Abstract
The switch from symmetric to asymmetric cell division is a key feature of development in many organisms, including Bacillus subtilis sporulation. Here we demonstrate that, prior to the onset of asymmetric cell division, the B. subtilis chromosome is partitioned into two unequally sized domains, with the origin-proximal one-third of the future forespore chromosome condensed near one pole of the cell. Asymmetric chromosome partitioning is independent of polar division, as it occurs in cells depleted of FtsZ but depends on two transcription factors that govern the initiation of sporulation, σH and Spo0A-P. It is also independent of chromosome partitioning proteins Spo0J and Soj, suggesting the existence of a novel mechanism controlling chromosome structure. Thus, our results demonstrate that, during sporulation, two separable events prepare B. subtilis for asymmetric cell division: the relocation of cell division sites to the cell poles and the asymmetric partitioning of the future forespore chromosome.
Separation of daughter chromosomes occurs prior to cell division in nearly all organisms. In eukaryotes, the importance of properly orchestrating chromosome segregation and cytokinesis is highlighted by the existence of checkpoints to arrest the cell cycle when chromosomes are damaged or incompletely segregated. In bacteria, chromosome separation and cytokinesis are regulated both temporally and spatially, so that, even during rapid exponential growth, daughter chromosomes are well separated prior to the onset of division. Bacterial chromosome segregation appears to consist of two distinct steps: the rapid movement of the duplicated origins apart from one another (9, 11, 38, 40) and the separate condensation of the replicated daughter chromosomes (12, 19, 35). This process results in vegetative bacterial chromosomes having a bilobed structure during replication, with the daughter chromosomes being well separated prior to the onset of cytokinesis, so the path of the invaginating septum appears free of chromosomal DNA (4, 14). When chromosome segregation fails, as it does in mutants defective in chromosome partitioning or condensation, the chromosomes can be sheared during daughter cell separation (24, 34, 39, 47).
How, then, are bacterial chromosomes prepared for asymmetrically positioned division events, such as that which occurs during the sporulation pathway of Bacillus subtilis? At the onset of sporulation, the chromosome is reorganized into a rod-like structure (the axial filament), which is readily distinguished from the bilobed chromosome of vegetative cells (31). Next, a septum is formed close to one cell pole, trapping the origin-proximal region of the chromosome in the smaller daughter cell (the forespore), while the remainder is translocated across the septum after division (43, 44). This translocation event requires the SpoIIIE protein (1, 43), without which the forespore receives only the origin-proximal 30% of a chromosome, while the remainder is located in the mother cell (44, 45). Thus, in the B. subtilis sporulation pathway, the relative order of division and chromosome segregation is reversed, with division occurring prior to chromosome segregation, rather than after.
Little is known about the architecture of the forespore chromosome before the sporulation septum is synthesized. Is the region of the invaginating septum cleared of DNA prior to asymmetric septation, as during vegetative growth, or is the septum required to partition the chromosome? Previous studies have supported the latter proposal, largely due to the failure to observe partitioned axial filaments in the absence of septation (45). The presence of a partitioned axial filament with a condensed region of the chromosome near one cell pole was therefore thought to indicate that a sporulation septum had formed (13, 21, 29, 45), implying that chromosome partitioning was a consequence of cell division. However, these studies preceded reliable methods to simultaneously observe the asymmetrically positioned sporulation septum and the chromosomes (27), leaving open the possibility that individual chromosomes are reorganized into two separate domains, creating a gap to accommodate the invaginating septum. Here, we address this issue using time-lapse deconvolution microscopy together with fluorescent membrane stains that allow visualization of septa and nucleic acid stains to reveal chromosome structure. We demonstrate that, during sporulation, the bacterial chromosome is partitioned into two domains of unequal size prior to asymmetric cell division. This asymmetric chromosome condensation event is independent of cell division protein FtsZ, providing evidence for an FtsZ-independent reorganization of cellular architecture that serves to prepare the bacterium for asymmetric cell division.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
PY79 was the wild-type strain used in these studies (46). The remaining strains were PY79 derivatives and contained the following additional mutations or genetic constructs: KP427 Pspac-spo0H (25), KP444 Pspac-ftsZ (2), KP527 Δspo0J-soj (33), and KP648 Δspo0A::erm (17). All experiments were performed at 37°C. Sporulation was induced by resuspension (37), with the time of resuspension being defined as the onset of sporulation (t0). Identical results were obtained when sporulation was induced by nutrient exhaustion (not shown). The IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Pspac promoter was used to allow depletion of FtsZ from strain KP444 during sporulation (27) and to deplete strain KP427 cells of σH. In the latter case, strain KP427 was grown and sporulated in media lacking IPTG. Luria-Bertani medium was used for exponential growth of PY79, and the culture was grown to an optical density at 600 nm of 0.300.
Fluorescent stains.
Fluorescent membrane stains FM 4-64, MitoTracker red CMXros, and MitoTracker green and DNA stains SYTO 16 and DAPI (4′,6′-diamidino-2-phenylindole) were obtained from Molecular Probes (Eugene, Oreg.) and resuspended as recommended by the manufacturer. Final concentrations of 2 μg of FM 4-64/ml, 0.2 μg of MitoTracker red CMXros/ml, 10 μg of MitoTracker green/ml, 0.2 μg of SYTO 16/ml, and 0.3 μg of DAPI/ml were used.
