacter sphaeroides.
FIG. 4.
HPLC traces of acetone-methanol extracts of Rhodobacter sphaeroides and Rhodospirillum rubrum strains. (A) Functional analysis of Rhodospirillum rubrum bchP. (i) BpheaGG standard; (ii) Rhodobacter sphaeroides bchP mutant DBCΩ:T6G5; (iii) DBCΩ:T6G5[pSK1RRbchP], the Rhodobacter sphaeroides bchP mutant complemented with Rhodospirillum rubrum bchP; (iv) DBCΩ:T6G5[pSK1bchP], the Rhodobacter sphaeroides bchP mutant complemented with Rhodobacter sphaeroides bchP; (v) BpheaP standard. (B) Functional analysis of modified versions of Rhodospirillum rubrum bchP via complementation of Rhodobacter sphaeroides bchP mutant DBCΩ:T6G5. (i) DBCΩ:T6G5[pSK1RRbchPΔx]; (ii) DBCΩ:T6G5[pSK1bchPR/PS]; (iii) DBCΩ:T6G5[pSK1bchPS/PR]; and (iv) DBCΩ:T6G5[pSK1bchPS/PS]. (C) Functional analysis of Rhodospirillum rubrum bchG. (i) Rhodobacter sphaeroides bchG mutant T6G1[pRKSK1]; (ii) T6G1[pSK1RRbchG], the Rhodobacter sphaeroides bchG mutant complemented with Rhodospirillum rubrum bchG; (iii) T6G1[pEB bchG], the Rhodobacter sphaeroides bchG mutant complemented with Rhodobacter sphaeroides bchG. (D) Analysis of a novel mutant strain of Rhodospirillum rubrum. (i) Wild-type Rhodospirillum rubrum[pRKSK1]; (ii) Rhodospirillum rubrum[pSK1bchP], the new, BchlaP-containing strain; (iii) Rhodobacter sphaeroides wild-type strain DBCΩ. The retention times of the four principal pigments are indicated by vertical dashed lines: 1, BchlaGG; 2, BchlaP; 3, BpheaGG; and 4, BpheaP. Other known peaks are individually labeled: a, dihydro-GG-esterified Bchla; b, tetrahydro-GG-esterified Bchla. Traces are normalized according to the maximum Bchl peak, with the exception of the trace for strain T6G1[pRKSK1], displayed at the same scale as that of strain T6G1[pSK1RRbchG]. The Bphe region of most traces is expanded vertically sixfold; this is indicated by a break in the trace at a retention time of 21.5 min.
Modification of the central region of Rhodospirillum rubrum BchP and interchange of the N- and C-terminal domains of the Rhodospirillum rubrum and Rhodobacter sphaeroides BchP proteins
As has already been noted, Rhodospirillum rubrum BchP differs from other similar polypeptides in the presence of an abnormal stretch of 17 amino acids towards the center of the sequence. We speculated that this difference may be responsible for the discrepancy in substrate specificity between the enzymes, and so the central region of the gene was modified to make it more homologous to the central region of the Rhodobacter sphaeroides gene by replacing codons encoding the sequence AAPAASAPTPATAAGPA with those coding for TSSVLQ; see Fig 3A. The modified Rhodospirillum rubrum gene, bchPΔx, was also cloned into vector pRKSK1, yielding plasmid pSK1RRbchPΔx.
Complementation of Rhodobacter sphaeroides mutant DBCΩ:T6G5 was again attempted (Fig. 4Bi), demonstrating that the modified gene still encoded a GG-Bphe reductase but that no GG-Bchl reductase activity had been acquired. Plasmid pSK1bchPR/PS, which contained a hybrid gene consisting of the 5′ half of Rhodospirillum rubrum bchP and the 3′ half of Rhodobacter sphaeroides bchP, with the fragments fused as for bchPΔx, was found to possess neither GG-Bphe reductase nor GG-Bchl reductase activity (Fig. 4Bii). However, when complementation of the mutant was attempted with a similar fusion between the 5′ half of Rhodobacter sphaeroides bchP and the 3′ half of Rhodospirillum rubrum bchP, expressed in plasmid pSK1bchPS/PR, not only was BpheaP biosynthesis restored, but approximately 10% of the Bchl present was found to be esterified with dihydro-GG (Fig. 4Biii). Complete restoration of both BchlaP and BpheaP synthesis was achieved by using the positive control plasmid, pSK1bchPS/PS, in which the two relevant Rhodobacter sphaeroides bchP fragments were reunited (Fig. 4Biv).
