Abstract
Vibrio anguillarum possesses at least two N-acylhomoserine lactone (AHL) quorum-sensing circuits, one of which is related to the luxMN system of Vibrio harveyi. In this study, we have cloned an additional gene of this circuit, vanT, encoding a V. harveyi LuxR-like transcriptional regulator. A V. anguillarum ΔvanT null mutation resulted in a significant decrease in total protease activity due to loss of expression of the metalloprotease EmpA, but no changes in either AHL production or virulence. Additional genes positively regulated by VanT were identified from a plasmid-based gene library fused to a promoterless lacZ. Three lacZ fusions (serA::lacZ, hpdA-hgdA::lacZ, and sat-vps73::lacZ) were identified which exhibited decreased expression in the ΔvanT strain. SerA is similar to 3-phosphoglycerate dehydrogenases and catalyzes the first step in the serine-glycine biosynthesis pathway. HgdA has identity with homogentisate dioxygenases, and HpdA is homologous to 4-hydroxyphenylpyruvate dioxygenases (HPPDs) involved in pigment production. V. anguillarum strains require an active VanT to produce high levels of an l-tyrosine-induced brown color via HPPD, suggesting that VanT controls pigment production. Vps73 and Sat are related to Vibrio cholerae proteins encoded within a DNA locus required for biofilm formation. A V. anguillarum ΔvanT mutant and a mutant carrying a polar mutation in the sat-vps73 DNA locus were shown to produce defective biofilms. Hence, a new member of the V. harveyi LuxR transcriptional activator family has been characterized in V. anguillarum that positively regulates serine, metalloprotease, pigment, and biofilm production.
Diverse gram-negative and gram-positive bacteria have been shown to use intercellular communication mechanisms to regulate the transcription of multiple target genes in concert with cell density. This type of communication, termed quorum sensing, is mediated through the production of diffusible signal molecules, termed autoinducers or pheromones, which effectively enable a bacterium to monitor its own population density (for reviews, see references 18, 24, 25, and 56). In gram-negative bacteria, the best-studied autoinducer molecules are N-acylhomoserine lactones (AHLs), which vary in the length, saturation state, and C3 substitutions of the N-acyl side chain (25).
The cell density-dependent regulation of bioluminescence in Vibrio (Photobacterium) fischeri (38, 39) is frequently used as the paradigm for quorum sensing. In this marine symbiont, as the bacterial cell population density increases, the level of the autoinducer N-(3-oxohexanoyl)homoserine lactone (3-oxo-C6-HSL) (19) accumulates until a critical threshold concentration is reached. 3-Oxo-C6-HSL then binds to the transcriptional activator LuxR, and the resulting LuxR/3-oxo-C6-HSL complex triggers transcription of the luminescence (lux) operon, resulting in the emission of light (39).
Similar to V. fischeri, the free-living marine bacterium Vibrio harveyi also regulates bioluminescence in a cell density-dependent manner through the production and sensing of N-(3-hydroxybutanoyl)-l-homoserine lactone (3-hydroxy-C4-HSL) (for a review, see reference 39). However, the regulation of light production in V. harveyi is very different and appears to be more complex than in V. fischeri. Based on genetic analyses, Freeman and Bassler (21, 22) have proposed a model for the regulation of bioluminescence in V. harveyi that involves two signaling systems and two autoinducer molecules. The first quorum-sensing system relies on 3-hydroxy-C4-HSL (10), the synthesis of which is directed by luxM. Interestingly, the gene product of luxM shows no homology to the LuxI family of AHL synthases (6). Regulation of bioluminescence via the second quorum-sensing system utilizes another, as yet unidentified, signal molecule (AI-2), which is chemically distinct from AHLs and is synthesized via LuxS (64).
The sensors for 3-hydroxy-C4-HSL and AI-2, named LuxN and LuxQ, respectively, resemble proteins belonging to two-component signaling systems (6, 8), and each possesses a conserved histidine kinase and a response regulator domain. At low cell densities, in the absence of signal molecules, LuxN and LuxQ are suggested to work in parallel by relaying the phosphates from their response regulator domains to a shared phosphorelay protein, LuxU (22). The phosphate from LuxU is then transferred to the response regulator domain of the σ54-dependent activator LuxO, which, when phosphorylated, represses bioluminescence, together with σ54, by activating the expression of an as yet unidentified repressor (7, 21, 31). In contrast, at high cell densities, the signal molecules accumulate and are believed to bind to their respective sensors (21, 22). It is thought that 3-hydroxy-C4-HSL may bind directly to LuxN, whereas AI-2 is postulated to bind to LuxQ via interaction with a putative periplasmic protein, LuxP. Binding of the signals is suggested to switch the sensor kinase activities of LuxN and LuxQ into phosphatases, leading to the dephosphorylation of LuxO and derepression of the lux operon. This then allows LuxR, the transcriptional activator, to positively activate the bioluminescence genes. LuxR from V. harveyi has no similarity to LuxR from V. fischeri.
The nonbioluminescent salmonid fish pathogen Vibrio anguillarum contains multiple quorum-sensing systems (46, 47). A V. fischeri LuxI homologue, VanI, was characterized and shown to be required for the production of N-(3-oxodecanoyl)-l-homoserine lactone (3-oxo-C10-HSL) (46). A null mutant for vanI showed a loss of 3-oxo-C10-HSL production. However, the vanI mutant was still capable of weakly activating bioluminescence gene biosensors in Escherichia coli. Two additional AHLs were identified, N-hexanoyl-l-homoserine lactone (C6-HSL) and N-(3-hydroxyhexanoyl)-l-homoserine lactone (3-hydroxy-C6-HSL), along with the gene, vanM, responsible for their synthesis (47). VanM was shown to be homologous to LuxM of V. harveyi. Moreover, a second gene, vanN, which encodes a V. harveyi LuxN homologue, was found downstream of vanM (47).
To further characterize the V. harveyi-like quorum-sensing system in V. anguillarum, we wanted to determine if a homologue of the V. harveyi LuxR transcriptional activator is also present in V. anguillarum. To date, three homologues (HapR, OpaR, and SmcR) of the V. harveyi LuxR transcriptional activator have been identified in Vibrio spp. HapR from V. cholerae is the positive regulator of a hemagglutinin protease encoded by the hap gene (27). OpaR from V. parahaemolyticus is the positive regulator of an opacity phenotype that appears to be correlated with capsular polysaccharide production (20, 33). SmcR from V. vulnificus is thought to positively regulate the metalloprotease gene (vvp) and to negatively regulate the cytolysin gene (vvhA), alkaline phosphatase production, motility, fimbria production, and biofilm formation (35, 36, 58).
This work describes the cloning and characterization of vanT from V. anguillarum, which codes for a homologue of V. harveyi LuxR. Functional analyses suggest that VanT positively regulates the biosynthesis of serine, glycine, and one-carbon-unit molecules, the expression of the metalloprotease gene empA, pigment production, and biofilm formation.
MATERIALS AND METHODS
Strains, phage, plasmids, and media.
