Abstract
Transcription of ResDE-controlled genes in Bacillus subtilis was induced by sodium nitroprusside and nitric oxide. This induction requires the sensor kinase ResE and the response regulator ResD. Among members of the ResDE regulon, only the flavohemoglobin gene was induced by nitrosative stress via both a ResDE-dependent mechanism and an unidentified ResDE-independent mechanism.
The ResD-ResE signal transduction system controls the transcription of various genes that function in aerobic and anaerobic respiration, as well as genes of the Pho regulon (for a review, see references 11 and 24). Direct interaction between the response regulator ResD and promoter DNA of ResDE regulon genes was demonstrated (23, 34), and transcriptional activation of one of the genes, ctaA, in vitro by ResD was also reported (27). ResD-dependent activation is heightened in vivo when the level of oxygen becomes limited (22). A previous study suggested that ResE kinase and phosphatase activities are reciprocally regulated in response to oxygen level (21). Oxygen limitation seems to shift the ResE enzyme activity from a phosphatase-dominant to a kinase-dominant form, thereby leading to higher levels of ResD phosphorylation. However, we have also shown that oxygen limitation alone is not sufficient to induce expression of the ResDE regulon and that full induction requires both anaerobiosis and the presence of nitrite (15, 20). It is unknown whether nitrite itself or a metabolite of nitrite is needed for the induction. Because nitrite is spontaneously converted to nitric oxide (NO) by protonation and NO is freely diffusible in and among cells, NO might be responsible for the observed stimulatory effect by nitrite. In this paper, I examined whether NO affects ResDE-dependent gene regulation under aerobic and anaerobic conditions.
A mutation in hmp affects oxygen-dependent regulation of the ResDE regulon.
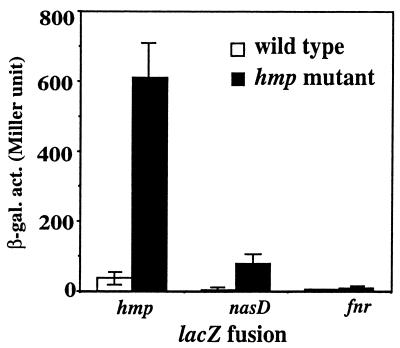
The hmp gene encodes a flavohemoglobin that is known to function in the detoxification of NO (4, 8, 10, 16, 17, 29). Studies of the flavohemoglobin of the enteric bacteria showed that it catalyzes conversion of NO to nitrate under aerobic conditions (7, 8) and to N2O under anaerobic conditions (13). If Bacillus subtilis Hmp functions in the detoxification of NO, and if NO is involved in induction of ResDE-dependent genes, a mutation in the flavohemoglobin gene (hmp) (15) may cause the accumulation of NO. This might result in the partial derepression of ResDE-controlled genes when hmp mutant cells are cultured in the presence of nitrite under aerobic conditions. Figure 1 shows that expression of three ResDE-dependent genes, hmp, nasD (nitrite reductase gene), and fnr (an anaerobic regulator gene), was low in cells of the wild-type strain grown under aerobic conditions. The expression of hmp and nasD in a hmp mutant was approximately 20-fold higher than that in wild-type cells. In contrast, aerobic fnr expression in the hmp mutant was only slightly elevated. The derepression by the hmp mutation was detected only in the presence of nitrite and not in the presence of nitrate (data not shown), consistent with a previous result that nitrate cannot be reduced to nitrite in the nitrogen-rich medium under aerobic conditions that repress expression of narGHJI and nasBC, encoding respiratory nitrate reductase and assimilatory nitrate reductase, respectively (9). This result suggests that accumulation of NO derived from nitrite in the hmp mutant leads to partial induction of nasD and hmp, while fnr expression is less responsive to NO.
FIG. 1.