Functional analysis of Rhodospirillum rubrum bchG.
The BchG proteins of Rhodobacter sphaeroides and Rhodobacter capsulatus attach either geranylgeranyl pyrophosphate (GGPP) or phytyl pyrophosphate (PPP) to bacteriochlorophyllide (1, 6, 27). In view of the novel properties of Rhodospirillum rubrum BchP, bchG from this organism was transferred to Rhodobacter sphaeroides in order to verify that it encoded a functional Bchl synthetase. Plasmid pSK1RRbchG was introduced into T6G1, a mutant of Rhodobacter sphaeroides deficient in GG-Bchl synthetase activity as a result of transposon insertion into the native bchG gene (1). While the negative control, T6G1[pRKSK1], remained devoid of Bchl (Fig. 4Ci), HPLC analysis of an acetone-methanol extract of T6G1[pSK1RRbchG] cells demonstrated that Bchl was now present and 63% of this Bchl was esterified with GG; the remainder was esterified with tetrahydro-GG, dihydro-GG, or phytol (Fig. 4Cii).
This profile is similar to that observed previously for Rhodobacter sphaeroides strain T6G1[pEB bchG] (Fig. 4Ciii), in which complementation of the same mutant was attempted with the native Rhodobacter sphaeroides gene in a related expression vector (1). Partial restoration of pigment protein complex levels was also evident in T6G1[pSK1RRbchG], but not in T6G1[pRKSK1] (data not shown). Complete restoration of a wild-type phenotype to mutant T6G1 cannot be achieved with a bchG gene alone, since the transposon insertion also causes disruption of downstream bchP gene expression (1). The presence of substantial quantities of Bchl in T6G1[pSK1RRbchG] demonstrated that the introduced Rhodospirillum rubrum gene did encode a functional Bchl synthetase.
Generation of a novel mutant strain of Rhodospirillum rubrum possessing Bchl esterified with phytol.
The data obtained show that Rhodospirillum rubrum can be thought of as a partially disabled bchP mutant, in which the encoded enzyme possesses one of the two activities present in the Rhodobacter sphaeroides homolog. Plasmid pSK1bchP, bearing the Rhodobacter sphaeroides bchP gene, was introduced into Rhodospirillum rubrum, as was the insert-free plasmid pRKSK1 as a negative control. HPLC analysis of the resulting strains showed that, while Rhodospirillum rubrum [pRKSK1] had only the BchlaGG normally found in Rhodospirillum rubrum (Fig. 4Di), this pigment was present at only a low level in the strain containing pSK1bchP; instead, there was a predominance of BchlaP (Fig. 4Dii). The biosynthesis of this pigment demonstrated that the only impediment to its formation in Rhodospirillum rubrum had been a mutation in the native gene.
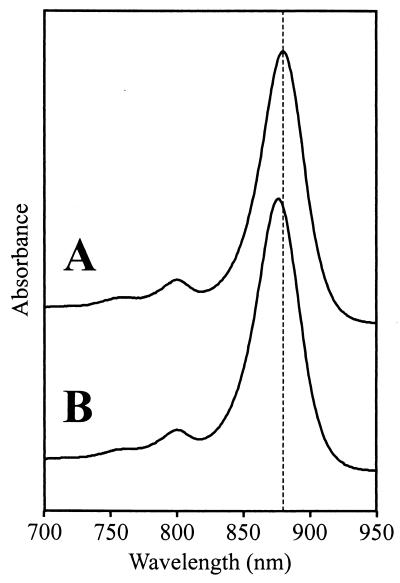
Preliminary analysis of the novel, BchlaP-containing strain by absorbance spectroscopy showed that the photosynthetic complexes were present in approximately wild-type levels, although a perturbation of the LH1 structure was indicated by a blue shift in the position of the 880-nm peak by approximately 3.5 nm (Fig. 5). As in Rhodospirillum rubrum[pRKSK1], the Bphe species present was BpheaP (Fig. 4D), and the strain retained the ability to grow photosynthetically.
FIG. 5.