Bacterial strains and plasmids are described in Table 1. E. coli SY327 (λpir) was used for transformation after subcloning fragments into either the pNQ705-1 or pDM4 suicide vector. All plasmids to be conjugated into V. anguillarum were transformed into E. coli S17-1 (λpir), which was used as the donor strain. Plasmid transfers from E. coli to V. anguillarum were done as previously described (45). E. coli XL1-Blue was used for bacteriophage lambda infections and for most transformations. E. coli DH5αPRO was used for the two-plasmid screening. E. coli JM109 was used to harbor the bioluminescence sensor plasmid pSB1075.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or relevant markers | Reference(s) or source |
|---|---|---|
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) | 57 |
| SY327 | Δ(lac-pro) argE(Am) rif malA recA56 λpir | 43 |
| S17-1 | thi pro hsdR hsdM+recA RP4-2-Tc::Mu-Km::Tn7 λpir | 61 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-pro) [F′ proAB lacIqlacZΔM15 Tn10(Tetr)] | Stratagene |
| DH5αPRO | deoR endA1 gyrA96 hsdR17(rK−mK+) recA1 relA1 supE44 thi-1 Δ(lacZYA-argFV169) φ80ΔlacZΔM15 F− λ−PN25/tetRPlacIq/lacI Spr | Clonetech |
| C. violaceum CV026 | Kmr; strain ATCC 31532, defective in C6-HSL production due to a mini-Tn5 insertion in the cviI gene | 29, 34, 69 |
| V. anguillarum | ||
| NB10 | Wild type, serotype O1, clinical isolate from the Gulf of Bothnia | 49 |
| AC10 | In-frame deletion of vanT | This study |
| DM70 | Cmr; polar mutation in sat gene | This study |
| Plasmids | ||
| pBluescript | Apr; ColE1 origin | Stratagene |
| pBSVanT-175 | Apr; pBluescript containing 175-bp PCR fragment from vanT (bp 463-640) | This study |
| pBSVanT3-2 | Apr; pBluescript containing cloned fragment with the vanT gene | This study |
| pDM4 | Cmr; suicide vector with an R6K origin (pir requiring) and sacBR genes from Bacillus subtilis | 45 |
| pDMVanT1 | Cmr; pDM4 derivative containing vanT bp 146-418 fused in-frame to bp 1028-1303 | This study |
| pNQ705-1 | Cmr; suicide vector that contains an R6K origin (pir requiring) | 37 |
| pNQSat1 | Cmr; pNQ705-1 derivative containing a 220-bp internal 5′ fragment of sat gene | This study |
| pSup202 | Cmr Tcr Apr; pBR325 derivative which carries mobilization genes (RP4 mob) | 61 |
| pMC1871 | Tcr; contains a promoterless lacZ gene that lacks a ribosome-binding site and first 8 amino acid codons | Pharmacia Biotech |
| pTL61T | Apr; contains a promoterless lacZ gene and the lacZ ribosome-binding site | 32 |
| pDM5 | Cmr Tcr; pSup202 derivative containing the promoterless lacZ gene cassette from pMC1871 used for translational gene fusion studies | This study |
| pDM8 | Cmr Tcr; pSup202 derivative containing the promoterless lacZ gene cassette from pTL61T used for transcriptional gene fusion studies | This study |
| pDM8-Vps73 | Cmr Tcr; pDM8 derivative containing a vps73::lacZ transcriptional gene fusion | This study |
| pDM8-EmpA | Cmr Tcr; pDM8 derivative containing an empA::lacZ transcriptional gene fusion | This study |
| pDM8-SerA | Cmr Tcr; pDM8 derivative containing a serA::lacZ transcriptional gene fusion | This study |
| pDM8-HpdA | Cmr Tcr; pDM8 derivative containing an hpdA::lacZ transcriptional gene fusion | This study |
| pDM5-S5 | Cmr Tcr; pDM5 derivative containing a vps73::lacZ translational gene fusion | This study |
| pDM5-S13 | Cmr Tcr; pDM5 derivative containing an hpdA::lacZ translational gene fusion | This study |
| pDM5-Ssp4 | Cmr Tcr; pDM5 derivative containing a serA::lacZ translational gene fusion | This study |
| pPROLar.A131 | Knr; p15A origin, protein expression vector using a Plac/ara-1 promoter | Clontech |
| pLar-VanT | Knr; derivative of pPROLar.A131 containing the vanT gene fused to the Plac/ara-1 promoter | This study |
| pAC-EmpA | Cmr; pACYC184 derivative containing the empA gene and its promoter | This study |
| pSB1075 | Apr; contains a fusion of lasRI′::luxCDABE in pUC18 | 69 |
E. coli was routinely grown in Luria broth, which contains Bacto-tryptone (10 g/liter), Bacto yeast extract (5 g/liter), and sodium chloride (10 g/liter). For V. anguillarum, Trypticase soy broth medium (TSB) from BBL was used for routine growth. For selection against E. coli after conjugation, two Vibrio selective media were used, TCBS agar (Difco Laboratories) and VAM medium (4). Biofilm growth medium was minimal M63 salts (60) supplemented with 1% (wt/vol) NaCl, 1.5% (wt/vol) Casamino Acids, 1% (wt/vol) glucose, 1 mM MgSO4, and 10 μg of thiamine per ml.
Antibiotic concentrations for all E. coli strains were ampicillin at 100 μg/ml, tetracycline at 10 μg/ml, kanamycin at 30 μg/ml, and chloramphenicol at 25 μg/ml. Antibiotic concentrations for V. anguillarum in the various media were: TSB and TCBS, tetracycline at 5 μg/ml and chloramphenicol at 5 μg/ml; VAM, chloramphenicol at 1 μg/ml.
Isolation and detection of AHLs.
AHLs were purified and characterized as described by Cámara et al. (9). Essentially, spent supernatants from stationary-phase cultures of V. anguillarum NB10 and AC10 grown overnight in TSB were extracted with dichloromethane, and solvent extracts were separated by thin-layer chromatography (TLC) as previously described (9, 34). For analysis of 3-hydroxy-C6-HSL and C6-HSL, reverse-phase aluminum-backed RP18 F254S TLC plates (20 cm by 20 cm; Merck) and a mobile phase of 60% (vol/vol) methanol in water were employed. In contrast, 3-oxo-C10-HSL was analyzed on aluminum-backed silica gel 60 F254 normal-phase TLC plates (20 cm by 20 cm; Merck) using a 45%:55% (vol/vol) hexane-acetone mix as the mobile phase. AHL synthetic standards (see below) were used as markers. Detection of 3-hydroxy-C6-HSL and C6-HSL was done by overlaying the TLC plates with soft top agar seeded with the Chromobacterium violaceum CV026 reporter using both the AHL activation and inhibition violacein assays (34). For the detection of 3-oxo-C10-HSL, a bioluminescent E. coli lux-based AHL biosensor termed E. coli JM109(pSB1075), which contains an intact lasR gene and the lasI promoter from Pseudomonas aeruginosa fused to luxCDABE from Photorhabdus luminescens, was used (70).
The AHL standards, 3-oxo-C10-HSL, 3-hydroxy-C6-HSL, and C6-HSL, were synthesized, purified, and characterized as described previously (9, 13). Each compound was subjected to mass spectrometry (MS), proton nuclear magnetic resonance spectroscopy, and infrared spectroscopy. For 3-hydroxy-C6-HSL and C6-HSL, spectroscopic data are provided by Chhabra et al. (13), and Milton et al. (46) give data for 3-oxo-C10-HSL.
DNA techniques and sequencing.
Oligonucleotides were synthesized using Applied Biosystems DNA/RNA synthesizer model 394. Unless otherwise stated, all conditions for the various DNA techniques were as described by Sambrook et al. (57). Reaction conditions for the DNA-modifying enzymes and DNA restriction enzymes were performed as suggested by the manufacturers. Double-stranded DNA sequencing was performed using the dideoxy chain termination method with T7 DNA polymerase (Pharmacia Biotech) and by primer walking in two directions from known regions of DNA sequence. The T7 and T3 primers were used for sequencing fragments in pBluescript. For sequencing the gene fusions to lacZ, a primer (5′-GATTAAGTTGGGTAACGC-3′) whose site is located at the 5′ end of the lacZ gene was used.
PCR conditions.
PCR was performed as previously described (37) except that instead of the standard reaction buffer, a buffer containing 1% Thesit was used (52). To obtain the initial 175-bp V. anguillarum PCR fragment using the degenerate primer, the PCR optimization kit from Stratagene (buffer 8) was employed. When a PCR fragment required minimal errors, the high-fidelity Pfu polymerase (Stratagene) was used.
Cloning of the vanT DNA locus.
Protein alignments of LuxR from V. harveyi (59), OpaR from Vibrio parahaemolyticus (33), and HapR from Vibrio cholerae (27) indicated that these proteins were most homologous at the amino terminus. Two oligonucleotides were designed based on complementation to the V. harveyi luxR sequence (59). HarveyiR-3 [5′-GGCACTAGT(CT)TGICGIACIAC(AG)TG(AG)TT-3′] is a degenerate, inosine-containing oligonucleotide that is complementary to V. harveyi residues 522 to 539 that encode amino acids NHVVRQF (59) and that has an SpeI site at its 5′ end. HarveyiR-4 [5′-GGTGAGCTCAAACGTAAACAGCAACTGATGGA-3′] was designed to be directly complementary to V. harveyi luxR residues 363 to 385 that encode amino acids KRKQQLME (59) and that has a SacI site at the 5′ end. A 175-bp fragment was amplified from the chromosome of V. anguillarum, purified from 1% agarose using Ultrafree-DA spin columns (Millipore), digested overnight with restriction enzymes SacI and SpeI, cloned into similarly digested pBluescript (Stratagene), creating pBSVanT-175, and sequenced. The deduced protein sequence of this fragment was 92% similar to LuxR of V. harveyi (59).
The 175-bp fragment was used as a probe to screen a previously described (44) gene bank of chromosomal DNA from V. anguillarum in the Lambda Zap II bacteriophage (Stratagene). Probe labeling, plaque hybridization, and excision of the recombinant plasmids were done as previously described (44, 45). For all recombinant plasmids, the chromosomal DNA inserts were unstable, as deletions occurred during excision from the bacteriophage (data not shown). To acquire a stable DNA fragment and an active vanT gene for sequencing and functional analyses, we used preliminary data suggesting that E. coli does not express the empA gene without VanT present in the cell. Thus, pAC-EmpA, which carries empA and its promoter, was transformed into XL1-Blue E. coli. pAC-EmpA was created by ligating an XbaI- and SalI-digested empA-containing fragment from pBS80 (44) to XbaI- and XhoI-digested pACYC184. Excised plasmids from the positive plaques were transformed into XL1-Blue carrying pAC-EmpA and plated onto TSA medium containing 2% skim milk. A colony that gave a reproducible zone of clearing was picked. The pBluescript plasmid pBSVanT3-2, containing the cloned DNA fragment, was purified and used for sequencing the vanT gene.
Construction of vanT mutant strain.