Effect of an hmp mutation on aerobic expression of ResDE-controlled genes. Construction of fnr-lacZ (25), nasD-lacZ (20), and hmp-lacZ (15) was previously reported. B. subtilis cells were cultured under aerobic conditions in 2× yeast extract-tryptone medium supplemented with 1% glucose and 10 mM KNO2. Samples were withdrawn during the exponential and stationary phases of the growth curve, and the maximal activity during growth is shown. The data are averages from more than three experiments with standard errors. β-gal. act., β-galactosidase activity.
Sodium nitroprusside stimulates aerobic expression of the ResDE regulon.
If the increased transcription observed in the hmp mutant is caused by endogenous accumulation of NO, exogenous NO would cause a similar effect. This possibility was examined by adding sodium nitroprusside (SNP) to the culture medium. SNP in solution is known to release NO when exposed to visible light, when exposed to various reducing agents, or by addition of eukaryotic tissues (1). It was also reported that treatment of soybean cells with SNP at concentrations of between 0.1 and 1.0 mM generated 0.25 to 2.0 μM NO (5). Although SNP has been widely used as an NO generator, an argument has been raised that SNP is an NO+ donor and not an NO releaser (18). Therefore, the following effect of SNP might be due to released NO, but the possibility that the effect is caused by the nitrosation of an unknown target is not excluded.
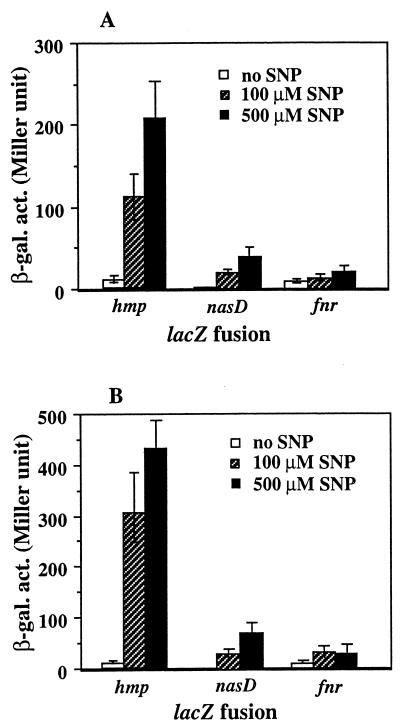
SNP was added at a concentration of 100 or 500 μM to early- to mid-log-phase aerobic cultures of wild-type cells carrying the lacZ fusions. The cultures were further incubated in the presence of SNP for 1 h with shaking. As shown in Fig. 2A, addition of SNP increased the level of ResDE-controlled genes compared to that in cultures grown in the absence of SNP. The induction ratio with 500 μM SNP varies depending on the fusion examined; that is, the induction ratios were 2.4-fold for fnr-lacZ, 16-fold for nasD-lacZ, and 20-fold for hmp-lacZ, which correspond well to the induction ratios of the fusions with the hmp mutation described above. The possibility that nitrite generated by autoxidation of NO (12) affected the gene expression is unlikely, because addition of nitrite at 500 μM did not alter the expression (data not shown). SNP is known to produce ferricyanide, but expression of the lacZ fusions was not changed in the presence of 500 μM ferricyanide (data not shown). The induction by SNP was further increased in the hmp mutant strain (Fig. 2B), suggesting accumulation of NO in the hmp mutant.
FIG. 2.
Induction of ResDE-dependent genes by SNP. Wild-type (A) and hmp mutant (B) strains carrying the various transcriptional lacZ fusions were grown aerobically in 2× yeast extract-tryptone medium. When cells reached early or mid-exponential growth (optical density at 600 nm of around 0.2), cells were treated with SNP at 100 or 500 μM. For a control, no SNP was added. The incubation was continued with shaking for 1 h before harvesting for measurement of β-galactosidase activity (β-gal. act.). The activities are averages from more than three experiments with standard errors.
Induction by SNP requires ResDE.