Preparation of samples for immunofluorescence microscopy.
Samples were prepared for immunofluorescence microscopy as described previously (28, 33) using rabbit antibodies specific for σF (a gift from Richard Losick) and affinity-purified chicken antibodies specific for FtsZ (a gift from Petra Levin). The secondary antibodies were Cy5-labeled goat anti-chicken (Jackson Immunolabs) and Oregon green-labeled goat anti-rabbit (Molecular Probes). DAPI and FM 4-64 were added to Slow Fade equilibration buffer (Molecular Probes), as previously described (33).
Microscopy.
Membrane and chromosome morphology was assessed using an Applied Precision optical sectioning microscope (27), with samples stained and slides prepared as described previously (27, 33). Briefly, the bacteria were stained with the appropriate fluorescent membrane stain and DAPI on a slide and immobilized with a poly-l-lysine (Sigma)-treated coverslip. However, MitoTracker red CMXros staining was more uniform if it was mixed with the cells in an Eppendorf tube prior to applying cells to a slide. Between 6 and 12 optical sections were immediately collected from each field, and the images were deconvolved using Delta Vision software.
We noted that the chromosomes of vegetatively growing B. subtilis cells are extremely sensitive to exposure to the UV light used to visualize DAPI-stained chromosomes. During time-lapse microscopy, DAPI-stained chromosomes undergo a cycle of decondensation and recondensation upon exposure to UV excitatory light. For example, after focusing on the cells using FM 4-64 fluorescence, the first image of DAPI-stained chromosomes collected during a 2-s exposure to UV light shows the nucleoids to be condensed; however, after an additional 2 s of exposure to UV light, the chromosomes decondense and appear to fill the entire cytoplasm. This effect is likely due to UV-induced nicking of DNA and the consequent relaxation of supercoils that normally contribute to chromosome condensation (15). After more-prolonged UV exposure (>90 s), the nucleoids recondense and appear similar to their original state. In contrast, nucleoids stained with SYTO 16 appear condensed and do not change appearance even after prolonged exposure to blue excitation light; however, after a 5-s exposure to UV light, they rapidly decondense. We therefore minimized exposure of the cells to UV light prior to image acquisition. This phenomenon may explain the somewhat variable structure of the chromosomes of growing B. subtilis, which can appear either well condensed (as shown here) or more diffuse and unstructured. The chromosomes of sporulating cells also decondensed, but only after prolonged exposure to UV.
Time-lapse microscopy.
Sporulating PY79 cells were stained with FM 4-64 (1 μg/ml) and SYTO 16 (0.2 μg/ml) at the time of resuspension, and time-lapse microscopy was performed as described previously (27). Briefly, 40 μl of culture was placed on a coverslip at 1.0 h after the onset of sporulation (t1.0), covered with a chambered slide, and heated to 30°C. Three optical sections spaced 0.15 μm apart were collected every 15 min for 1 h. Under these conditions, the frequency of polar septation was low, likely due to fluorescence toxicity: only about 5% of the cells that were in focus throughout the experiment synthesized a polar septum by the end of the experiment. Of these sporangia, about 50% had a clearly partitioned forespore chromosome prior to the onset of septal biogenesis. The remaining sporangia had a partial sporulation septum at the start of the experiment and a partitioned chromosome.
Quantitation of the DNA content of the polar condensed regions.
The DNA content of the polar condensed regions of the forespore chromosome (or future forespore chromosome) was quantified using Delta Vision, version 2.10, software (Applied Precision). Identical results were obtained when DNA content was determined both before and after deconvolution. Images were collected from 3 to 6 focal planes of DAPI- and FM 4-64-stained bacteria at either t1.5 for sporangia with partial septa or at both t1.0 and t1.5 for sporangia lacking septa. Separate stacks of images were taken for each fluorophore, and the DAPI images were collected before the FM 4-64 images. Exposure times were adjusted to be within the linear range of the camera yet to contain sufficient data for accurate deconvolution and quantitation. The average maximum pixel values were ∼3,000 counts/pixel. Following the completion of DNA translocation into the forespore, measurements of DAPI staining intensity underestimate the amount of DNA in the forespore, probably due to the higher concentration of DNA in the small forespore than in the larger mother cell and consequently the greater quenching of the DAPI fluorescence in the forespore (32). Because the measurements presented here are of incompletely translocated forespore chromosomes, in which the concentration of the DNA should be lower than after translocation is complete, these measurements may be less affected by quenching. Nonetheless, our measurements may somewhat underestimate the proportion of DNA in the polar condensed region.
The DNA content of the polar condensed region relative to that of the remaining portion was determined by using the “edit polygon” function of the Delta Vision, version 2.10, software. We defined one polygon that enclosed the small condensed region (polar DNA) and a second that enclosed the entire chromosome (total DNA). The “polygon statistics function” was used to determine the integrated intensity of each of these regions, a measure that reflects the total fluorescence intensity within each region (DNA content). At this stage of sporulation, the total DNA content of the cell is expected to be equivalent to two complete chromosomes. Therefore, to determine the proportion of the future forespore chromosome that was within the polar condensed region, we multiplied the ratio of polar DNA content to total DNA content by 2. A total of 18 wild-type sporangia (from t1.5) were quantified, 10 that lacked a partial or complete sporulation septum by FM 4-64 staining and 8 that had a partial sporulation septum as shown by FM 4-64 staining that failed to completely traverse the cell.