For functional analyses, an in-frame deletion was made in vanT by allelic exchange as previously described (45). To create the new deletion allele, pBSVanT3-2 was used as the template with PCR primers VanT-A (5′-GGAAGATCTGCCAATACGCGAACC-3′) and VanT-B (5′-AAGTTAAGCCTAGTGCATGAGTTGTTAATC-3′) to create a fragment from bp 0 to 65 and with primers VanT-C (5′-CACTAGGCTTAACTTCCTAGTCTC-3′) and VanT-D (5′-CTCGAGCTCGGTTTGGCTATCGAG-3′) to create a fragment from bp 675 to 950. These two fragments contained an overlap of similar sequence and were used as templates in a second PCR using primers VanT-A and VanT-D. Using BglII and SacI, the fragment was cloned into the BglII and SacI sites of pDM4, creating pDMVanT1. After allelic exchange using pDMVanT1, the start codon of vanT was fused to the last amino acid codon of the gene, and the strain containing this deletion was called AC10. The in-frame deletion in AC10 was confirmed by PCR amplification of this locus using primers VanT-10B (5′-GGACTAGTAATTCAACAGTCATTGGCA-3′) and VanT-16B (5′-CTCGAGCTCCTTTGACGATTCATGCTCA-3′) and subsequent DNA sequencing.
Construction of a polar mutation within the sat-vps73 locus.
A mutant carrying a polar mutation in the sat-vps73 locus (DM70) was made by the integration of a suicide plasmid, pNQSat1, into the sat gene as described previously (44). A 220-bp PCR fragment complementary to the 5′ end of the sat gene was amplified from the V. anguillarum chromosome using primers Sat1-A (5′-GGAAGATCTCATTGATCCCAAATGTTA-3′) and Sat1-B (5′-GGGACTAGTATACATCATTTCCTATCT-3′). Using BglII and SpeI, the PCR fragment was cloned into similar sites of pNQ705-1, creating pNQSat1. The entire pNQSat1 was inserted 54 bp downstream of the sat start site. Since the PCR fragment did not contain the sat promoter or start codon, the plasmid insertion created a null mutation of sat and will have a polar effect on vps73 and other possible uncharacterized downstream genes. The insertion of the plasmid was checked by PCR analysis as previously described (44). Stability of the insertion mutation was tested by growth for 30 generations in the absence of chloramphenicol.
Two-plasmid screening for genes regulated by VanT.
A method was designed that used two plasmids in an E. coli background to identify V. anguillarum genes that are either positively or negatively regulated by VanT. The first plasmid, pLar-VanT, is a pPROLar.A131 (Clontech Laboratories) derivative that contains the vanT gene fused to a hybrid regulatory unit (Plac/ara-1) between the lac promoter and the araBAD promoter. To create pLar-VanT, a PCR fragment that contains a promoterless vanT gene (bp 63 to 770) was amplified using primers VanT-LarA (5′-CGGGGTACCCATGGAAACATCGATAGAA-3′) and VanT-LarB (5′-CGCGGATCCGTTCATTAAATTGCTGGT-3′). The fragment was digested with KpnI and BamHI and ligated into pPROLar.A131 similarly digested. Due to its fusion to the hybrid promoter, Plac/ara-1, the expression of vanT was highly inducible by l-arabinose and IPTG (isopropylthiogalactopyranoside) and tightly repressed by AraC and LacI in E. coli DH5αPRO.
The second plasmid, pDM5, is a pSUP202 (61) derivative that contains a promoterless lacZ gene for translational gene fusion studies. The promoterless lacZ gene for pDM5 lacks a ribosome-binding site and the first eight nonessential amino-terminal amino acid codons. This 3.1-kb promoterless lacZ gene cartridge was removed from pMC1871 (Pharmacia Biotech) using PstI and ligated into the PstI site within the ampicillin gene of pSUP202. A unique SmaI site is located just upstream of lacZ that allows promoter/gene fusions to the amino-terminal part of the promoterless lacZ gene. For randomization of gene fusions, V. anguillarum chromosomal DNA was digested with DraI, SspI, or EcoRV, and the resulting chromosomal fragments were ligated to SmaI-digested pDM5. The ligation was transformed into DH5αPRO containing pLar-VanT and plated on LB medium containing 50 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml but no inducers. After incubation at 37°C, the colonies were replica plated onto LB medium containing 50 μg of X-Gal per ml, 1 mM IPTG, and 0.2% l-arabinose. Any colonies that appeared blue on one plate but white or lighter blue on the other within 48 h were picked for further analysis. As pDM5 contains mobilization genes, plasmids carrying the gene fusions were mobilized into V. anguillarum strains for further gene regulation studies using β-galactosidase assays.
Construction of transcriptional β-galactosidase fusions to empA, vps73, serA, and hpdA.
Transcriptional gene fusions were constructed between the reporter gene lacZ from E. coli and four V. anguillarum promoters, serA, vps73, hpdA-hgdA, and empA. For these studies, the vector, pDM8, was designed to contain the entire lacZ gene and its ribosome-binding site but to lack the lacZ promoter. pDM8 is a derivative of pDM5 and pTL61T which carries a promoterless lacZ gene designed for transcriptional gene fusion studies (32). pDM5 was digested with SmaI and SacI, and the 5′ region of the promoterless lacZ gene was removed and replaced with a 2,200-bp SmaI-SacI fragment from the 5′ region of the promoterless lacZ gene from pTL61T. A unique SmaI site just upstream of the lacZ gene is used for the fusion of promoter regions from other genes.
For the empA transcriptional fusion, a PCR fragment containing the empA promoter (276 bp upstream) but lacking the possible ribosome-binding site was obtained using primers EmpA-βgal-1 (5′-TCCCCCGGGTTATATTGATAGTTATGT-3′) and EmpA-βgal-3 (5′-TCCCCCGGGGAGAGTTATTATTAGCAT-3′). For the serA transcriptional fusion, a PCR fragment containing the serA promoter (230 bp upstream) was obtained using primers SerA-βgal-A (5′-TCCCCCGGGTTTCATTACAAAGCGCAC-3′) and SerA-βgal-C (5′-TCCCCCGGGAAAATGGGAAGGTGGGCA-3′). For the vps73 transcriptional fusion, a PCR fragment containing the possible vps73 promoter found in the 3′ end of the sat gene (295 bp upstream) was obtained using primers Vps73-βgal-1 (5′-TCCCCCGGGGAGGGATAGGATGTGTCA-3′) and Vps73-βgal-3 (5′-TCCCCCGGGCTTGCCAAATAGAATGT-3′). For the hpdA-hgdA transcriptional fusion, a PCR fragment containing the hpdA-hgdA promoter (318 bp upstream) was obtained using primers VllY-βgal-1 (5′-TCCCCCGGGTGTCGGTTTTTAATG-3′) and VllY-βgal-2 (5′-TCCCCCGGGGTTAACGCTTGTATT-3′). All PCR fragments were digested with SmaI and ligated to pDM8 that was similarly digested, creating pDM8-EmpA, pDM8-SerA, pDM8-Vps73, and pDM8-HpdA. These plasmids were then conjugated into the V. anguillarum wild type and the vanT mutant.
β-Galactosidase assays.
V. anguillarum cultures were grown overnight at 24°C in TSB containing tetracycline (2 μg/ml). Cell cultures were diluted to an optical density at 600 nm (OD600) of 0.05 in the same medium and further incubated at 24°C with shaking. Samples were taken at the various time points, and β-galactosidase assays were performed in triplicate according to Miller (42).
Measurement of protease activity.
Culture supernatants were assayed for protease activity as described by Denkin and Nelson (15) and Windle and Kelleher (68). Cells were grown overnight at 24°C in TSB with shaking, then diluted to an OD600 of 0.05 in TSB, and incubated further at 24°C with shaking. For comparative analysis between strains, a 2-h time period for incubation of the protease reaction mixture at 30°C was chosen.
Measurement of pigment production.
V. anguillarum strains were grown overnight at 24°C in TSB. Bacterial cell cultures were then diluted to an OD600 of 0.05 in either TSB or TSB containing 5 mM l-tyrosine and incubated at 24°C with shaking. At various time points, 1-ml samples were taken from the culture, and the supernatants were collected by centrifugation (12,000 × g, 2 min). Pigment production was estimated in the supernatant by measuring the absorbance at 400 nm. Cell densities were determined by counting CFU on Trypticase soy agar or by measuring the OD600.
Northern analysis.
The V. anguillarum wild type and vanT mutant were grown for 30 h at 24°C in TSB. Total RNA was then isolated using the RNeasy Minikit from Qiagen GmbH (Germany), and 10 μg of each sample was applied to a 2.2 M formaldehyde1% agarose gel and electrophoresed according to Sambrook et al. (57). Transfer of the RNA to ZetaProbe GT membranes (Bio-Rad), prehybridization, hybridization, and washing of the filter were performed as described in the ZetaProbe GT Blotting Membranes instruction manual. DNA fragments used as probes were generated by PCR amplification using primers within each of the genes: empA (bp 641 to 1374), serA (bp 200 to 372), vps73 (bp 959 to 1237), and hpdA (bp 289 to 1556). The hpdA DNA fragment was labeled using [α-32P]dCTP and random oligonucleotide priming (57), and the empA, serA, and vps73 DNA fragments were labeled using similar reaction conditions for random oligonucleotide priming, but instead of random hexamers, a specific primer within the DNA fragment was used. The radioactive bands were visualized using a Phosphorimager and the ImageQuant version 3.2 software from Molecular Dynamics.
