Abstract
Xer-mediated dimer resolution at the mwr site of plasmid pJHCMW1 is osmoregulated in Escherichia coli. Whereas under low-salt conditions, the site-specific recombination reaction is efficient, under high-salt conditions, it proceeds inefficiently. Regulation of dimer resolution is independent of H-NS and is mediated by changes in osmolarity rather than ionic effects. The low level of recombination at high salt concentrations can be overcome by high levels of PepA or by mutating the ARG box to a sequence closer to the E. coli ARG box consensus. The central region of the mwr core recombination site plays a role in regulation of site-specific recombination by the osmotic pressure of the medium.
Plasmids tend to form dimers by homologous recombination, a process that leads to multimer formation, which reduces the number of molecules in the cell and leads to plasmid loss (39, 40). The Xer site-specific recombination system ensures that dimeric plasmids are converted to monomers prior to cell division (4). Xer recombination was first identified through its role in the resolution of ColEl plasmid multimers (40) and has subsequently been demonstrated to mediate resolution in other related plasmids and pSC101 as well as the Escherichia coli chromosome (6, 16, 33). The tyrosine family recombinases XerC and XerD act at specific recombination sites present in plasmids and chromosomes to mediate a recombination reaction that proceeds via a Holliday junction intermediate (6, 7, 9, 12, 16, 17, 22, 37, 40). XerCD-mediated recombination at the chromosomal dif site requires a 28-bp core site, which includes two 11-bp binding sites for the recombinases XerC and XerD and a 6-bp central region (23, 42). In contrast, recombination at sites present in plasmids, e.g., psi (pSC101) and cer (ColE1), requires a core site plus about 180-bp additional accessory sequences, which are bound by accessory proteins, PepA and ArgR (cer) or PepA and ArcA (psi), and ensure that the reaction is exclusively intramolecular (14, 15, 17, 35, 36, 38). The recombination reaction at psi occurs by sequential strand exchanges mediated by XerC and then XerD, while for cer, only one pair of strand exchanges mediated by XerC to form a Holliday junction has been observed (15). The recombination reaction at cer seems to be completed by still unidentified XerD-independent cellular processes (2).
The multiresistance plasmid pJHCMW1, originally isolated from Klebsiella pneumoniae (18, 44, 45), contains the recombination site mwr. Like cer and psi, this site contains a core site and accessory sequences, which appear to be related to those of cer and which interact with PepA and ArgR. As is the case for cer, a reporter plasmid harboring two directly repeated mwr sites formed Holliday junctions in vitro in the presence of ArgR and PepA, but did not complete the recombination reaction under the assay conditions (43). On the other hand, resolution of dimers of a recombinant clone consisting of pUC18 and the pJHCMW1 mwr site in vivo when E. coli cells were growing in L broth (which contains 0.5% NaCl) was inefficient. Stability experiments showed that this low level of resolution efficiency was not enough to prevent loss of the recombinant clone in E. coli JC8679 (43). In this work, we show that Xer recombination in E. coli between direct repeats of mwr is osmoregulated. Resolution of dimers was inefficient in regular L broth, which contains 0.5% NaCl, while it was efficient in cells cultured in L broth lacking NaCl. The low level of recombination observed at high salt concentrations could be overcome by high levels of PepA or by mutating the ARG box to a sequence closer to the E. coli ARG box consensus. Mutagenesis experiments showed that the mwr central region of the recombination core site plays a role in regulation of site-specific recombination by the osmotic pressure of the medium.