Absorbance spectra of intracytoplasmic membranes from (A) wild-type Rhodospirillum rubrum[pRKSK1] and (B) Rhodospirillum rubrum complemented with bchP in plasmid pSK1bchP. Spectra are normalized at their LH1 absorbance maxima. It can be seen that the 880-nm peak is slightly blue-shifted in Rhodospirillum rubrum[pSK1bchP].
DISCUSSION
Identification of part of a PGC of Rhodospirillum rubrum.
In common with other purple bacteria and heliobacteria, such as Rhodobacter capsulatus, Rhodospirillum centenum, and Heliobacillus mobilis, photosynthesis-related genes of Rhodobacter sphaeroides are clustered on the genome within a region of approximately 40 kb (4, 25, 38, 39). Within the Rhodobacter sphaeroides and Rhodobacter capsulatus PGCs, the bchP and bchG genes are found in close proximity (1, 3), and a similar situation has now been found to exist in Rhodospirillum rubrum. The DNA flanking the newly identified bchP homolog included a bchG gene, demonstrated to encode a functional GG-Bchl synthetase by successful complementation of a bchG mutant of Rhodobacter sphaeroides; previously published data placed a bchG gene within a Rhodospirillum rubrum PGC (5). Also present in the newly sequenced region were homologs of the Rhodobacter sphaeroides PGC genes ppsR and ORF427, the Rhodospirillum rubrum homolog of the latter being ORF449. The gene order bchG-ORF449-bchP present in Rhodospirillum rubrum is identical to that in both Rhodobacter sphaeroides and Rhodobacter capsulatus.
BchP of Rhodobacter sphaeroides converts BpheaGG to BpheaP, as well as BchlaGG to BchlaP, whereas BchP of Rhodospirillum rubrum is restricted to the former of these activities
Both the Bchl and Bphe components of the Rhodobacter sphaeroides photosynthetic apparatus are principally esterified with phytol. In contrast, the isoprenoid moieties of these two pigments in Rhodospirillum rubrum differ; that of Bphe is again phytol, but that of Bchl is the less-reduced alcohol GG. GG-esterified Bchl appears to be a precursor of BchlaP in both Rhodobacter sphaeroides and Rhodobacter capsulatus, although direct esterification with phytol and the intermediates dihydro-GG and tetrahydro-GG may also occur.
The gene responsible for the hydrogenation of the isoprenoid moiety of Bchl in these species is bchP (3, 6). The present work shows that BchP of Rhodobacter sphaeroides has, in effect, two substrates, converting BpheaGG to BpheaP as well as BchlaGG to BchlaP. In contrast, the BchP of Rhodospirillum rubrum is restricted to the former of these activities. Rhodobacter sphaeroides bchP was heterologously expressed in wild-type Rhodospirillum rubrum; the resultant strain possessed BchlaP, further demonstrating that the enzymes encoded by the bchP genes differ in their biosynthetic abilities.
From the available data, certain deductions can be made concerning the activity of the GG-Bphe reductase enzyme of Rhodospirillum rubrum in relation to the dual-function enzyme of the Rhodobacter species. It appears that GG-Bphe reductase must be highly specific, in being able only to reduce attached isoprenoid moieties of Bphe, and not free GG or GGPP. If GG-Bphe reductase was able to reduce free GG or GGPP, then any PPP formed would be available to the Bchl synthetase present. Not only is the Bchl synthetase of Rhodobacter capsulatus able to utilize PPP as a substrate, giving BchlaP, but it is preferred over GGPP in vitro (27). Given that the Rhodobacter sphaeroides Bchl synthetase is likely to have similar substrate requirements, it would be expected that BchlaP would be detectable in Rhodobacter sphaeroides strain DBCΩ:T6G5[pSK1RRbchP], a bchP mutant complemented with Rhodospirillum rubrum bchP; however, no BchlaP was found.
On the basis of the observed substrate preference of the Rhodobacter capsulatus Bchl synthetase, Oster et al. (27) proposed that the bchP gene product was able to reduce GG or GGPP prior to esterification. Interestingly, Keller et al. (21) have demonstrated that a homolog of bchP from Arabidopsis thaliana encodes a multifunctional GG reductase, catalyzing the reduction of GGPP to PPP as well as that of ChlaGG to ChlaP. We conclude that the GG-Bphe reductase of Rhodospirillum rubrum differs in two respects from previously studied BchP homologs. First, it requires attached isoprenoid moieties of Bphe as substrates, and second, it can reduce BpheaGG to BpheaP, but it cannot reduce BchlaGG to BchlaP.