Biofilm formation assays.
Two types of biofilm formation assays were performed, one on glass and the other on untreated polyvinylchloride (PVC) plastic. For biofilm formation on PVC plastic, an assay similar to that described previously for P. aeruginosa, E. coli, and Staphylococcus (50, 53, 63) was used with a few modifications. V. anguillarum strains were grown overnight in biofilm growth medium and then diluted to an OD600 of 1. Five microliters of the diluted bacteria was transferred to wells of a PVC microtiter dish (Falcon 35117) containing 200 μl of biofilm growth medium. Bacteria were allowed to grow without agitation for 6, 8, 10, 12, and 14 h at 24°C. Biofilm formation was quantified as described previously (63). Each strain was tested in three separate experiments, and for each experiment five wells were used for each strain. The values recorded are averages of these experiments.
For biofilm formation on glass, microscope glass slides were cleaned by soaking in a solution of 10% HCl and 90% methanol for 1 h at room temperature, rinsed in distilled water, and dried. Four sterile glass slides were suspended from the top of a 400-ml beaker into 300 ml of sterile biofilm growth medium. An overnight bacterial culture, grown in biofilm growth medium, was diluted in the same medium to an OD600 of 1. Three milliliters was used to inoculate the beaker. The culture was allowed to grow with stirring at “moderate speed” at room temperature. After 4 h, one slide was removed, bacteria were wiped away from one side, and the opposite side was stained immediately with 1 ml of 0.1% acridine orange in potassium phosphate buffer (pH 7.4) (48) for 5 min at room temperature. After 5 min, the stain was removed and the slide was washed twice in distilled water before air-drying. To prevent nutrient depletion of the growth medium, the remaining slides were removed, dipped carefully into 200 ml of fresh biofilm growth medium, suspended into another beaker containing 300 ml of fresh biofilm growth medium, and incubated as above for an additional 12 h. One slide was removed every 4 h and stained with acridine orange. Biofilm samples were taken at 4, 8, 12, and 16 h, and all experiments were done in triplicate.
The slides were analyzed for biofilm formation by viewing under a Zeiss Axioplan fluorescent microscope coupled to a charge-coupled device camera. Biofilm formation was quantified by determining the percent area coverage. Five representative images were taken for each biofilm and stored as computer images using the Adobe Photoshop 5.0 program. The images were converted into a grey scale using the Adobe Photoshop program and then converted to a binary mode using the Image Tool Software, version 2, from the University of Texas Health Science Center at San Antonio. This software makes all pixels in the image either black (background) or white (biofilm). The area coverage was then given as the percentage of white pixels. This was done for all five images taken and averaged.
Computer analysis.
Database searches were done using the sequence analysis software (17) of the Genetics Computer Group, Inc. (University of Wisconsin).
Fish infections.
Rainbow trout (Oncorhynchus mykiss) weighing approximately 10 to 15 g were infected with V. anguillarum either by intraperitoneal injections or by immersion in seawater containing V. anguillarum as previously described (45). The immersion and intraperitoneal infections were done at least twice. Five fish were infected for each bacterial dilution used. The 50% lethal doses (LD50s) were calculated as described by Reed and Muench (54). The LD50s recorded are an averaged value for all infections for each strain.
Nucleotide sequence accession numbers.
The complete vanT DNA sequence and the partial DNA sequences of the serA, sat-vps73, and hpdA-hgdA loci have been submitted to GenBank. Accession numbers were assigned to vanT (AF457643), serA (AF457644), hpdA-hgdA (AF457645), and sat-vps73 (AF457646).
RESULTS
Characterization and mutagenesis of the vanT gene.
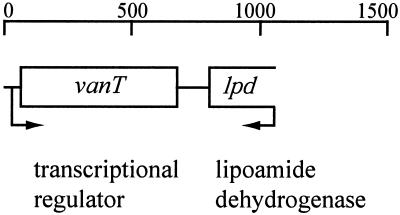
To determine whether V. anguillarum carries a gene homologous to the luxR gene of V. harveyi, degenerate primers based on the luxR coding sequence were designed. Using these primers, a 175-bp PCR fragment was generated. DNA sequence analysis of this fragment revealed the presence of an open reading frame (ORF), the deduced protein product of which showed 92% identity to LuxR. Using this PCR fragment as a probe, a recombinant clone containing the V. anguillarum gene was obtained from a V. anguillarum genomic library. The V. anguillarum DNA fragment was sequenced, and its genetic organization is shown in Fig. 1.
FIG. 1.
Genetic organization of the vanT DNA locus. Arrows indicate the direction of transcription.
One complete ORF was identified and named vanT. The deduced VanT protein sequence was 81% identical to LuxR of V. harveyi (59), 74% identical to HapR of V. cholerae (27), 82% identical to SmcR of Vibrio vulnificus (35, 58), and 82% identical to OpaR of V. parahaemolyticus (33). The amino terminus (residues 4 to 72) of VanT was also 30% identical to the amino terminus of the TetR family of repressor proteins, which contains a helix-turn-helix motif characteristic of DNA-binding proteins (51). A putative helix-turn-helix motif was identified in VanT at amino acid residues 39 to 58 and was similarly located in all LuxR homologues. Using the TopPred-2 search program (65), a putative transmembrane region was identified near the carboxy terminus (amino acid residues 160 to 180).
In addition, one partial ORF was also found (Fig. 1). The deduced protein-coding sequence of this partial ORF is very similar to the lpd gene of V. parahaemolyticus, V. cholerae, and V. vulnificus. This gene encodes a putative dihydrolipoamide dehydrogenase and is also located downstream of the luxR homologues characterized in these Vibrio spp.
To determine the function of VanT, an in-frame mutation was made that deleted the entire vanT gene, leaving only the first and last amino acid codons. This was done by allelic exchange with the altered gene carried on a suicide vector as described previously (45). The mutant strain was called AC10.
VanT positively regulates expression of the metalloprotease gene empA.
Since the EmpA metalloprotease of V. anguillarum is similar to the hemagglutinin protease of V. cholerae and since HapR regulates hap expression, it was thought that VanT may regulate empA expression. To test this idea, the promoter of empA was fused to a promoterless E. coli lacZ gene to generate a transcriptional fusion carried on plasmid pDM8-EmpA. This plasmid was conjugated into the wild-type V. anguillarum and the vanT mutant strain, and LacZ assays were done over the growth curve (Fig. 2A). For the wild type, LacZ activity was delayed until the cell density was high and then increased steadily throughout stationary-phase growth. In the vanT mutant, the empA promoter was virtually inactive compared to the wild type, suggesting that VanT is essential for expression of empA under these growth conditions.
FIG. 2.
Analyses of metalloprotease activity in the wild type (Wt) and the vanT mutant. (A) Growth curve and expression of empA::lacZ transcriptional gene fusion. Overnight cultures were diluted to an OD600 nm of 0.05 and then incubated with shaking at 24°C. Samples were taken at various times and analyzed for growth (OD600, open symbols) and β-galactosidase expression (Miller units, solid symbols). Miller units for the vector control were between 100 and 200 U. (B) Protease activity. Overnight cultures were diluted to an OD600 of 0.05 and then incubated with shaking at 24°C. Samples were taken at various times and analyzed for growth (open symbols) and for azocasein degradation (OD442, solid symbols). For protease activity, 100 μl of filtered-sterilized culture supernatant was mixed with 100 μl of azocasein solution and incubated at 30°C for 2 h. The reaction was stopped by the addition of trichloroacetic acid, the unreacted azocasein was removed by centrifugation, and the absorbance at 442 nm was determined. (C) Northern analysis. Total RNA was isolated from 30-h cultures of the wild type and vanT mutant grown at 24°C and hybridized to a DNA fragment complementary to the empA gene.
For further confirmation, a quantitative protease assay (Fig. 2B) previously employed to assay EmpA (15) was used. The protease activity of the vanT mutant was negligible compared with that of the wild type. In addition, Northern analysis was done using total RNA from the wild-type and the vanT mutant strains. Figure 2C shows that the amount of RNA transcript hybridizing to a probe from the empA gene was significantly reduced in the vanT mutant compared to the wild type. Taken together, these results suggest a requirement for VanT in the expression of the empA gene under the conditions of growth used in this study, although we cannot rule out the possibility that VanT may also regulate additional proteases previously detected in an empA mutant (44).
AHL analysis in the vanT mutant.