MATERIALS AND METHODS
E. coli strains and plasmids.
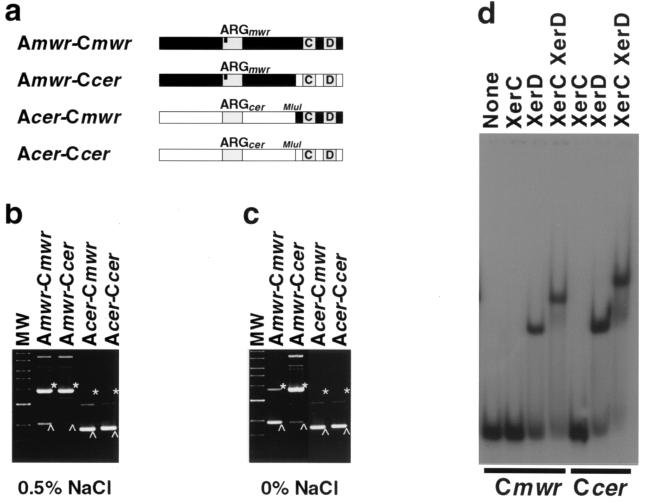
The E. coli strains and plasmids are described in Table 1. Plasmids pMET cm and pMET pm are pUC18 carrying hybrid sites consisting of the accessory sequences from cer or psi, respectively, and the mwr core recombination site. To generate pMET cm and pMET pm, the plasmids pLN9 (which contains the cer site) and pLN10 (which contains the psi site) (15) were treated with MluI and EcoRI fragments to delete the core recombination sites. This treatment resulted in plasmids carrying DNA fragments consisting of the cer or psi accessory sequences, respectively, ending in an MluI site. These plasmids were treated with MluI and EcoRI and ligated to a synthetic fragment containing the mwr core region flanked by MluI and EcoRI sites. The pMET mc plasmid was generated by site-directed mutagenesis of pES to replace the appropriate nucleotides (43) and transform the mwr core into the cer core region. Plasmid pHP2.3 was generated by site-directed mutagenesis of pMET mc to modify the central region of the cer core recombination site. Throughout the paper, the sites are described according to the following general nomenclature: A, accessory sequence; C, core recombination site (e.g., A mwr-C cer describes a hybrid site with the accessory sequences from mwr and the core recombination region from cer) (Fig. 1). When necessary, further clarification is provided. The wild-type sites are mostly identified by the single mention of the name, but when considered helpful for clarity, the A and C names have been detailed (e.g., A mwr-C mwr).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic or genotypea | Source or reference |
|---|---|---|
| E. coli strains | ||
| DS941 | AB1157 recF143 lacIqlacZΔM15 | 41 |
| DS956 | DS941 argR::fol (Tmpr) | 5 |
| DS957 | DS941 pepA::Tn5 (Kanr) | 35 |
| DS981 | DS941 xerC2::aph (Kanr) | 16 |
| DS9012 | DS941 hns::Tn 10 | 21 |
| JC8679 | DS945 recBC sbcA (hyperrecombinogenic) | 40 |
| Plasmids | ||
| pES | EcoRI-SacI fragment containing the pJHCMW1 mwr site (A mwr-C mwr) in pUC18 | This work |
| pKD3 | pES with substitution C to T at the ArgR binding site (ARG box) | This work |
| pMET pm | pUC18 derivative carrying A psi-C mwr (psi accessory sequence-mwr core site) | This work |
| pMET cm | pUC18 derivative carrying A cer-C mwr | This work |
| pMET mc | pUC18 derivative carrying A mwr-C cer | This work |
| pHP23 | pMET mc with the mwr central region (A mwr-C hp23) (cr CAGATG) | This work |
| pMIG103 | pUC18 derivative with directly repeated psi sites | M. Bregu |
| pMIG104 | pUC18 derivative with directly repeated modified psi sites; modification by flipping order of XerC and XerD binding sites with respect to accessory sequence [A psi-C psi(DC)] | M. Bregu |
| pMIG107 | pUC18 derivative with directly repeated psi core sites (C psi) | M. Bregu |
| pSDC124 | pUC18 derivative carrying two directly repeated dif sites (A dif-C dif) | S. Colloms |
| pSDC164 | pUC18 derivative with directly repeated cer sites with a GATCCA central region [A cer-C cer (cr, GATCCA)] | S. Colloms |
| pSDC165 | pUC18 derivative with directly repeated A psi-C psi (cr, GCTCCA) | S. Colloms |
| pSDC167 | pUC18 derivative with directly repeated A psi-C psi (cr, GACCCA) | S. Colloms |
| pSDC169 | pUC18 derivative with directly repeated A psi-C psi (cr, GATTCA) | S. Colloms |
| pLN9 | pUC18 derivative with directly repeated A cer-C psi | 15 |
| pLN10 | pUC18 derivative with directly repeated A psi-C cer | 15 |
| pCS349 | argR gene cloned under the control of plac (includes its own promoter); λdv replicon; Cmr | 34, 36 |
| pCS119 | pepA gene cloned under the control of plac (includes its own promoter); λdv replicon; Chlr | 34, 36 |
| pCS118 | pepA gene cloned in opposite orientation with respect to plac (includes its own promoter); λdv replicon; Chlr | 34, 36 |
| pKS492 | pUC18 containing 280-bp HpaI-TaqI cer fragment | 36 |
A, accessory sequence; C, core site; cr, central region; Chl, chloramphenicol; Kan, kanamycin; Tmp, trimethoprim.