RESULTS AND DISCUSSION
Asymmetric chromosome condensation precedes asymmetric division.
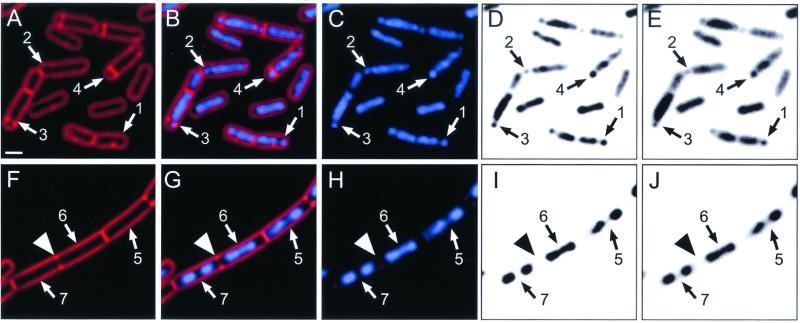
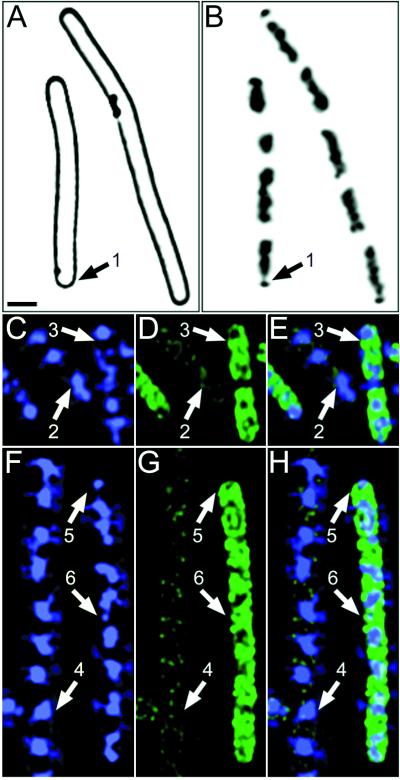
We have previously shown that vital membrane stain FM 4-64 readily detects both complete (Fig. 1A to E, arrows 4) and incomplete (Fig. 1A to E, arrows 3) sporulation septa, which form very close to the cell pole (27). We therefore stained wild-type B. subtilis at t1.5 with FM 4-64 and the DNA-specific stain DAPI and observed that many sporangia containing an incomplete sporulation septum also contained a small, condensed region of one chromosome near the cell pole (Fig. 1A to E, arrows 3). Furthermore, many cells in which there was no evidence of a polar septum contained a small portion of the chromosome condensed near one or both poles (Fig. 1A to E, arrows 1 and 2). These condensed DNA regions are in the same location as those in cells containing a partial sporulation septum (Fig. 1A to E, arrows 3) and are readily seen before (Fig. 1E, arrows 1 and 3) or after deconvolution (Fig. 1C and D, arrows 1 and 3), as well as in fixed cells. We estimate that the gap between the two chromosome domains is approximately 5 to 10 times the space necessary to accommodate two lipid bilayers and the intervening cell wall material of the future sporulation septum.
FIG. 1.
Asymmetric chromosome condensation during sporulation precedes asymmetric cell division and is morphologically distinct from the chromosome structure of symmetrically dividing cells. Samples were harvested and stained with FM 4-64 to visualize membranes (red; A, B, F, and G) and with DAPI to infer chromosome structure (blue; B, C, G, and H and D, E, I, and J). The inverted gray-scale images in panels D, E, I, and J enhance visualization of chromosome structure. Panels E and J show chromosome structure prior to deconvolution; panels D and I show the same fields after deconvolution. (A to E) Sporulating cells of wild-type B. subtilis at t1.5. Arrows 1 and 2, cells that lack sporulation septa (A) and that contain an asymmetrically condensed forespore chromosome (C to E); arrows 3, cell with a sporulation septum that is incomplete (A), as it fails to completely traverse the cell, and with an asymmetrically condensed forespore chromosome (C to E). Arrows 4, sporangium with a sporulation septum that appears to completely traverse the cell (A), creating the larger mother cell and smaller forespore. (F to J) Exponentially growing cells of wild-type B. subtilis, which tend to remain attached following completion of division, thereby forming long chains of individual cells. Arrowheads, partial septum that is forming in a single, long cell (F). One of the two nascent daughter cells contains a bilobed nucleoid (arrows 6), while the second contains two distinct nucleoids that are already well separated (arrows 7). Arrows 5, second nascent daughter cell in which the two visible nucleoids are well separated. Bar (A), 1 μm.