Previously, a mutation in the AHL synthase gene vanM resulted in the complete abolition of AHL production, including 3-oxo-C10-HSL, which is synthesized via VanI (47). One likely explanation for this observation is that V. anguillarum may contain a quorum-sensing circuit similar to that of V. harveyi, regulating the production of 3-oxo-C10-HSL via VanI. This possibility suggests that VanT may regulate 3-oxo-C10-HSL production. To test this hypothesis, AHL production was assayed in the wild type and the vanT mutant. TLC analysis of spent cell-free supernatants combined with AHL biosensor overlays were carried out to determine whether there were any changes in the AHL profile with respect to C6-HSL, 3-hydroxy-C6-HSL, and 3-oxo-C10-HSL in the vanT mutant compared with the wild type. No differences were observed, suggesting that regulation of 3-oxo-C10-HSL production in V. anguillarum is not likely to occur via VanT (data not shown).
Virulence analysis.
For several bacterial pathogens, quorum sensing has been shown to regulate the production of virulence determinants (for reviews, see references 67 and 71). We have previously shown that mutations in vanM and vanN did not alter the virulence of V. anguillarum (47). However, no conclusion could be made from these results because a parallel quorum-sensing system, similar to the luxSPQ system of V. harveyi, may also exist in V. anguillarum, activating or inactivating the same repressor as the vanMN circuit. Hence, a mutation in vanT, located downstream of the parallel circuits, would aid in the interpretation of additional virulence studies. The LD50s were therefore determined in rainbow trout for the wild type and the vanT mutant strain. Both immersion in infected seawater and intraperitoneal infection were used. For the immersion route, there was only a sixfold difference between the LD50s for the wild type (3 × 103 bacteria per ml of seawater) and the vanT mutant (5 × 102 bacteria per ml of seawater). For the intraperitoneal route, again the LD50s were similar for the wild type and the vanT mutant, 21 and 24 bacterial cells, respectively. These data suggest that VanT is not essential for virulence.
Screening for genes regulated by VanT.
Since V. anguillarum contains β-galactosidase activity that utilizes X-Gal but not ONPG (o-nitrophenyl-β-d-galactopyranoside) as a substrate, we could not use transposons that contained promoterless lacZ genes for promoter characterization in V. anguillarum. Thus, to identify additional genes regulated by VanT, we designed a two-plasmid system for the random screening in E. coli of V. anguillarum genes regulated by VanT. Both plasmids were maintained within the same E. coli strain, DH5αPRO. The first plasmid, pLar-VanT, contains a hybrid arabinose and lactose promoter, Plac/ara-1, that was fused to the vanT gene. This promoter tightly regulates the expression of the vanT gene by allowing gene expression only when arabinose and IPTG are present. The second plasmid is a derivative of pDM5 (Table 1) which carries a promoterless lacZ gene used in translational gene fusion studies.
To create random V. anguillarum promoter-gene fusions, we inserted chromosomal DNA fragments digested with either DraI, SspI, or EcoRV into a unique SmaI restriction site located just upstream of the lacZ gene. For some DNA fragments, this ligation resulted in in-frame promoter-gene fusions to the promoterless lacZ gene of pDM5. To screen for promoters regulated by VanT, the ligated products were transformed into E. coli DH5αPRO(pLar-VanT), and transformants were spread onto plates containing only X-Gal. After overnight growth, the colonies were replica plated onto X-Gal plates containing arabinose and IPTG to induce Plac/ara-1-vanT. Bacterial colonies containing promoter-lacZ fusions that were positively activated by VanT were darker blue in the presence of arabinose and IPTG and white or lighter blue in the absence of these inducers. Three colonies with the highest expression of LacZ were chosen for further analysis (pDM5-Ssp4, pDM5-S5, and pDM5-S13). To confirm similar regulatory effects in V. anguillarum, the three plasmids were mobilized, utilizing mobilization genes carried on pDM5 derivatives, into the V. anguillarum wild type and the vanT mutant. Since the β-galactosidase of V. anguillarum does not respond to ONPG as a substrate, LacZ activity was measured (data not shown). All three constructs showed a two- to fivefold lower LacZ expression in the vanT mutant compared to the wild-type strain.
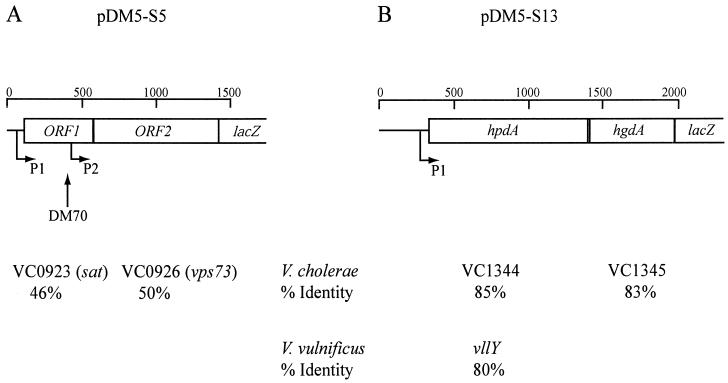
The V. anguillarum DNA inserts from pDM5-Ssp4, pDM5-S5, and pDM5-S13 were sequenced. The DNA sequence of pDM5-Ssp4 indicated that a small 34-codon ORF was fused in-frame to the lacZ gene. The 34 amino acids encoded by the DNA sequence showed 85% identity to 3-phosphoglycerate dehydrogenase from V. cholerae, which is encoded by the VC2481 locus on chromosome I, which is similar to the serA gene of E. coli (26). In E. coli, 3-phosphoglycerate dehydrogenase oxidizes 3-phosphoglycerate to 3-phosphohydroxypyruvate, the first step in the biosynthesis of serine, glycine, and one-carbon-unit molecules. No additional ORF was seen in the DNA sequence 300 bp upstream of the serA peptide. Thus, it is likely that this 300 bp contains a promoter.
The 1,377-bp insert from pDM5-S5 (Fig. 3A) contained a partial ORF encoding 275 amino acids that was fused in-frame with the lacZ gene. These 275 amino acids showed 50% identity to the protein encoded by the VCO926 locus (also called vps73) on chromosome I of V. cholerae (26). Vps73 is involved in the production of extracellular polysaccharides (EPS) for biofilm formation (73). Just upstream of ORF2 (vps73), a complete ORF encoding a 150-amino-acid protein homologous to numerous bacterial serine acetyltransferases was found. When the V. cholerae genome database was searched, these 150 amino acids showed 46% identity with the serine acetyltransferase-related protein encoded by region VCO923 of chromosome I (26).
FIG. 3.
Genetic organization of the cloned DNA fragments fused to the E. coli lacZ gene carried on pDM5-S5 and pDM5-S13. The horizontal arrows indicate the putative promoters and direction of transcription. The vertical arrow indicates the site of the plasmid insertion within the sat gene that created the polar mutation in the sat-vps73 DNA locus (strain DM70).
Thus, ORF1 (sat) and ORF2 (vps73) of V. anguillarum are located in a similar DNA locus as the coding regions VCO923 and VCO926 in V. cholerae. However, the genetic organization appears to be different between these two vibrios, since there are two additional genes between these two coding regions in V. cholerae. In addition, a very highly conserved consensus σ70 promoter sequence was found within the 3′ end of ORF1 just upstream of ORF2 (vps73), and a less conserved σ70 promoter sequence was found upstream of ORF1 (sat) (Fig. 3A).
For pDM5-S13, 2 kb upstream of the lacZ gene were sequenced, and the genetic organization is shown in Fig. 3B. Within this 2 kb, one complete ORF and a partial ORF were found. The partial ORF, encoding 194 amino acids, was fused in-frame to the lacZ gene. A GenBank database search revealed similarity between these 194 amino acids and numerous homogentisic acid oxidases. Hence, this ORF was designated hgdA. A V. cholerae genome search with these 194 amino acids showed 82.5% identity to the coding region VC1345, encoding a putative oxidoreductase. The complete ORF upstream of hgdA encodes a 357-amino-acid protein that showed similarity to numerous prokaryotic and eukaryotic HPPDs and was thus designated hpdA. The HPPDs that gave the highest identity (80%) were VllY from V. vulnificus (11) and the encoded protein from VC1344 of V. cholerae (26). VllY has been shown to confer hemolytic and pigment-producing activities when expressed in E. coli.
Interestingly, hpdA from V. anguillarum also appears to induce pigment production. Figure 4A shows a spectrophotometric profile of colored supernatant from the wild-type and the vanT mutant strains containing pDM5-S13 at various times during growth. The V. anguillarum strains were grown in the presence or absence of l-tyrosine, which has been shown to enhance pigment production in V. cholerae (55). As shown in Fig. 4A, at 48 h of growth, pigment production decreased fourfold for the vanT mutant in the absence of l-tyrosine and twofold in the presence of l-tyrosine, compared to the wild type. Moreover, pigment production was enhanced two- to threefold in the presence of l-tyrosine. There was no obvious ORF in the 300 bp upstream of the hpdA gene, indicating that a possible promoter upstream of hpdA is likely driving the expression of lacZ.
FIG. 4.