The cofactor requirement of Rhodospirillum rubrum BchP is likely to be identical to that of Rhodobacter sphaeroides BchP; this has been putatively identified as NADPH (3). The origin of the proposed substrate of GG-Bphe reductase, BpheaGG, is not certain, but loss of the Mg2+ ion from Bchl is the most likely source of the Bphe that Rhodospirillum rubrum and other species of photosynthetic bacteria require. In their experiments with the Rhodobacter capsulatus Bchl synthetase enzyme, Oster et al. (27) found bacteriopheophorbide a, the nonesterified equivalent of Bphea, to be an unsuitable substrate for esterification with either GGPP or PPP, and yet no separate Bphe synthetase gene appears to be present within the known PGCs. Magnesium is lost from chlorophylls relatively easily; this may be nonenzymatic, or may perhaps be catalyzed by a dechelatase enzyme, similar to those postulated to exist in plants (see reference 22 for a review).
Attempts to identify the basis for differences in activity between the BchP homologs.
Sequence analysis shows that bchP from Rhodospirillum rubrum differs from Rhodobacter sphaeroides bchP and other known genes encoding GG-(b)chl reductases in the presence of an additional stretch of 11 to 12 codons close to the center of the coding region. Insertion of a fragment of Rhodospirillum rubrum DNA at some stage could have disrupted a once fully functional bchP gene, resulting in partial loss of function. The observation that the DNA encoding the extra amino acids displays atypical codon usage for Rhodospirillum rubrum genes but typical codon usage in another reading frame supports the occurrence of such an insertion event. Manipulation of this region to eliminate the extra codons and to make it resemble that present in Rhodobacter sphaeroides bchP failed to produce a gene capable of restoring BchlaP biosynthesis to Rhodobacter sphaeroides. However, further mutations may have occurred over time, reinforcing the enzyme's inability to utilize substrates other than BpheaGG.
In an attempt to localize such changes, two complementary hybrid genes were constructed and tested for their ability to complement a Rhodobacter sphaeroides bchP mutant. Synthesis of BchlaP was not restored in either case; indeed, when the 5′ half of Rhodospirillum rubrum bchP was fused to the 3′ half of Rhodobacter sphaeroides bchP, no activity of either kind resulted. However, a fusion between the 5′ half of Rhodobacter sphaeroides bchP and the 3′ half of Rhodospirillum rubrum bchP resulted in a functional GG-Bphe reductase and the presence of a small amount of dihydro-GG-Bchla, demonstrating that the hybrid enzyme was at least capable of catalyzing the first step in the reduction of the GG moiety of Bchl. Interestingly, a recently described purple bacterium, Roseospirillum parvum, has been found to possess tetrahydro-GG-esterified Bchla as 20% of its Bchl content, with the remainder being almost entirely BchlaP; no Bphe species other than BpheaP were identified (29). This pigment composition may indicate a related defect in a GG-Bchl reductase enzyme.
Relationship between GG-Bphe reductase activity and assembly and function of the photosynthetic complexes.
Although the Bchl of Rhodospirillum rubrum is esterified with GG, the Bphe within the RC is, in contrast, esterified with phytol. We infer a preference for phytylated Bphe over BpheaGG during the assembly of the RC and a requirement for the phytylated pigment for optimal functioning of the complex. The RC of Rhodobacter sphaeroides can assemble with BpheaGG instead of the usual BpheaP, for example, in the bchP mutant DBCΩ:T6G5, but upon expression of Rhodospirillum rubrum bchP from plasmid pSK1RRbchP, the only pigment to become phytylated is the Bphea of the RC. Mutation of bchP in Rhodospirillum rubrum may result in a strain possessing RCs assembled with BpheaGG, but the survival of such a strain under laboratory conditions may not be representative of the situation in the natural environment.
Although a requirement for BpheaP in Rhodospirillum rubrum has not been proven, this pigment must engender some selective advantage for a GG-Bphe reductase gene to exist at all; the source of this advantage is, however, uncertain. Interestingly, it has been inferred from crystallographic data for the RC that the phytol tails of the Bchl and Bphe molecules are more tightly packed in the active branch than in the inactive branch (10).