This chromosome structure was distinct from that of exponentially growing cells, which showed symmetrically condensed (or bilobed) chromosomes that were well separated prior to the onset of cell division (Fig. 1F to J). A large cell in the process of medial cell division is shown in Fig. 1F to J. Within this dividing cell there is a single bilobed nucleoid on the right side of the nascent septum (arrows 6) and two well-separated nucleoids on the left side (arrows 7).
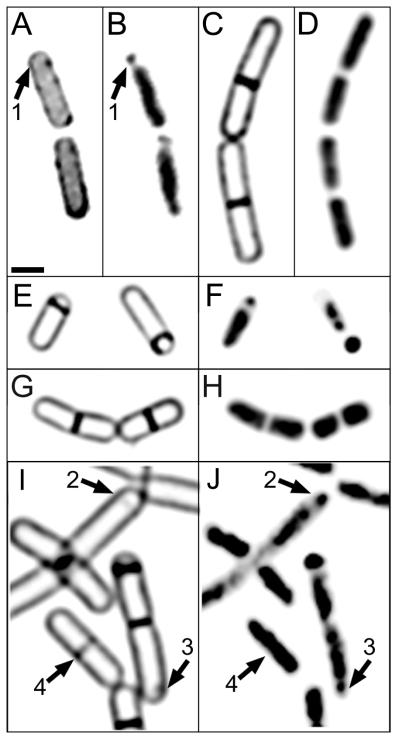
Biological membranes can be partitioned into domains differing in lipid composition, which might affect the ability of FM 4-64 to stain certain regions of the cell, such as the sporulation septum. We therefore used two structurally distinct membrane-permeable stains to examine septal structure: MitoTracker red CMXros (Fig. 2A) and MitoTracker green (not shown). Identical results were obtained with these two stains, as many sporangia showed a condensed region of DNA near one pole in the absence of a sporulation septum (Fig. 2B, arrow 1). Thus, regardless of the fluorescent membrane stain used, forespore chromosome condensation was observed prior to the onset of asymmetric cell division.
FIG. 2.
Cells of the wild type and several mutants stained with membrane stain MitoTracker red CMXros (A) or FM 4-64 (C, E, G, and I) and DNA stain DAPI (B, D, F, H, and J). Sporulation was induced by resuspension, and samples were harvested at t1.5. More than 300 sporangia were examined for each strain; representative examples are shown. (A and B) Wild-type sporangia (PY79) stained with MitoTracker red CMXros (A) and DAPI (B). Both cells lack a sporulation septum and contain an asymmetrically condensed forespore chromosome (arrows 1). (C and D) In the absence of σH (strain KP427), polar septation is blocked and the chromosomes appear decondensed. More than 400 cells were examined by fluorescence microscopy; none showed polar chromosome condensation or polar septa. (E and F) In the presence of IPTG, σH is expressed in strain KP427. Under these conditions, polar septation and chromosome condensation are identical to those in wild-type cells. (G and H) Asymmetric chromosome condensation does not occur in a strain (KP648) lacking the spo0A gene. More than 1,000 cells were examined; representative examples are shown. (I and J) In Δ(spo0J-soj) strain KP527, some cells lacking a polar septum show clear asymmetric chromosome condensation (arrows 2 and 3). Arrows 4, cell with a medial nascent septum without separated nucleoids.
Quantitation of the DAPI staining intensity showed that the amount of DNA in the asymmetrically condensed region in cells completely lacking a sporulation septum was identical to the DNA content in forespores of sporangia in which synthesis of the sporulation septum had started but was incomplete (30% ± 4% versus 29% ± 4% of the total forespore chromosome content, measured as described in Materials and Methods). Thus, the proportion of the forespore chromosome present in the condensed region prior to the formation of the polar septum is similar to the DNA content of forespores in mutants that lack a functional copy of spoIIIE (30% [44, 45]) and that are defective in postseptational chromosome translocation (43). These observations suggest that partitioning of the origin-proximal 30% of the future forespore chromosome precedes asymmetric cell division.
Timing and frequency of asymmetric chromosome condensation.
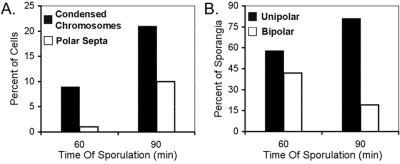
The timing of chromosome condensation relative to septation was quantitatively assessed by comparing the frequencies of each event at t1.0 and t1.5 (Fig. 3). At t1.0, when very few (1%) cells had formed a polar septum, 9% had asymmetrically condensed chromosomes (Fig. 3A). Thirty minutes later, 10% of cells had polar septa and condensed chromosomes, while an additional 11% had condensed chromosomes but no polar septa. At both time points, we could readily detect condensation at both poles of the sporangium, in which cases it appeared that about 30% of each chromosome was condensed near each pole. Approximately 42% of sporangia containing partitioned chromosomes were partitioned at both poles at t1.0, whereas, by t1.5, only 19% displayed bipolar condensation (Fig. 3B). The decreased occurrence of bipolar condensed chromosomes at later times during sporulation suggests that the mother cell chromosome may decondense as sporulation proceeds (a point to which we will return later). Furthermore, some condensed chromosomes may not be observed due to the limited resolution of light microscopy.
FIG. 3.