Measurement of pigment production and promoter activity of the hpdA gene. (A) For pigment production, overnight cultures of the wild type (Wt) and the vanT mutant containing pDM5-S13, which carries the hpdA gene and its promoter, were diluted to an OD600 of 0.05 into TSB medium with (solid symbols) and without 5 mM l-tyrosine (open symbols) and incubated with shaking at 24°C. Samples were taken from the culture at various times, and the supernatant was filter sterilized to remove bacteria. Pigment production in the supernatant was estimated by measuring the absorbance at 400 nm. (B) For hpdA promoter activity, an hpdA-lacZ transcriptional gene fusion was made and contained on pDM8-HpdA in the wild type and the vanT mutant. Overnight cultures were diluted into fresh TSB medium to an OD600 of 0.05 and then incubated with shaking at 24°C. Samples were taken at various times and analyzed for growth (OD600, open symbols) and β-galactosidase expression (Miller units, solid symbols). Miller units for the vector control were between 100 and 200 U. (C) Northern analysis. Total RNA was isolated from 30-h cultures of the wild type and vanT mutant grown at 24°C and hybridized to a DNA fragment complementary to the hpdA gene.
To confirm that VanT regulates the putative promoters upstream of the vps73, serA, and hpdA genes, transcriptional gene fusions were made between these promoters and a promoterless E. coli lacZ gene and carried on plasmids pDM8-Vps73, pDM8-SerA, and pDM8-HpdA, respectively. These constructs were mobilized into the V. anguillarum wild type and vanT mutant, and LacZ assays were done. The results are shown in Fig. 4B and Fig. 5A and 5C. As the bacteria entered stationary phase, a two- to threefold decrease in LacZ activity was seen for each of the three promoters. To confirm the lacZ data, Northern analyses were done using total RNA from the wild type and the vanT mutant. The amount of RNA transcript hybridizing to a DNA probe from the hpdA gene (Fig. 4C), the vps73 gene (Fig. 5B), and the serA gene (Fig. 5D) was significantly decreased in the vanT mutant compared to the wild type. These data suggest that VanT positively regulates pigment production, serine biosynthesis, and genes possibly involved in EPS production.
FIG. 5.
β-Galactosidase activity of vps73::lacZ and serA::lacZ transcriptional gene fusions. (A) vps73::lacZ was carried on pDM8-Vps73 and (C) serA::lacZ was carried on pDM8-SerA in the wild type (Wt) and vanT mutant. Overnight cultures were diluted into fresh TSB medium to an OD600 of 0.05 and then incubated with shaking at 24°C. Samples were taken at various times and analyzed for growth (OD600, open symbols) and β-galactosidase expression (Miller units, solid symbols). Miller units for the vector control were between 100 and 200 U. (B and D) Northern analysis. Total RNA was isolated from 30-h cultures of the wild type and vanT mutant grown at 24°C and hybridized to a DNA fragment complementary to either the vps73 gene (B) or the serA gene (D).
VanT and the sat-vps73-containing DNA locus are involved in biofilm formation.
Yildiz and Schoolnik (73) have shown that the vps73-containing region of V. cholerae is involved in EPS production and biofilm formation. To determine if the sat-vps73-containing DNA locus in V. anguillarum is involved in biofilm formation, a polar mutation was made by inserting a suicide plasmid into ORF1 (sat) of the wild-type strain (Fig. 3), resulting in mutant strain DM70. This insertion resulted in a null mutation of ORF1 (sat) and should exert a polar effect on the downstream ORF2 (vps73) and any other genes that may be downstream of ORF2 on the chromosome. This mutant, DM70, was then tested for its ability to form a biofilm on a glass surface compared to the wild type.
Biofilms were formed by suspending glass slides into biofilm growth medium, inoculated with a bacterial strain, and allowing cells to grow on the glass at room temperature with gentle stirring. Spent medium was replaced with fresh medium after 4 h, and the biofilm was allowed to continue growing for 16 h. A glass slide was withdrawn from the medium at 4 h, 8 h, 12 h, and 16 h and stained with acridine orange, and the percent area coverage was determined. These data are shown in Fig. 6. Both the vanT mutant and the mutant containing a polar mutation in the sat-vps73 DNA locus attached to the glass surface. By 16 h, the vanT mutant was unable to develop any biofilm structure, whereas the mutant carrying a polar mutation in the sat-vps73 DNA locus appeared to begin forming a biofilm, although it was not as extensive as the wild-type biofilm.
FIG. 6.
Biofilm attachment to a glass surface for the wild type (wt), the vanT mutant, and the mutant carrying a polar mutation in the sat-vps73 DNA locus. (A) Typical view of the progression of biofilm formation for each strain at 4 h, 8 h, 12 h, and 16 h. Slides were stained with acridine orange and then viewed with a fluorescent microscope. Bar, 20 μm. (B) Biofilm formation was quantified by determining the percent area coverage on a glass slide. For each strain, five images were taken at each time point and saved as a computer image. These images were converted into black (background) and white (biofilm) pixels using the Image Tool Software, version 2, and the percentage of white pixels is given as the percentage of biofilm covering the glass surface.
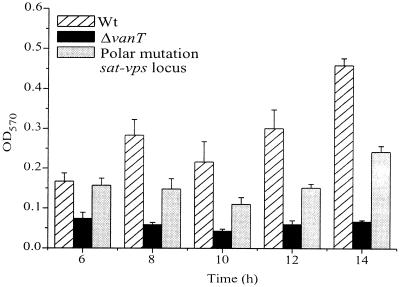
To further confirm the biofilm data, the same V. anguillarum strains were also tested for biofilm formation on a PVC plastic surface. A previously described method using a 96-well PVC microtiter dish was used (50, 53, 63). The wild type, the vanT mutant, and the mutant carrying a polar mutation in the sat-vps73 DNA locus were grown for 6 h, 8 h, 10 h, 12 h, and 14 h in biofilm growth medium in a 96-well PVC microtiter plate at 24°C without shaking. The wells were rinsed, fixed, and stained with crystal violet. To quantitate the bacteria present in the biofilms, the crystal violet was removed using 33% acetic acid, and the OD570 was determined. Figure 7 shows that the in-frame vanT mutant produced very little biomass that adhered to the plastic after 14 h, whereas the mutant carrying a polar mutation in the sat-vps73 DNA locus produced a reduced level of biomass compared to the wild type. Taken together, these data suggest that the V. anguillarum sat-vps73 DNA locus is involved in biofilm formation and that VanT is a positive regulator of genes found at this DNA locus. Moreover, VanT may also regulate additional genes involved in biofilm formation.
FIG. 7.
Quantification of bacteria in biofilms formed on PVC plastic by the wild-type (Wt) NB10, the vanT mutant, and the mutant carrying a polar mutation in the sat-vps73 DNA locus. Biofilms were allowed to form in a 96-well PVC microtiter dish. The microtiter dish was incubated for 6 h, 8 h, 10 h, 12 h, and 14 h at 24°C. The unattached bacteria were removed, and the biofilms were stained with crystal violet. The crystal violet was solubilized, and the absorbance, which correlated to the biofilm mass, was determined.
DISCUSSION
V. anguillarum has been shown to contain multiple quorum-sensing circuits (46, 47), incorporating VanIR, which is similar to the LuxIR system from V. fischeri, and VanMN, which is similar to the LuxMN system of V. harveyi. However, the role of these quorum-sensing circuits in V. anguillarum remains unclear. To investigate the possibility of a dual quorum-sensing channel system in this organism similar to that of V. harveyi, we cloned and characterized the vanT gene, encoding a homologue of the LuxR transcriptional activator of bioluminescence in V. harveyi.
During studies characterizing the LuxR homologues from V. cholerae (HapR) and V. vulnificus (SmcR), a hapR DNA probe was shown to hybridize weakly to chromosomal DNA from V. anguillarum, suggesting that V. anguillarum may also contain a LuxR homologue (27, 35). Both HapR and SmcR were shown to positively regulate homologous genes, hap and vvp, respectively, which encode metalloproteases (27, 58). EmpA of V. anguillarum is a homologue to the Hap and Vvp metalloproteases (12, 44), and in the present study, we have shown that this organism possesses a LuxR homologue, VanT, which positively regulates empA expression. These results suggest that this family of regulatory proteins, thus far only found in Vibrio spp. (27, 33, 35, 58, 59), may have very similar regulatory functions.
In a previous study (47), a mutation in vanM was shown to abolish production of C6-HSL and 3-hydroxy-C6-HSL as well as 3-oxo-C10-HSL, which is synthesized via VanI (46). This suggests that the VanMN circuit in some way regulates the expression of VanIR. Interestingly, a vanN mutant produced wild-type levels of AHLs, suggesting that V. anguillarum possesses an alternative mechanism for the regulation of 3-oxo-C10-HSL production, different from the dual sensory channel model of V. harveyi. To further support this idea, we show in this study that VanT, the possible regulator of the dual sensory channel model, produces wild-type levels of AHLs. This suggests that the vanM mutation may affect the production of 3-oxo-C10-HSL via a sensory circuit different from that used by V. harveyi.
To find genes in addition to empA that are regulated by VanT, a random approach was taken. A lacZ reporter gene fusion screening system was designed to allow the identification of V. anguillarum genes activated or repressed by VanT. In this study, three DNA loci were characterized that were positively regulated by VanT. The first V. anguillarum DNA locus contained genes encoding homologues of homogentisate 1,2-dioxygenase (HGD) and an HPPD. Both of these enzymes are involved in tyrosine catabolism. l-Tyrosine undergoes transamination to 4-hydroxypyruvate, which is converted to homogentisate by HPPD. HGD then converts homogentisate to maleylacetoacetate, which, after additional enzymatic steps, is finally converted to fumarate and acetoacetate. Both continuous expression of HPPD and inactivation of HGD results in accumulation of homogentisate, which can be oxidized, polymerizing into the red-brown pigment pyomelanin (40).