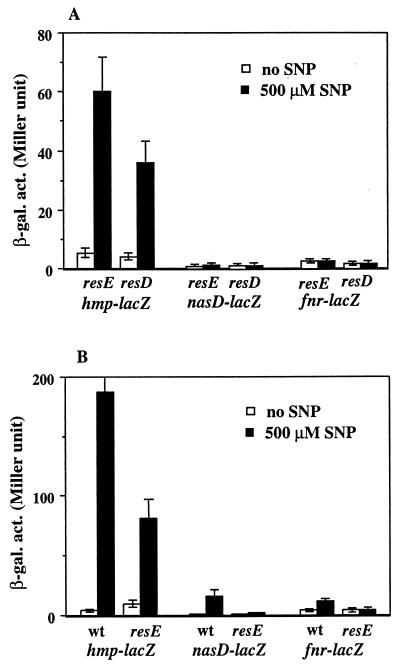
Because expression of all ResDE-controlled genes so far tested was more or less induced by SNP (data not shown), it is likely that the induction is dependent on ResD and ResE. The effects of resD and resE mutations on the induction by SNP were examined. The induction of the ResDE-dependent genes by SNP was completely abolished except in the case of hmp-lacZ, where a reduced but significant induction (approximately 10-fold) was observed in either the resD or resE mutant (Fig. 3A). The resD and resE genes constitute an operon together with their upstream genes, resA, resB, and resC. resD and resE are transcribed mainly from the res operon promoter, which is positively regulated by ResD and ResE (30). Therefore, we cannot exclude the possibility that the resE mutant could not respond to SNP because resD is not fully expressed in the absence of resE. In other words, nitrosative stress might stimulate phosphorylation of ResD independently of ResE, either by interaction with another sensor kinase or through a low-molecular-weight, high-energy phosphodonor such as acetyl phosphate. To examine this possibility, a strain in which the resABCDE genes are transcribed from an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible P spac promoter (30) was used. A resE mutation was also introduced in the Pspac-resABCDE strain. The lacZ fusions in the Pspac-resABCDE strain were induced by SNP in the presence of IPTG, and the induction was impaired in the Pspac-resABCDΔE strain, which lacks resE (Fig. 3B). Again, induction of hmp-lacZ by SNP was reduced (60-fold in resE+ versus 9-fold in resE) but not completely abolished. These results indicate that ResE perceives a signal, either NO itself or a signal derived from NO, leading to phosphorylation of ResD which activates transcription of the ResDE regulon. In addition, a ResDE-independent mechanism induces hmp transcription in response to NO.
FIG. 3.
Effects of resD and resE on induction of ResDE-dependent genes by SNP. (A) Wild-type and resD and resE mutant strains carrying lacZ fusions were grown aerobically and treated with 500 μM SNP as described in the legend to Fig. 2. (B) Expression of lacZ in Pspac-resABCDE (wild type [wt]) and Pspac-resABCDΔE (resE) strains. The activities are averages from two or three experiments with standard errors. β-gal. act., β-galactosidase activity.
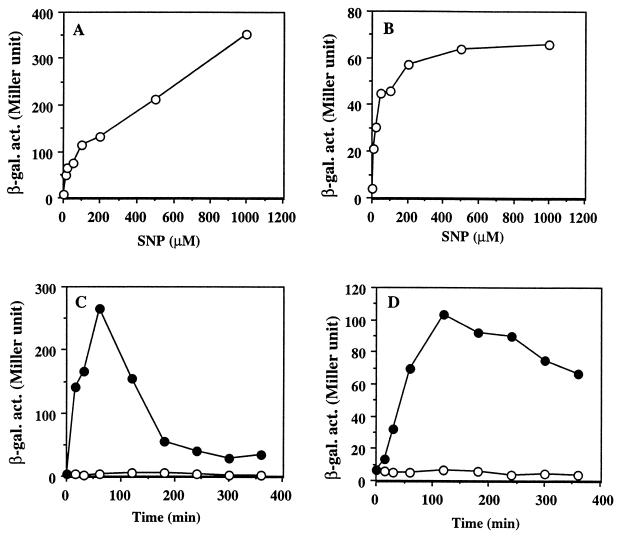
The response of the hmp promoter activity to SNP was further examined in the wild type and the resDE mutant (Fig. 4). The concentration of SNP that induces maximal promoter activity in wild-type cells was higher than 1 mM (Fig. 4A). In contrast, the expression in the resDE mutant reached a plateau in the presence of 50 μM SNP (Fig. 4B). β-Galactosidase activities were also measured after treatment with 500 μM SNP for a period of 6 h. An increase in β-galactosidase activity was detected within 15 min. hmp-lacZ expression in wild-type cells was highest at around 1 h and then sharply decreased (Fig. 4C). In contrast, the activity level persisted longer in the resDE mutant (Fig. 4D). This suggests different kinetics of ResDE-dependent and -independent activation by SNP.