Timing of polar chromosome condensation. Sporulating cultures of PY79 were stained with FM 4-64 and DAPI, and the numbers of cells containing septa and condensed chromosomes were determined for samples collected 60 (334 cells) and 90 min (285 cells) after the onset of sporulation. (A) The percentage of total cells containing a polar condensed chromosome is plotted next to the percentage of cells containing a polar septum. (B) Percentages of sporulating cells with unipolar or bipolar condensed chromosomes.
Visualization of chromosome dynamics and asymmetric septation with time-lapse microscopy.
The results presented above suggested that asymmetric chromosome condensation preceded formation of the asymmetrically positioned sporulation septum; we used time-lapse microscopy to provide qualitative support for this proposal. A sporulating culture was stained with vital DNA stain SYTO 16 (Molecular Probes) and membrane stain FM 4-64, and deconvolution time-lapse microscopy was performed as described in Materials and Methods, with images collected every 15 min. Figure 4 shows two sporangia from this time-lapse experiment (of the nine that synthesized a polar septum during the experiment). The first sporangium (Fig. 4A to C) shows a clear partitioning of the future forespore chromosome into two domains, with the smaller domain near the cell pole. By the next time point (Fig. 4A′ to C′), a partial sporulation septum was synthesized in the gap between the chromosome domains, and by the next time point (Fig. 4A" to C"), the septum was complete.
FIG. 4.
Time-lapse microscopy: asymmetric DNA condensation precedes formation of the asymmetrically positioned sporulation septum. Sporulating cultures of PY79 were stained with vital DNA stain SYTO 16 (green; B, C, E, and F) and membrane stain FM 4-64 (red; A, C, D, and F) and prepared for time-lapse deconvolution microscopy as described in Materials and Methods. Images were acquired every 15 min. Shown are images taken 0, 15 (primes), and 30 min (double primes) after the start of the time-lapse experiment. The predivisional sporangium (A to C) shows a condensed region of the future forespore chromosome near one cell pole. Arrowheads, sites of synthesis of the sporulation septum. (D to F) Sporangium that contains a sporulation septum at the beginning of the experiment (arrowheads) and an asymmetrically condensed chromosome at the forespore distal pole (arrows), where a second partial septum is synthesized (D′) and later degraded (D") in an abortive division event that we have previously described (27). Bar, 1 μm.
The second sporangium demonstrates that asymmetric chromosome partitioning also occurs at the forespore distal pole of the mother cell (Fig. 4D to F), preceding the abortive cell division event that often occurs at the second potential site of polar septation (Fig. 4D′ to F′) (27). When this second septum regressed, the partitioned domain in the mother cell decondensed (Fig. 4E, E′, and E"). Thus, asymmetric chromosome condensation precedes asymmetric septation and is reversible.
Polar chromosome condensation depends on transcription factors σH and Spo0A.
Spo0A-P is required for the switch from medial to bipolar FtsZ ring assembly (21); thus mutants lacking Spo0A fail to relocalize the FtsZ rings and continue to divide medially. They also maintain the bilobed chromosome structure typical of vegetative growth (Fig. 2G and H) (21). In contrast, σH is not required for the bipolar localization of FtsZ, although, for reasons that are unclear, mutants that lack σH fail to form polar septa (21).
We tested the dependence of asymmetric chromosome condensation on σH by utilizing a strain in which expression of the gene encoding σH (spo0H) was under the control of an IPTG-inducible promoter (18). In the presence of IPTG, polar septa and polar condensed chromosomes appeared at the same rate and frequency as in the wild type (Fig. 2E and F). However, in the absence of σH, neither polar septa nor polar chromosome domains were formed (Fig. 2C and D). Indeed, the chromosomes of σH-deficient cells were poorly condensed, filling the cytoplasm to a much greater extent than the wild type or any of the other mutants examined, even in fixed cells. It is possible that this chromosome condensation defect inhibits polar septation, which could be governed by a checkpoint monitoring asymmetric chromosome condensation. Alternatively, the septation defect may be caused by the reduced expression of cell division proteins such as FtsZ and FtsA in the absence of σH (8, 10) or by the decreased Spo0A-directed gene expression in the absence of σH (36).
Chromosome-partitioning proteins Spo0J and Soj are not required for asymmetric chromosome condensation.
Two candidates for proteins that control chromosome architecture during sporulation are Spo0J and Soj, homologues of chromosome- and plasmid-partitioning proteins (6). Spo0J and Soj are required for efficient chromosome segregation (16) and together regulate the transcription of genes required for polar septation: Soj acts as a transcriptional repressor, whereas Spo0J inhibits Soj, allowing sporulation-specific gene expression and polar septation (3, 30). Spo0J and Soj provide attractive candidates for proteins involved in modulating chromosome architecture, since Spo0J binds to the origin-proximal region of the chromosome likely to be in the polar condensed region and assembles into foci near the cell poles (9, 23). Furthermore, it has been suggested that Soj compacts Spo0J foci (26), an activity that could cause dramatic changes in chromosome architecture, such as asymmetric chromosome partitioning.