The homogentistate pathway for the production of melanin-like pigments has been shown for several bacteria (16, 41, 55), and homogentistate is the primary precursor of melanin-like pigments for marine bacteria, including V. cholerae, Shewanella colwelliana, and a Hyphomonas strain (28). The V. anguillarum HPPD enzyme was shown to be highly homologous to that of V. cholerae (VC1344) (26) and S. colwelliana (23). In addition, the V. anguillarum HPPD appears to be positively regulated by VanT and to be responsible for the production of a red-brown pigment in stationary phase when expressed from a plasmid-borne copy of hpdA. Moreover, the presence of l-tyrosine enhanced the production of this pigment, as has been shown for V. cholerae (55). This suggests that, in V. anguillarum, pigment production likely occurs via the homogentistate pathway and that the pigment is likely of the pyomelanin type.
Interestingly, the HPPD enzymes of V. vulnificus (11) and of Legionella pneumophila (70) have both hemolytic and pigment production activities. However, bacteria defective in this hemolytic activity have not yet been tested for virulence in animal models. Melanin pigments have previously been shown to be free-radical traps (2), and in one bacterial pathogen, Burkholderia cepacia, melanin pigment is thought to protect it from the bactericidal effects of the respiratory burst via free-radical-scavenging properties (74). V. cholerae has been shown to induce melanin production in response to stressful physiological conditions encountered in the host as well as in the environment. This suggests that melanin production may play a role in survival of this bacterium during stress (14). In Sinorhizobium meliloti, the tyrosine catabolism pathway has also been suggested to play a role in the starvation response (41). Whether the V. anguillarum HPPD enzyme has hemolytic activity, free-radical-scavenging properties, or stress-related properties is not known, but it is interesting to speculate that the role of melanin could be similar to that in other bacteria.
The second DNA locus regulated by VanT carried genes similar to VCO926 (vps73) and VCO923 (serine acetyltransferase) on chromosome I of V. cholerae (26). Yildiz and Schoolnik (73) have implicated this region of the V. cholerae genome in the production of EPS for biofilm formation. In addition, two transposon insertion sites in the V. cholerae genome that lead to a defect in rugose polysaccharide production have been mapped to the vps region (3). These similarities suggested that the sat-vps73 DNA locus in V. anguillarum may be involved in biofilm production. In support of this hypothesis, a mutant carrying a polar mutation within the sat-vps73 locus produced a very reduced biofilm.
These observations prompted us to determine whether VanT was also involved in the formation of biofilms. Under the growth conditions used in this study, a vanT isogenic mutant was unable to form a biofilm. Thus, V. anguillarum may have a set of vps-like genes similar to those of V. cholerae that are required for EPS production during biofilm formation. VanT, and possibly quorum sensing, may regulate the expression of these genes. Characterization of the possible vps-like locus in V. anguillarum is in progress.
Interestingly, the V. harveyi-like LuxR transcriptional regulators HapR and OpaR have been suggested to regulate the rugose colony morphology of V. cholerae (27) and the opaque colony morphology of V. parahaemolyticus (33), respectively. In addition, EPS is necessary for the rugose phenotype, which plays a role in biofilm formation (3, 67, 73), and for the opaque phenotype (20). Furthermore, LuxR from V. harveyi has been speculated to be involved in regulation of rugose colony morphology (31), and SmcR, the LuxR homologue of V. vulnificus, was shown to negatively regulate biofilm formation, as determined by microtiter plate assays (36). Hence, it is likely that all five characterized LuxR homologues regulate colony morphology or biofilm production by controlling EPS production.
Regulation of EPS production is likely to be a common function for this family of transcriptional regulators among the vibrios. However, these LuxR homologues probably differ in the way in which they regulate EPS production. VanT and OpaR appear to be activators of EPS production, whereas HapR, SmcR, and possibly LuxR negatively regulate either biofilm formation or a rugose phenotype and hence appear to repress EPS production.
The third DNA locus positively regulated by VanT contained the serA gene, which, in E. coli, catalyzes the first step in biosynthesis of serine, glycine, and one-carbon-unit molecules. Furthermore, the gene for lipoamide dehydrogenase (lpd), located downstream of all luxR homologues characterized thus far, is part of the glycine cleavage (GCV) enzyme system of E. coli (62), which is used in the final step of this biosynthesis pathway. The GCV enzyme complex in E. coli is composed of four protein complexes, one of which is lipoamide dehydrogenase. The GCV enzyme complex is suggested to maintain proper levels of glycine and C1-unit-molecule concentrations by converting excess glycine to C1-unit molecules. The GCV system is suggested to be highly regulated, as is probably the biosynthesis of serine and glycine. The global regulator, the leucine-responsive regulatory protein, regulates both the serA gene and the GCV system. It is likely that other global regulators are also involved in the regulation of this pathway.
Serine, glycine, and C1-unit-molecule biosynthesis, tyrosine catabolism, and EPS production are all regulated by environmental conditions, which lead to a physiological response by the bacterium. A bacterium's response to its environment can lead to numerous adaptive physiological changes that are tightly controlled by a number of regulatory proteins or signal molecules. Recently, quorum sensing has been suggested to be an integral component of gene regulatory networks that control bacterial responses such as entry into stationary phase, nutrient limitation, and stress response (30, 72). Results presented in this study suggest that the role of quorum sensing in V. anguillarum may be that of a global regulator.
For many pathogenic bacteria, the production of proteases and pigments as well as the capacity to form biofilms contributes to virulence. Thus, it is surprising that vanT is not essential for the pathogenesis of V. anguillarum infections. Several factors are known to contribute to the virulence of V. anguillarum (for reviews, see references 1 and 5). The well-characterized siderophore-based iron uptake system carried on the plasmid pJM1 is essential for promoting growth in vivo and therefore virulence. While lipopolysaccharides contribute to serum resistance, motility is important for entry into the fish host. In addition, although not genetically proven, extracellular products such as hemolysins, lipases, proteases, and a neurotoxic acetylcholinesterase have all been suggested to play a role in the pathology of vibriosis.
Although it is not clear why mutation of vanT does not reduce the virulence of V. anguillarum, it is possible that VanT may only be required for regulation of metalloprotease, pigment production, and biofilm formation outside of the fish host.
Acknowledgments
We thank David Kirke for some of the TLC analysis and Ram Chhabra and Chris Harty for the synthesis of AHLs.
This work was supported by a grant from the Swedish Council for Forestry and Agricultural Research (to D.M.), by a grant from the Carl Tryggers Foundation, Sweden (to D.M.), by a grant from the Swedish Research Council for Engineering Sciences (to D.M.), and by a grant and a studentship from the Biotechnology and Biological Sciences Research Council, United Kingdom (to A.H., P.W., and M.C.), which are gratefully acknowledged.
REFERENCES
- 1.Actis, L. A., M. E. Tomalsky, and J. H. Crosa. 1999. Vibriosis, p. 523-557. In P. T. K. Woo and E. W. Bruno (ed.), Fish diseases and disorders, vol. 3: viral, bacterial and fungal infections. Cab International Publishing, Wallingford, United Kingdom. [Google Scholar]
- 2.Agodi, A., S. Stefani, C. Corsaro, F. Campanile, S. Gribaldo, and G. Sichel. 1996. Study of a melanic pigment of Proteus mirabilis. Res. Microbiol. 147:167-174. [DOI] [PubMed] [Google Scholar]
- 3.Ali, A., Z. H. Mahmud, J. G. Morris, Jr., S. Sozhamannan, and J. A. Johnson. 2000. Sequence analysis of Tn phoA insertion sites in Vibrio cholerae mutants defective in rugose polysaccharide production. Infect. Immun. 68:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alsina, M., J. Mart&ıacute;nez-Picado, J. Jofre, and A. R. Blanch. 1994. A medium for presumptive identification of Vibrio anguillarum. Appl. Environ. Microbiol. 60:1681-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin, B., and D. A. Austin. 1999. Bacterial fish pathogens: disease of farmed and wild fish, p. 272-275. Springer and Praxis Publishing Ltd., Chichester, United Kingdom..