FIG. 4.
Induction of hmp-lacZ by SNP. (A and B) Various concentrations of SNP were added to aerobic cultures, which were then incubated for 1 h. (C and D) SNP (500 μM) was added at time zero, and cultures were incubated for up to 6 h. Open circles, without SNP; closed circles, with SNP. The β-galactosidase activity (β-gal. act.) in the wild type (A and C) and in the resDE mutant (B and D) was measured.
Exogenous NO can also induce expression of the ResDE regulon under anaerobic conditions.
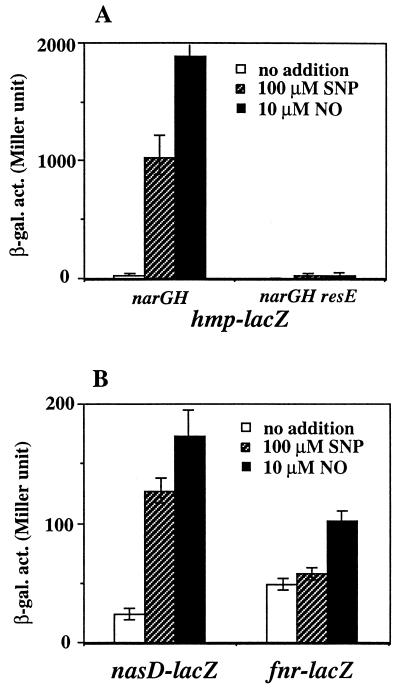
The results described here demonstrated that the aerobic expression of the ResDE regulon is induced by SNP. Cells grown anaerobically by nitrate respiration produce nitrite, from which NO is nonenzymatically derived. This normally would be sufficient to induce gene expression. In order to determine whether exogenous NO induces gene expression under anaerobic conditions, the endogenous production of NO was blocked by introducing a mutation in narGH, which encodes subunits of the respiratory nitrate reductase (9). The mutant strain is unable to grow by anaerobic respiration; however, it grows anaerobically by fermentation in the presence of glucose and pyruvate (19). Therefore, the narGH mutant was grown anaerobically by fermentation to examine the effect of SNP. Figure 5 shows that anaerobic expression of these genes in the narGH mutant in the absence of SNP (19) was up-regulated only 2- to 8-fold (compared to aerobic expression [Fig. 2A]), confirming that oxygen limitation alone is not sufficient to induce expression. When cells were treated with 100 μM SNP, the activity of hmp-lacZ (Fig. 5A) and nasD-lacZ (Fig. 5B) increased 5.5- and 57-fold compared to the untreated cells, respectively. There was little, if any, observable effect on fnr expression by SNP addition (Fig. 5B). A more marked induction was observed when 10 μM NO was added. Expression of fnr and nasD increased approximately 2- and 7.5-fold, respectively, and a greater-than-100-fold increase in hmp expression was detected after NO addition (Fig. 5). β-Galactosidase activities of hmp-lacZ, nasD-lacZ, and fnr-lacZ strains during aerobic growth in the absence of SNP were 10.5, 2.5, and 8.3 Miller units, respectively (Fig. 2A). The activities of the same fusions in cells challenged with NO during anaerobic growth were 1,886, 173, and 101 Miller units (Fig. 5), showing 180-, 69-, and 12-fold increases over the basal levels, respectively. The increase in hmp-lacZ induced by SNP or NO is dependent primarily on ResE, and only a low level of induction (approximately 6-fold) was detected in a narGH resE mutant (Fig. 5A).