We therefore investigated the structure of the chromosome in the spo0J soj double mutant, which is able to express early-sporulation-specific genes and thereby to synthesize the polar septum. If either Spo0J or Soj is required for asymmetric chromosome condensation, then, in the double mutant, polar septation should occur before chromosome partitioning. This was not the case, as we observed chromosome partitioning before polar septation in some sporangia (Fig. 2I and J, arrows 2), although in many cases the polar chromosome domain was less clearly separated from the remainder of the chromosome than in the wild type (Fig. 2I and J, arrows 3). This could suggest that Spo0J or Soj or both stabilize the partitioned chromosome or that the multiple copies of the chromosome found in the spo0J soj double mutant (41) preclude efficient packing. In keeping with the importance of these proteins for chromosome partitioning, we noted that the spo0J soj double mutant occasionally formed medial septa directly through a chromosome (Fig. 2I and J, arrows 4) and that it produced anucleate cells and minicells (not shown). Thus neither Spo0J or Soj is essential for asymmetric chromosome condensation, although it remains possible that they stabilize the partitioned chromosome domain.
Depletion of FtsZ during sporulation does not prevent asymmetric chromosome condensation.
An early event in the asymmetric division pathway of B. subtilis is the relocalization of FtsZ from the midpoint to the cell poles (21), an event likely to be necessary to localize other proteins to polar division sites (5, 20, 22). The results described above demonstrate that asymmetric chromosome condensation occurs prior to the onset of septal biogenesis but did not address whether it is independent of FtsZ ring assembly, which precedes cell division. We therefore depleted cells of FtsZ during sporulation by utilizing a strain in which the ftsZ gene is under the control of the IPTG-inducible Pspac promoter (2). When cells are grown without IPTG, FtsZ levels fall, ultimately preventing cell division, but not cell growth, thereby causing the production of long, multinucleate cells (filaments) lacking detectable FtsZ. Under sporulation conditions, these filaments fail to synthesize the sporulation septum, although the cells express early sporulation-specific proteins (2).
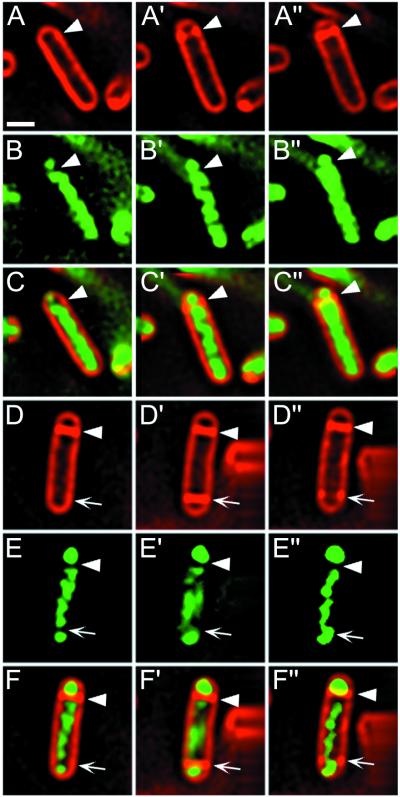
Following FtsZ depletion, we readily observed small condensed regions of the chromosomes during sporulation, both near the cell poles (Fig. 5A and B, arrows 1) and, less frequently, within the filaments (Fig. 5F to H, arrows 6). Nearly 50% of the filaments in the population contained at least one asymmetrically condensed chromosome by t2. We confirmed that the filaments with asymmetrically condensed chromosomes had entered the sporulation pathway by observing that they contained the early-sporulation-specific protein σF by immunofluorescence microscopy (Fig. 5F to H, arrows 5 and 6). In contrast, filaments without asymmetrically condensed chromosomes showed no σF immunostaining (Fig. 5F to H, arrows 4). Filaments with asymmetrically condensed chromosomes lacked FtsZ rings, as assessed by immunofluorescence microscopy (not shown); indeed, under these depletion conditions, we were unable to detect the FtsZ protein in Western blots (not shown). Thus, the polar chromosome condensation machinery may function independently of FtsZ ring formation. If so, then condensation would be independent of the entire cell division apparatus, whose assembly at future division sites is abolished in the absence of FtsZ.
FIG. 5.
Chromosome condensation and expression of sporulation-specific protein σF in wild-type and FtsZ-depleted cells. (A and B) Following depletion of FtsZ (strain KP444; see Materials and Methods), cell division is inhibited, producing long cells (A; FM 4-64 staining) that contain asymmetrically condensed chromosomes (B; DAPI staining) near the cell pole (arrows 1). (C to H) Cells were harvested at t2, prepared for immunofluorescence microscopy, and stained with anti-σF antibodies (green; D, E, G, and H) and DAPI (blue; C, E, F, and H). (C to E) Wild-type cells that show no σF immunostaining lack polar condensed regions of their chromosomes (arrows 2), whereas those with bright σF immunostaining show condensed chromosomes in various stages of translocation into the forespore (arrows 3). (F to H) Cells of strain KP444 (Pspac-ftsZ) were depleted of FtsZ by growth without IPTG (as described Materials and Methods), blocking cell division. Cells that show no σF immunostaining have symmetrically condensed chromosomes (arrows 4), whereas those with bright σF immunostaining show polar condensed regions (arrows 5 and 6). Scale bar, (A), 1 μm.
A novel determinant of cellular asymmetry.