- 6.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 7.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol. 12:403-412. [DOI] [PubMed] [Google Scholar]
- 8.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 9.Cámara, M., M. Daykin, and S. R. Chhabra. 1998. Detection, purification and synthesis of N-acylhomoserine lactone quorum sensing molecules. Methods Microbiol. Bacterial Pathogenesis 27:319-330. [Google Scholar]
- 10.Cao, J.-G., and E. A. Meighen. 1989. Purification and structural identification of an autoinducer for the luminescence system of V. harveyi. J. Biol. Chem. 264:21670-21676. [PubMed] [Google Scholar]
- 11.Chang, T. M., Y. C. Chuang, J. H. Su, and M. C. Chang. 1997. Cloning and sequence analysis of a novel hemolysin gene (vllY) from Vibrio vulnificus. Appl. Environ. Microbiol. 63:3851-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, J. C., C. P. Shao, and L. I. Hor. 1996. Cloning and nucleotide sequencing of the protease gene of Vibrio vulnificus. Gene 183:255-257. [DOI] [PubMed] [Google Scholar]
- 13.Chhabra, S. R., P. Stead, N. J. Bainton, G. P. C. Salmond, G. S. A. B., Stewart, P. Williams, and B. W. Bycroft. 1993. Autoregulation of carbapenem biosynthesis in Erwinia carotovora by analogues of N-(3-oxohexanoyl)-l-homoserine lactone. J. Antibiot. 46:441-449. [DOI] [PubMed] [Google Scholar]
- 14.Coyne, V. E., and L. al-Harthi. 1992. Induction of melanin biosynthesis in Vibrio cholerae. Appl. Environ. Microbiol. 58:2861-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denkin, S. M., and D. R. Nelson. 1999. Induction of protease activity in Vibrio anguillarum by gastrointestinal mucus. Appl. Environ. Microbiol. 65:3555-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denoya, C. D., D. D. Skinner, and M. R. Morgenstern. 1994. A Streptomyces avermitilis gene encoding a 4-hydroxyphenylpyruvic acid dioxygenase-like protein that directs the production of homogentisic acid and an ochronotic pigment in Escherichia coli. J. Bacteriol. 176:5312-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunny, G. M., and S. C. Winans. 1999. Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 19.Eberhard, A., A. L. Burlingame, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 20.Enos-Berlage, J. L., and L. L. McCarter. 2000. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J. Bacteriol. 182:5513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibiro harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 22.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuqua, W. C., V. E. Coyne, D. C. Stein, C.-M. Lin, and R. M. Weiner. 1991. Characterization of melA, a gene encoding melanin biosynthesis from the marine bacterium Shewanella colwelliana. Gene 109:131-136. [DOI] [PubMed] [Google Scholar]
- 24.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 25.Hardman, A. M., G. S. A. B. Stewart, and P. Williams. 1998. Quorum sensing and the cell-cell communication dependent regulation of gene expression in pathogenic and nonpathogenic bacteria. Antonie van Leeuwenhoek J. Microbiol. 74:199-210. [DOI] [PubMed] [Google Scholar]
- 26.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harvey luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 28.Kotob, S. I., S. L. Coon, E. J. Quintero, and R. M. Weiner. 1995. Homogentisic acid is the primary precursor of melanin synthesis in Vibrio cholerae, a Hyphomonas strain, and Shewanella colwelliana. Appl. Environ. Microbiol. 61:1620-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. A. B. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PA01. Mol. Microbiol. 17:333-343. [DOI] [PubMed] [Google Scholar]
- 30.Lazazzera, B. A. 2000. Quorum sensing and starvation: signals for entry into stationary phase. Curr. Opin. Microbiol. 3:177-182. [DOI] [PubMed] [Google Scholar]
- 31.Lilly, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 32.Linn, T., and R. St. Pierre. 1990. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J. Bacteriol. 172:1077-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarter, L. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180:3166-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. A. B. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition of the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 35.McDougald, D., S. A. Rice, and S. Kjelleberg. 2000. The marine pathogen Vibrio vulnificus encodes a putative homologue of the Vibrio harveyi regulatory gene, luxR: a genetic and phylogenetic comparison. Gene 248:213-221. [DOI] [PubMed] [Google Scholar]
- 36.McDougald, D., S. A. Rice, and S. Kjelleberg. 2001. SmcR-dependent regulation of adaptive phenotypes in Vibrio vulnificus. J. Bacteriol. 183:758-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGee, K., P. Hörstedt, and D. Milton. 1996. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J. Bacteriol. 178:5188-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meighen, E. A. 1991. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55:123-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meighen, E. A. 1994. Genetics of bacterial bioluminescence. Annu. Rev. Genet. 28:117-139. [DOI] [PubMed] [Google Scholar]
- 40.Menon, I. A., S. D. Persad, H. F. Haberman, P. K. Basu, J. F. Norfray, C. C. Felix, and B. Kalyanaraman. 1991. Characterization of the pigment from homogentisic acid and urine and tissue from alkaptonuria patient. Biochem. Cell Biol. 69:260-273. [DOI] [PubMed] [Google Scholar]
- 41.Milcamps, A., and F. J. de Bruijn. 1999. Identification of a novel nutrient-deprivation-induced Sinorhizobium meliloti gene (hmgA) involved in the degradation of tyrosine. Microbiology 145:935-947. [DOI] [PubMed] [Google Scholar]
- 42.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1992. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174:7235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milton, D. L., R. O'Toole, P. Hörstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milton, D. L., A. Hardman, M. Camara, S. R. Chhabra, B. W. Bycroft, G. S. A. B. Stewart, and P. Williams. 1997. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J. Bacteriol. 179:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milton, D. L., V. J. Chalker, D. Kirke, A. Hardman, M. Cámara, and P. Williams. 2001. The LuxM homologue, VanM, from Vibrio anguillarum directs the synthesis of N-(3-hydroxyhexanoyl)-l-homoserine lactone and N-hexanoyl-l-homoserine lactone. J. Bacteriol. 183:3537-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mittelman, M. 1999. Recovery and characterization of biofilm bacteria associated with medical devices. Methods Enzymol. 310:534-551. [DOI] [PubMed] [Google Scholar]
- 49.Norqvist, A., Å. Hagström, and H. Wolf-Watz. 1989. Protection of rainbow trout against vibriosis and furunculosis by the use of attenuated strains of Vibrio anguillarum. Appl. Environ. Microbiol. 55:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 51.Pan, W., and B. G. Spratt. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11:769-775. [DOI] [PubMed] [Google Scholar]
- 52.Ponce, M. R., and J. L. Micol. 1992. PCR amplification of long DNA fragments. Nucleic Acids Res. 20:623.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis, and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 54.Reed, L. J., and J. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 55.Ruzafa, C., A. Sanchez-Amat, and F. Solano. 1995. Characterization of the melanogenic system in Vibrio cholerae ATCC 14035. Pigment Cell Res. 8:147-152. [DOI] [PubMed] [Google Scholar]
- 56.Salmond, G. P. C., B. W. Bycroft, G. S. A. B. Stewart, and P. Williams. 1995. The bacterial ′enigma': cracking the code of cell-cell communication. Mol. Microbiol. 16:615-624. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 58.Shao, C.-P., and L.-I. Hor. 2001. Regulation of metalloprotease gene expression in Vibrio vulnificus by a Vibrio harveyi LuxR homologue. J. Bacteriol. 183:1369-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Showalter, R. E., M. O. Martin, and M. R. Silverman. 1990. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J. Bacteriol. 172:2946-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silhavy, T., M. Berman, and L. Enquist. 1984. Experiments with gene fusions . Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 61.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:787-796. [Google Scholar]
- 62.Stauffer, G. V. 1996. Biosynthesis of serine, glycine, and one-carbon units, p. 506-513. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 63.Stepanovic, S., D. Vukovic, I. Dakic, B. Savic, and M. Svabic-Vlahovic. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40:175-179. [DOI] [PubMed] [Google Scholar]
- 64.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Excherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.von Heijne, G. 1992. Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 66.Wai, S. N., Y. Mizunoe, A. Takade, S-I. Kawabata, and S-I Yoshida. 1998. Vibrio cholerae 01 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams, P., M. Camara, A. Hardman, S. Swift, D. Milton, V. J. Hope, K. Winzer, B. Middleton, D. I. Pritchard, and B. W. Bycroft. 2000. Quorum sensing and the population dependent control of virulence. Phil. Trans. R. Soc. Lond. Sect. B 355:667-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Windle, H. J. P., and D. Kelleher. 1997. Identification and characterization of a metalloprotease activity from Helicobacter pylori. Infect. Immun. 65:3132-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. A. N. B. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acylhomoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
- 70.Wintermeyer, E., U. Rdest, B. Ludwig, A. Debes, and J. Hacker. 1991. Characterization of legiolysin (lly), responsible for haemolytic activity, colour production and fluorescence of Legionella pneumophila. Mol. Microbiol. 5:1135-1143. [DOI] [PubMed] [Google Scholar]
- 71.Winzer, K., and P. Williams. 2001. Quorum sensing and the regulation of gene expression in pathogenic bacteria. Int. J. Med. Microbiol. 291:131-143. [DOI] [PubMed] [Google Scholar]
- 72.Withers, H., S. Swift, and P. Williams. 2001. Quorum sensing as an integral component of gene regulatory networks in Gram-negative bacteria. Curr. Opin. Microbiol. 4:186-193. [DOI] [PubMed] [Google Scholar]
- 73.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae 01 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zughaier, S. M., H. C. Ryley, and S. K. Jackson. 1999. A melanin pigment purified from an epidemic strain of Burkholderia cepacia attenuates monocyte respiratory burst activity by scavenging superoxide anion. Infect. Immun. 67:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]