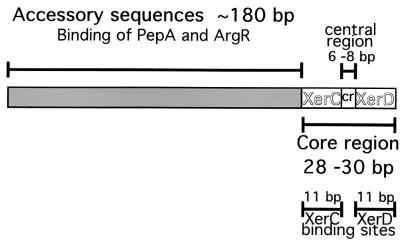
FIG. 1.
Schematic structure of plasmid Xer site-specific recombination sites. The sites contain a core recombination region that includes the XerC and XerD binding sites (11 bp each) and a central region (6 to 8 bp), as well as the accessory sequences (∼180 bp) with which ArgR and PepA interact (in the case of psi, ArcA and PepA). The diagram is not to scale. The hybrid sites utilized in this paper consist of accessory sequences (A) and core recombination regions (C) from different sites.
Bacterial growth media and general DNA procedures.
Growth of bacteria was in Lennox L broth (32) (1% tryptone, 0.5% yeast extract, 0.5% NaCl [called “high osmolarity” throughout the text]) or medium containing the same concentrations of tryptone and yeast extract with either no NaCl added or with other NaCl concentrations indicated in the text. In the case of solid medium, 2% agar was added. Transformations were carried out as described by Cohen et al. (13). Restriction endonuclease and ligase treatments were carried out as recommended by the suppliers. Plasmid DNA preparations and DNA gel extractions were performed with the QIAspin miniprep kit and QIAquick gel extraction kit, respectively (Qiagen). Nucleotide sequencing was performed at the DNA sequencing facility, Department of Biochemistry, University of Oxford. Site-directed mutagenesis was carried out with the Quikchange site-directed mutagenesis kit (Stratagene). Osmolality values were determined in a vapor pressure osmometer (Wescor 5500).
In vivo resolution assay.
To prepare dimers, E. coli JC8679 was transformed with plasmid DNA. The transformed strains were cultured in L broth medium in the presence of 100 μg of ampicillin per ml for 20 generations, and plasmid DNA was purified and electrophoresed in a 0.7% agarose gel. DNA of the correct size to be a plasmid dimer was purified from agarose gels with the QIAquick gel extraction kit (Qiagen). Since dimers run close to the position of open circular monomer DNA, the isolated samples were used to transform the XerC-deficient E. coli DS981. In this strain, dimers will not be resolved by Xer recombination allowing the isolation of transformants that had obtained a plasmid dimer. Purified plasmid dimers were transformed into E. coli DS941 to determine the efficiency of Xer-mediated dimer resolution.
DNA binding assays.
Gel mobility shift assays were performed as described by Blakely et al. (7). The oligonucleotides used had the following structure (the putative binding regions are underlined): cer, 5′GATCCGCGGTGCGTACAATTAAGGGATTATGGTAAATACG and 5′AATTCGTATTTACCATAATCCCTTAATTGTACGCACCGCG; and mwr, 5′GATCCGGCGGTGCACGCAACAGATGTTATGGTAAATACG and 5′AATTCGTATTTACCATAACATCTGTTGCGTGCACCGCCG.