FIG. 5.
Induction of ResDE-dependent genes by SNP and NO under anaerobic conditions. narGH or narGH resE mutant cells carrying an hmp-lacZ fusion (A) and narGH mutant cells carrying a nasD-lacZ or fnr-lacZ fusion (B) were cultured anaerobically in 2× yeast extract-tryptone medium supplemented with 0.5% glucose and 0.5% pyruvate. Mid-exponential-phase cells were treated with 500 μM SNP or 10 μM NO and were further incubated for 1 h. The concentration of NO was determined by using the concentration of saturated solutions at room temperature (approximately 1.8 mM). Saturated NO solutions were prepared in distilled water in a sealed serum vial by first sparging with argon gas to remove oxygen and then bubbling with NO gas. The NO solution was withdrawn with gas-tight Hamilton syringes. Error bars indicate standard errors. β-gal. act., β-galactosidase activity.
Significance of NO and NO-related species in bacterial gene regulation.
Although numerous studies have shown a versatile role for NO as a signal molecule in mammalian physiological processes (2), only recently has the importance of NO in bacterial gene regulation been recognized. The flavohemoglobin genes (hmp) from Escherichia coli (28), and Salmonella enterica serovar Typhimurium (3) are induced by NO. The induction of E. coli hmp is dependent on MetR, a DNA binding protein of the LysR family (18), and inactivation of the iron-dependent Fur repressor by NO is responsible for activation of S. enterica hmp (3). NO also activates an oxidative-stress response in E. coli (26) through nitrosylation of the [2Fe-2S] centers in SoxR, the redox-sensitive transcriptional regulator (6). Endogenous and exogenous NO induces nitrite reductase and nitric oxide reductase genes that function in denitrification in Rhodobacter sphaeroides 2.4.3 (14) and Paracoccus denitrificans (33). The induction requires NnrR or NNR (the nitrite and nitric oxide reductase regulator), a transcriptional activator of the FNR/CAP family. NnrR (31) and NNR (32) lack cysteine clusters present in FNR, which provide coordination for a redox-sensitive [4Fe-4S] cluster. If NO interacts directly with these regulators, it does so in a manner that does not involve nitrosylation of an iron-sulfur center.
To my knowledge, this is the first case demonstrating that NO and NO-related species are involved in global gene regulation in a gram-positive bacterium. In soil, its natural environment, B. subtilis constantly encounters NO produced by denitrifiers. NO is also generated endogenously during nitrate respiration. In addition, B. subtilis has a protein (YflM) that shows significant homology to the N-terminal oxygenase domain of inducible nitric oxide synthase of mammals (http://genolist.pasteur.fr/SubtiList/). Whether YflM is involved in NO synthesis remains to be tested, and regulation of yflM expression has yet to be examined. Future studies are required to determine if NO directly interacts with the sensor kinase ResE. Because ResE has no cysteine residue, it is not susceptible to S nitrosylation by NO, and there is no evidence at present suggesting that ResE utilizes a cofactor containing metal that may be another target of NO. It is unknown whether NO is the sole signal for the ResDE signal transduction pathway. Oxygen limitation and NO may synergistically activate the ResDE regulon. Alternatively, NO-dependent gene induction might be a property of anaerobically grown cells. Since NO reacts rapidly with oxygen under aerobic conditions, oxygen limitation ensures the efficacy of the signal. Induction of fnr expression by NO or SNP is much lower than that of hmp and nasD. An induction ratio for fnr by oxygen limitation (expression ratio during anaerobiosis versus aerobiosis) is also at least 10-fold lower than those of hmp and nasD, which is consistent with a hypothesis that the ResDE regulon is activated by the same signal derived by either nitrosative stress or oxygen limitation.