Phytol tails are predicted to be less rigid than GG tails and may therefore be important for this tight packing. However, published data on bchP mutants of both Rhodobacter sphaeroides and Rhodobacter capsulatus appear to be directly contradictory to the importance of phytol over GG as the esterifying alcohol of the Bphe molecules of the RC. In the Rhodobacter sphaeroides mutant, reduction of the photosynthetic growth rate in relation to the wild type was found to be in almost direct proportion to the reduction in the total level of Bchl present in the photosynthetic complexes, suggesting that it was the formation or stability of the complexes that was affected, rather than their electron transfer capability (3). Bollivar et al. (6) found that neither RC function nor energy transfer from light-harvesting to RC complexes was impaired in a bchP mutant of Rhodobacter capsulatus, which presumably also possessed BpheaGG in place of the usual BpheaP. All of these findings may, however, simply reflect limitations in the experimental procedures performed. Further detailed analyses of mutant and wild-type RCs from Rhodospirillum rubrum and Rhodobacter sphaeroides may lead to an answer. Particularly useful may be Rhodobacter sphaeroides strain DBCΩ:T6G5[pSK1RRbchP], the porphyrin pigment and photosynthetic complex compositions of which mirror those found in wild-type Rhodospirillum rubrum.
Higher plants, green algae, and cyanobacteria possess a pigment-protein complex, PS2, the RC of which is believed to originate from the purple bacterial RC (23). Among the structural and functional similarities, they each have a related series of redox components arranged within their core proteins (30, 40). In place of the Bphe present in the bacterial RC, PS2 contains pheophytin. Phytol is the most common esterifying alcohol of Chls, and it is likely that this is also the case for this pheophytin, essentially the only pheophytin found in chloroplasts. The identification of a second role for the fully functional BchP, in catalyzing the synthesis of BpheaP as well as that of BchlaP, strongly suggests that homologs of this enzyme in PS2-containing organisms may be similarly responsible for the synthesis of phytylated pheophytin.
Finally, if, as we suggest in the introduction, the absence of LH2 and BchlaP from wild-type Rhodospirillum rubrum is linked, this raises the intriguing possibility that ancestors of this bacterium may have possessed an LH2-type complex and that it was the loss of this complex which eased the pressure to maintain GG-Bchl reductase activity.
Acknowledgments
We thank M. R. Jones, University of Bristol, for helpful comments on reaction centers.
This work was supported by the Biotechnology and Biological Sciences Research Council (United Kingdom).
REFERENCES
- 1.Addlesee, H. A., L. Fiedor, and C. N. Hunter. 2000. Physical mapping of bchG, orf427, and orf177 in the photosynthesis gene cluster of Rhodobacter sphaeroides: functional assignment of the bacteriochlorophyll synthetase gene. J. Bacteriol. 182:3175-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addlesee, H. A., L. C. D. Gibson, P. E. Jensen, and C. N. Hunter. 1996. Cloning, sequencing and functional assignment of the chlorophyll biosynthesis gene, chlP, of Synechocystis sp. PCC 6803. FEBS Lett. 389:126-130. [DOI] [PubMed] [Google Scholar]
- 3.Addlesee, H. A., and C. N. Hunter. 1999. Physical mapping and functional assignment of the geranylgeranyl-bacteriochlorophyll reductase gene, bchP, of Rhodobacter sphaeroides. J. Bacteriol. 181:7248-7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberti, M., D. H. Burke, and J. E. Hearst. 1995. Structure and sequence of the photosynthesis gene cluster, p. 1083-1106. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 5.Bauer, C. E., D. W. Bollivar, and J. Y. Suzuki. 1993. Genetic analyses of photopigment biosynthesis in eubacteria—a guiding light for algae and plants. J. Bacteriol. 175:3919-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollivar, D. W., S. J. Wang, J. P. Allen, and C. E. Bauer. 1994. Molecular genetic analysis of terminal steps in bacteriochlorophyll a biosynthesis: characterization of a Rhodobacter capsulatus strain that synthesizes geranylgeraniol-esterified bacteriochlorophyll a. Biochemistry 33:12763-12768. [DOI] [PubMed] [Google Scholar]