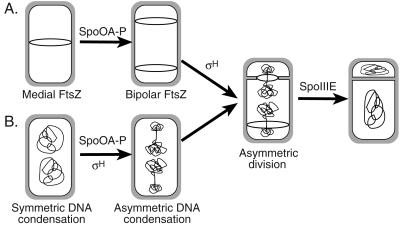
Our results demonstrate that clearing DNA from the path of an invaginating septum normally occurs during both symmetric and asymmetric cell division (Fig. 6). However, during the asymmetric cell division of sporulation, a single chromosome is partitioned into two separate domains of unequal sizes, whereas, during vegetative growth, the individual daughter chromosomes are separately and symmetrically condensed following their replication. While the mechanism by which bacteria partition a single chromosome into two distinct domains is presently unclear, our results dramatically illustrate that bacteria share with eukaryotes the ability to precisely control their chromosomal architecture.
FIG. 6.
A model for steps in the establishment of asymmetry during sporulation in B. subtilis. (A) The switch in the assembly of FtsZ (ring) from the cell midpoint to positions near both poles requires transcription factor Spo0A-P, and the onset of polar septation requires σH. (B) The reorganization of the chromosome condensation machinery from the symmetrically condensed bilobed structure of growing cells to form polar condensed chromosomes requires transcription factors Spo0A-P and σH. Asymmetric division then commences at one of the two sites of FtsZ assembly, and the remaining forespore chromosome is translocated across the sporulation septum by SpoIIIE. In cells lacking Spo0A-P, symmetric chromosome condensation is seen, whereas cells lacking σH show decondensed chromosomes that appear distinct from both the bilobed chromosomes of growing cells and the more-condensed chromosomes of sporulating bacteria.
Surprisingly, asymmetric chromosome condensation not only precedes asymmetric cell division but also occurs when asymmetric division is completely abolished by the depletion of cell division protein FtsZ. This suggests that asymmetric chromosome condensation is independent of the known members of the cell division machinery, since FtsZ is likely to be required for the recruitment of these proteins to potential division sites (7, 20, 42). Our results imply the existence of a novel determinant of cellular asymmetry that is independent of the known cell division machinery and that directs the reorganization of the chromosome in preparation for asymmetric division.
Acknowledgments
We thank Richard Losick for his gift of σF-specific antibodies and strains and Petra Levin and Richard Losick for the FtsZ-specific antibodies. We thank Aileen Rubio, Petra Levin, and Richard Losick for comments on the manuscript.
This work was supported by awards from the Arnold and Mabel Beckman Foundation and the Searle Scholars Program/The Chicago Community Trust and by an NSF predoctoral fellowship awarded to M.D.S.
REFERENCES
- 1.Bath, J., L. J. Wu, J. Errington, and C. Robinson. 2000. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290:995-997. [DOI] [PubMed] [Google Scholar]
- 2.Beall, B., and J. Lutkenhaus. 1991. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 5:447-455. [DOI] [PubMed] [Google Scholar]
- 3.Cervin, M., G. Spiegelman, B. Raether, K. Ohlsen, M. Perego, and J. Hoch. 1998. A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol. Microbiol. 29:85-95. [DOI] [PubMed] [Google Scholar]
- 4.Cooper, S. 1996. Segregation of cell structures, p. 1652-1660. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 5.Daniel, R. A., E. J. Harry, V. L. Katis, R. G. Wake, and J. Errington. 1998. Characterization of the essential cell division gene ftsL (yllD) of Bacillus subtilis and its role in the assembly of the division complex. Mol. Microbiol. 29:593-604. [DOI] [PubMed] [Google Scholar]
- 6.Gerdes, K., J. Moller-Jensen, and R. B. Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 7.Ghigo, J., D. S. Weiss, J. C. Chen, J. C. Yarrow, and J. Beckwith. 1999. Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol. 31:725-737. [DOI] [PubMed] [Google Scholar]
- 8.Gholamhoseinian, A., Z. Shen, J. J. Wu, and P. Piggot. 1992. Regulation of transcription of the cell division gene ftsA during sporulation of Bacillus subtilis. J. Bacteriol. 174:5647-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaser, P., M. E. Sharpe, B. Raether, M. Perego, K. Ohlsen, and J. Errington. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11:1160-1168. [DOI] [PubMed] [Google Scholar]
- 10.Gonzy-Treboul, G., C. Karmazyn-Campelli, and P. Stragier. 1992. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J. Mol. Biol. 224:967-979. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, G. S., D. Sitnikov, C. D. Webb, A. Teleman, A. Straight, R. Losick, A. W. Murray, and A. Wright. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90:1113-1121. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, G. S., and A. Wright. 1998. DNA segregation: putting chromosomes in their place. Curr. Biol. 8:925-927. [DOI] [PubMed] [Google Scholar]
- 13.Hauser, P. M., and J. Errington. 1995. Characterization of cell cycle events during the onset of sporulation in Bacillus subtilis. J. Bacteriol. 177:3923-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmstetter, C. E. 1996. Timing of synthetic activities in the cell cycle, p. 1627-1639. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 15.Holmes, V. F., and N. R. Cozzarelli. 2000. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl. Acad. Sci. USA 97:1322-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ireton, K., N. W. Gunther, and A. D. Grossman. 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ireton, K., D. Z. Rudner, K. J. Siranosian, and A. D. Grossman. 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 7:283-294. [DOI] [PubMed] [Google Scholar]