Acknowledgments
This work was supported by grants (MCB9996014 and MCB0110513) from the National Science Foundation and a Medical Research Foundation Seed Grant from the Oregon Health Sciences Foundation.
I am grateful to F. Marion Hulett for a kind gift of a strain and to Peter Zuber, James P. Shapleigh, and Pierre Moënne-Loccoz for invaluable discussions and advice.
REFERENCES
- 1.Bates, J. N., M. T. Baker, R. Guerra, Jr., and D. G. Harrison. 1991. Nitric oxide generation from nitroprusside by vascular tissue. Evidence that reduction of the nitroprusside anion and cyanide loss are required. Biochem. Pharmacol. 42:S157-S165. [DOI] [PubMed] [Google Scholar]
- 2.Bogdan, C. 2001. Nitric oxide and the regulation of gene expression. Trends Cell Biol. 11:66-75. [DOI] [PubMed] [Google Scholar]
- 3.Crawford, M. J., and D. E. Goldberg. 1998. Regulation of the Salmonella typhimurium flavohemoglobin gene. J. Biol. Chem. 273:34028-34032. [DOI] [PubMed] [Google Scholar]
- 4.Crawford, M. J., and D. E. Goldberg. 1998. Role for Salmonella flavohemoglobin in protection from nitric oxide. J. Biol. Chem. 273:12543-12547. [DOI] [PubMed] [Google Scholar]
- 5.Delledonne, M., J. Zeier, A. Marocco, and C. Lamb. 2001. Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 98:13454-13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding, H., and B. Demple. 2000. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc. Natl. Acad. Sci. USA 97:5146-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner, P. R., A. M. Gardner, L. A. Martin, Y. Dou, T. Li, J. S. Olson, H. Zhu, and A. F. Riggs. 2000. Nitric-oxide dioxygenase activity and function of flavohemoglobins. Sensitivity to nitric oxide and carbon monoxide inhibition. J. Biol. Chem. 275:31581-31587. [DOI] [PubMed] [Google Scholar]
- 8.Gardner, P. R., A. M. Gardner, L. A. Martin, and A. L. Salzman. 1998. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. USA 95:10378-10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaser, P., A. Danchin, F. Kunst, P. Zuber, and M. M. Nakano. 1995. Identification and isolation of a gene required for nitrate assimilation and anaerobic growth of Bacillus subtilis. J. Bacteriol. 177:1112-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hausladen, A., A. J. Gow, and J. S. Stamler. 1998. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. USA 95:14100-14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulett, F. M. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933-939. [DOI] [PubMed] [Google Scholar]
- 12.Kharitonov, V. G., A. R. Sundquist, and V. S. Sharma. 1994. Kinetics of nitric oxide autoxidation in aqueous solution. J. Biol. Chem. 269:5881-5883. [PubMed] [Google Scholar]
- 13.Kim, S. O., Y. Orii, D. Lloyd, M. N. Hughes, and R. K. Poole. 1999. Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 445:389-394. [DOI] [PubMed] [Google Scholar]
- 14.Kwiatkowski, A. V., and J. P. Shapleigh. 1996. Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J. Biol. Chem. 271:24382-24388. [DOI] [PubMed] [Google Scholar]
- 15.LaCelle, M., M. Kumano, K. Kurita, K. Yamane, P. Zuber, and M. M. Nakano. 1996. Oxygen-controlled regulation of flavohemoglobin gene in Bacillus subtilis. J. Bacteriol. 178:3803-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, L., M. Zeng, A. Hausladen, J. Heitman, and J. S. Stamler. 2000. Protection from nitrosative stress by yeast flavohemoglobin. Proc. Natl. Acad. Sci. USA 97:4672-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Membrillo-Hernández, J., M. D. Coopamah, M. F. Anjum, T. M. Stevanin, A. Kelly, M. N. Hughes, and R. K. Poole. 1999. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J. Biol. Chem. 274:748-754. [DOI] [PubMed] [Google Scholar]