- 7.Brown, A. E., and J. Lascelles. 1972. Phytol and bacteriochlorophyll synthesis in Rhodopseudomonas sphaeroides. Plant Physiol. 50:747-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunisholz, R. A., and H. Zuber. 1992. Structure, function and organization of antenna polypeptides and antenna complexes from the three families of Rhodospirillaneae. J. Photochem. Photobiol. B Biol. 15:113-140. [DOI] [PubMed]
- 9.Coomber, S. A., M. Chaudhri, A. Connor, G. Britton, and C. N. Hunter. 1990. Localized transposon Tn5 mutagenesis of the photosynthetic gene cluster of Rhodobacter sphaeroides. Mol. Microbiol. 4:977-989. [DOI] [PubMed] [Google Scholar]
- 10.Ermler, U., G. Fritzsch, S. K. Buchanan, and H. Michel. 1994. Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.65 Å resolution: cofactors and protein-cofactor interactions. Structure 2:925-936. [DOI] [PubMed] [Google Scholar]
- 11.Francke, C., and J. Amesz. 1995. The size of the photosynthetic unit in purple bacteria. Photosynth. Res. 46:347-352. [DOI] [PubMed] [Google Scholar]
- 12.Hess, S., M. Chachisvilis, K. Timpmann, M. R. Jones, G. J. S. Fowler, C. N. Hunter, and V. Sundström. 1995. Temporally and spectrally resolved subpicosecond energy transfer within the peripheral antenna complex (LH2) and from LH2 to the core antenna complex in photosynthetic purple bacteria. Proc. Natl. Acad. Sci. USA 92:12333-12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoff, A. J., and J. Deisenhofer. 1997. Photophysics of photosynthesis. Structure and spectroscopy of reaction centers of purple bacteria. Phys. Rep. Rev. Sect. Phys. Lett. 287:2-247. [Google Scholar]
- 14.Hunter, C. N., B. S. Hundle, J. E. Hearst, H. P. Lang, A. T. Gardiner, S. Takaichi, and R. J. Cogdell. 1994. Introduction of new carotenoids into the bacterial photosynthetic apparatus by combining the carotenoid biosynthetic pathways of Erwinia herbicola and Rhodobacter sphaeroides. J. Bacteriol. 176:3692-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter, C. N., P. McGlynn, M. K. Ashby, J. G. Burgess, and J. D. Olsen. 1991. DNA sequencing and complementation/deletion analysis of the bchA-puf operon region of Rhodobacter sphaeroides: in vivo mapping of the oxygen-regulated puf promoter. Mol. Microbiol. 5:2649-2661. [DOI] [PubMed] [Google Scholar]
- 16.Hunter, C. N., and G. Turner. 1988. Transfer of genes coding for apoproteins of reaction center and light-harvesting LH1 complexes to Rhodobacter sphaeroides. J. Gen. Microbiol. 134:1471-1480. [Google Scholar]
- 17.Jiang, Z. Y., and C. E. Bauer. 2001. Component of the Rhodospirillum centenum photosensory apparatus with structural and functional similarity to methyl-accepting chemotaxis protein chemoreceptors. J. Bacteriol. 183:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, M. R., G. J. S. Fowler, L. C. D. Gibson, G. G. Grief, J. D. Olsen, W. Crielaard, and C. N. Hunter. 1992. Mutants of Rhodobacter sphaeroides lacking one or more pigment-protein complexes and complementation with reaction-centre, LH1, and LH2 genes. Mol. Microbiol. 6:1173-1184. [DOI] [PubMed] [Google Scholar]
- 19.Karrasch, S., P. A. Bullough, and R. Ghosh. 1995. The 8.5 Å projection map of the light-harvesting complex I from Rhodospirillum rubrum reveals a ring composed of 16 subunits. EMBO J. 14:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz, J. J., H. H. Strain, A. L. Harkness, M. H. Studier, W. A. Svec, T. R. Janson, and B. T. Cope. 1972. Esterifying alcohols in the chlorophylls of purple photosynthetic bacteria. A new chlorophyll, bacteriochlorophyll (gg), all-trans-geranylgeranyl bacteriochlorophyllide a. J. Am. Chem. Soc. 94:7938-7939. [DOI] [PubMed] [Google Scholar]