- 18.Jaacks, K. J., J. Healy, R. Losick, and A. D. Grossman. 1989. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 171:4121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen, R. B., and L. Shapiro. 1999. Chromosome segregation during the prokaryotic cell cycle. Curr. Opin. Cell Biol. 11:726-731. [DOI] [PubMed] [Google Scholar]
- 20.Katis, V. L., R. G. Wake, and E. J. Harry. 2000. Septal localization of the membrane-bound division proteins of Bacillus subtilis DivIB and DivIC is codependent only at high temperatures and requires FtsZ. J. Bacteriol. 182:3607-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin, P. A., and R. Losick. 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 10:478-488. [DOI] [PubMed] [Google Scholar]
- 22.Levin, P. A., R. Losick, P. Stragier, and F. Arigoni. 1997. Localization of the sporulation protein SpoIIE in Bacillus subtilis is dependent upon the cell division protein FtsZ. Mol. Microbiol. 25:839-846. [DOI] [PubMed] [Google Scholar]
- 23.Lin, D. C. H., P. A. Levin, and A. D. Grossman. 1997. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 94:4721-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, G., G. C. Draper, and W. D. Donachie. 1998. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol. Microbiol. 29:893-903. [DOI] [PubMed] [Google Scholar]
- 25.Margolis, P. S., A. Driks, and R. Losick. 1993. Sporulation gene spoIIB from Bacillus subtilis. J. Bacteriol. 175:528-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marston, A. L., and J. Errington. 1999. Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol. Cell 4:673-683. [DOI] [PubMed] [Google Scholar]
- 27.Pogliano, J., N. Osborne, M. Sharp, A. Abanes-DeMello, A. R. Perez, Y.-L. Sun, and K. Pogliano. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pogliano, K., L. Harry, and R. Losick. 1995. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol. 18:459-470. [DOI] [PubMed] [Google Scholar]
- 29.Pogliano, K., A. E. M. Hofmeister, and R. Losick. 1997. Disappearance of the σE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to the establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 179:3331-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quisel, J. D., D. C. Lin, and A. D. Grossman. 1999. Control of development by altered localization of a transcription factor in B. subtilis. Mol. Cell 4:665-672. [DOI] [PubMed] [Google Scholar]
- 31.Ryter, A. 1965. Etude morphologie de la sporulation de Bacillus subtilis. Ann. Inst. Pasteur (Paris) 108:40-60. [PubMed] [Google Scholar]
- 32.Setlow, B., N. Magill, P. Febbroriello, L. Nakhimousky, D. E. Koppel, and P. Setlow. 1991. Condensation of the forespore nucleoid early in sporulation of Bacillus species. J. Bacteriol. 173:6270-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp, M. D., and K. Pogliano. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA 96:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharpe, M. E., and J. Errington. 1995. Postseptational chromosome partitioning in bacteria. Proc. Natl. Acad. Sci. USA 92:8630-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharpe, M. E., and J. Errington. 1999. Upheaval in the bacterial nucleoid. An active chromosome segregation mechanism. Trends Genet. 15:70-74. [DOI] [PubMed] [Google Scholar]
- 36.Siranosian, K. J., and A. D. Grossman. 1994. Activation of spo0A transcription by sigma H is necessary for sporulation but not for competence in Bacillus subtilis. J. Bacteriol. 176:3812-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teleman, A. A., P. L. Graumann, D. C. H. Lin, A. D. Grossman, and R. Losick. 1998. Chromosome arrangement within a bacterium. Curr. Biol. 8:1102-1109. [DOI] [PubMed] [Google Scholar]
- 39.Wang, L., and J. Lutkenhaus. 1998. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol. 29:731-740. [DOI] [PubMed] [Google Scholar]
- 40.Webb, C. D., P. L. Graumann, J. A. Kahana, A. A. Teleman, P. A. Silver, and R. Losick. 1998. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol. 28:883-892. [DOI] [PubMed] [Google Scholar]
- 41.Webb, C. D., A. Teleman, S. Gordon, A. Straight, A. Belmont, D. C.-H. Lin, A. D. Grossman, A. Wright, and R. Losick. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]
- 42.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, L. J., and J. Errington. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572-575. [DOI] [PubMed] [Google Scholar]
- 44.Wu, L. J., and J. Errington. 1998. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol. Microbiol. 27:777-786. [DOI] [PubMed] [Google Scholar]
- 45.Wu, L. J., P. J. Lewis, R. Allmansberger, P. M. Hauser, and J. Errington. 1995. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in B. subtilis. Genes Dev. 9:1316-1326. [DOI] [PubMed] [Google Scholar]
- 46.Youngman, P., J. B. Perkins, and R. Losick. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn 917 insertions. Mol. Gen. Genet. 195:424-433. [DOI] [PubMed] [Google Scholar]
- 47.Yu, X., E. Weihe, and W. Margolin. 1998. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J. Bacteriol. 180:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]