- 18.Membrillo-Hernández, J., M. D. Coopamah, A. Channa, M. N. Hughes, and R. K. Poole. 1998. A novel mechanism for upregulation of the Escherichia coli K-12 hmp (flavohemoglobin) gene by the ‘NO releaser’, S-nitrosoglutathione: nitrosation of homocysteine and modulation of MetR binding to the glyA-hmp intergenic region. Mol. Microbiol. 29:1101-1112. [DOI] [PubMed] [Google Scholar]
- 19.Nakano, M. M., Y. P. Dailly, P. Zuber, and D. P. Clark. 1997. Characterization of anaerobic fermentative growth in Bacillus subtilis: identification of fermentation end products and genes required for the growth. J. Bacteriol. 179:6749-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano, M. M., T. Hoffmann, Y. Zhu, and D. Jahn. 1998. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J. Bacteriol. 180:5344-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano, M. M., and Y. Zhu. 2001. Involvement of the ResE phosphatase activity in down-regulation of ResD-controlled genes in Bacillus subtilis during aerobic growth. J. Bacteriol. 183:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano, M. M., Y. Zhu, K. Haga, H. Yoshikawa, A. L. Sonenshein, and P. Zuber. 1999. A mutation in the 3-phosphoglycerate kinase gene allows anaerobic growth of Bacillus subtilis in the absence of ResE kinase. J. Bacteriol. 181:7087-7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakano, M. M., Y. Zhu, M. LaCelle, X. Zhang, and F. M. Hulett. 2000. Interaction of ResD with regulatory regions of anaerobically induced genes in Bacillus subtilis. Mol. Microbiol. 37:1198-1207. [DOI] [PubMed] [Google Scholar]
- 24.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 25.Nakano, M. M., P. Zuber, P. Glaser, A. Danchin, and F. M. Hulett. 1996. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J. Bacteriol. 178:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nukoshiba, T., T. deRojas-Walker, J. S. Wishnok, S. R. Tannenbaum, and B. Demple. 1993. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophage. Proc. Natl. Acad. Sci. USA 90:9993-9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul, S., X. Zhang, and F. M. Hulett. 2001. Two ResD-controlled promoters regulate ctaA expression in Bacillus subtilis. J. Bacteriol. 183:3237-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole, R. K., M. F. Anjum, J. Membrillo-Hernández, S. O. Kim, M. N. Hughes, and V. Stewart. 1996. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 178:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole, R. K., and M. N. Hughes. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36:775-783. [DOI] [PubMed] [Google Scholar]
- 30.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 178:1374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tosques, I. E., J. Shi, and J. P. Shapleigh. 1996. Cloning and characterization of nnrR, whose product is required for the expression of proteins involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 178:4958-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Spanning, R. J., A. P. De Boer, W. N. Reijnders, S. Spiro, H. V. Westerhoff, A. H. Stouthamer, and J. van der Oost. 1995. Nitrite and nitric oxide reduction in Paracoccus denitrificans is under the control of NNR, a regulatory protein that belongs to the FNR family of transcriptional activators. FEBS Lett. 360:151-154. [DOI] [PubMed] [Google Scholar]
- 33.Van Spanning, R. J. M., E. Houben, W. N. Reijnders, S. Spiro, H. V. Westerhoff, and N. Saunders. 1999. Nitric oxide is a signal for NNR-mediated transcription activation in Paracoccus denitrificans. J. Bacteriol. 181:4129-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, X., and F. M. Hulett. 2000. ResD signal transduction regulator of aerobic respiration in Bacillus subtilis: cta promoter regulation. Mol. Microbiol. 37:1208-1219. [DOI] [PubMed] [Google Scholar]