- 21.Keller, Y., F. Bouvier, A. d'Harlingue, and B. Camara. 1998. Metabolic compartmentation of plastid prenyllipid biosynthesis: evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur. J. Biochem. 251:413-417. [DOI] [PubMed] [Google Scholar]
- 22.Matile, P., S. Hortensteiner, H. Thomas, and B. Krautler. 1996. Chlorophyll breakdown in senescent leaves. Plant Physiol. 112:1403-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel, H., and J. Deisenhofer. 1988. Relevance of the photosynthetic reaction center from purple bacteria to the structure of photosystem II. Biochemistry 27:1-7. [Google Scholar]
- 24.Michotey, V., B. Toussaint, P. Richaud, and P. M. Vignais. 1996. Characterisation of the mcpA and mcpB genes capable of encoding methyl-accepting type chemoreceptors in Rhodobacter capsulatus. Gene 170:73-76. [DOI] [PubMed] [Google Scholar]
- 25.Naylor, G. W., H. A. Addlesee, L. C. D. Gibson, and C. N. Hunter. 1999. The photosynthesis gene cluster of Rhodobacter sphaeroides. Photosynth. Res. 62:121-139. [Google Scholar]
- 26.Olsen, J. D., G. D. Sockalingum, B. Robert, and C. N. Hunter. 1994. Modification of a hydrogen bond to a bacteriochlorophyll a molecule in the light harvesting 1 antenna of Rhodobacter sphaeroides. Proc. Natl. Acad. Sci. USA 91:7124-7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oster, U., C. E. Bauer, and W. Rüdiger. 1997. Characterization of chlorophyll a and bacteriochlorophyll a synthases by heterologous expression in Escherichia coli. J. Biol. Chem. 272:9671-9676. [DOI] [PubMed] [Google Scholar]
- 28.Perkins, H. J., and D. W. A. Roberts. 1962. Purification of chlorophylls, pheophytins and pheophorbides for specific activity determinations. Biochim. Biophys. Acta 58:486-498. [DOI] [PubMed] [Google Scholar]
- 29.Permentier, H. P., S. Neerken, K. A. Schmidt, J. Overmann, and J. Amesz. 2000. Energy transfer and charge separation in the purple non-sulfur bacterium Roseospirillum parvum. Biochim. Biophys. Acta 1460:338-345. [DOI] [PubMed] [Google Scholar]
- 30.Rögner, M., E. J. Boekema, and J. Barber. 1996. How does photosystem 2 split water? The structural basis of efficient energy conversion. Trends Biochem. Sci. 21:44-49. [DOI] [PubMed] [Google Scholar]
- 31.Saegesser, R., R. Ghosh, and R. Bachofen. 1992. Stability of broad host range cloning vectors in the phototrophic bacterium Rhodospirillum rubrum. FEMS Microbiol. Lett. 95:7-12. [Google Scholar]
- 32.Scheer, H. 1991. Structure and occurrence of chlorophylls, p. 3-30. In H. Scheer (ed.), Chlorophylls. CRC Press Inc., Cleveland, Ohio.
- 33.Schoch, S., and W. Schäfer. 1978. Tetrahydrogeranylgeraniol, a precursor of phytol in the biosynthesis of chlorophyll a—localization of the double bonds. Z. Naturforsch. 33c:408-412.
- 34.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 35.Walz, T., and R. Ghosh. 1997. Two-dimensional crystallization of the light-harvesting I reaction centre photounit from Rhodospirillum rubrum. J. Mol. Biol. 265:107-111. [DOI] [PubMed] [Google Scholar]
- 36.Walz, T., S. J. Jamieson, C. M. Bowers, P. A. Bullough, and C. N. Hunter. 1998. Projection structures of three photosynthetic complexes from Rhodobacter sphaeroides: LH2 at 6 Å, LH1 and RC-LH1 at 25 Å. J. Mol. Biol. 282:833-845. [DOI] [PubMed] [Google Scholar]
- 37.Wierenga, R. K., P. Terpstra, and W. G. Hol. 1986. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187:101-107. [DOI] [PubMed] [Google Scholar]
- 38.Xiong, J., K. Inoue, and C. E. Bauer. 1998. Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis. Proc. Natl. Acad. Sci. USA 95:14851-14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yildiz, F. H., H. Gest, and C. E. Bauer. 1992. Conservation of the photosynthesis gene cluster in Rhodospirillum centenum. Mol. Microbiol. 6:2683-2691. [DOI] [PubMed] [Google Scholar]
- 40.Zouni, A., H. T. Witt, J. Kern, P. Fromme, N. Krauss, W. Saenger, and P. Orth. 2001. Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature 409:739-743. [DOI] [PubMed] [Google Scholar]