Approximately 10 pmol of oligonucleotide was end labeled with 50 μCi of [γ-32P]ATP and phage T4 polynucleotide kinase (5 U) in kinase buffer (50 mM Tris HCl [pH 7.5], 10 mM MgCl2, 5 mM dithiothreitol [DTT], 0.1 mM spermidine) in a final volume of 20 μl. The labeled oligonucleotide was purified with a Nuctrap Probe Purification column (Stratagene) followed by ethanol precipitation. The radiolabeled oligonucleotide was dissolved in 15 μl of H2O and then made double stranded by annealing with 50 pmol of the complementary oligonucleotide. The mixture was heated to 75°C for a few minutes and then allowed to cool to room temperature overnight. The annealed double-stranded radiolabeled oligonucleotides were purified by electrophoresis on an 8% polyacrylamide gel in Tris borate buffer (100 mM Tris [pH 8], 100 mM boric acid, 2 mM EDTA) as described before (7). The radiolabeled oligonucleotides were mixed with 0.1 mg of poly(dI-dC) per ml and the appropriate protein(s). The binding reaction was carried out for 10 min at 37°C and immediately transferred to ice. The samples were analyzed by electrophoresis in a polyacrylamide gel as described above. The radioactive complexes were detected by exposure to X-ray film. To determine the dissociation constant (Kd), we followed the procedure described by Robinson and Sligar (31). Purified XerD at different concentrations (ranging from 5 μM to 0.25 nM) was incubated with radiolabeled DNA under the conditions described above and the bands corresponding to bound and unbound DNA were quantified. The values were utilized to calculate the fraction of bound DNA (θ), which can be expressed as θ ≈ 1/(1 + Kd/Pt), where Pt is the amount of XerD and Kd is the equilibrium dissociation constant: Kd = [P][DNA]/[P − DNA]. The calculated fraction of bound DNA values was fitted to the equation by nonlinear least-square analysis by using Excel (Microsoft).
In vitro aminopeptidase assays.
The enzymatic determinations were carried out basically as described by McCulloch et al. (26). Cells were harvested from overnight cultures; washed with a solution containing 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 10 mM β-mercaptoethanol; resuspended in a mixture containing 50 mM Tris-HCl (pH 8.2), 1 M NaCl, 10 mM MgCl2, 0.1 mM EDTA, and 0.1 mM DTT; and lysed by sonication. The soluble fraction was separated by centrifugation and heated at 75°C for 10 min. This fraction was used to determine aminopeptidase A activity. The reactions were performed in a buffer containing 20 mM Tris-HCl (pH 8.2), 100 mM KCl, 1 mM MnCl2, 0.1 mM EDTA, 1 mM l-leucine-p-nitroanilide, and 5 μg of protein at 37°C. Generation of p-nitroanilide was determined by measuring the A400.
RESULTS
Dimer resolution at mwr is osmoregulated.
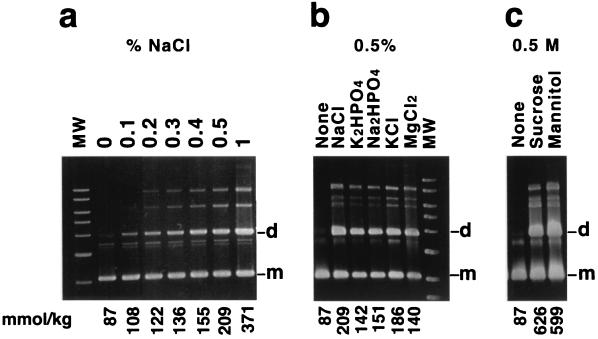
The efficiency of resolution of pES dimers, which contain directly repeated mwr sites, was strongly dependent on the NaCl concentration in the bacterial growth medium. Figure 2a shows the plasmid content of E. coli DS941 transformed with pES dimers and cultured at increasing NaCl concentrations (osmolality range, 87 to 371 mmol/kg). While resolution was almost complete in the absence of NaCl, it became more inefficient as the concentration of salt in the medium was increased. Increasing the osmolality of the medium by addition of other salts also resulted in the same inhibition of resolution of dimers carrying mwr (Fig. 2b). Therefore, this is a general effect of the concentration of the salts in the medium and not a specific inhibition by sodium or chloride ions. Resolution of dimers of pES in medium supplemented with 0.5 M sucrose or mannitol also led to a decrease in dimer resolution (Fig. 2c), suggesting that Xer site-specific recombination at mwr is inhibited by an increase in the external osmolarity and not by an ionic effect.
FIG. 2.
Resolution of pES dimers. Dimers were introduced by transformation into E. coli DS941. The cells were cultured in L medium with the addition of increasing NaCl concentrations (a); 0.5% NaCl, K2HPO4, Na2HPO4, KCl, and MgCl2 (b); or 0.5 M mannitol or sucrose (c). The osmolality values (millimoles per kilogram) are shown below each lane. The cultures were carried out in the presence of 100 μg of ampicillin per ml for 20 generations. Plasmid DNA was isolated and subjected to agarose gel electrophoresis. The slowly moving bands correspond to multimers (supercoiled and open circular). MW, linear molecular weight standards (10, 8, 6, 5, 4, 3, and 2 kb). d, dimer; m, monomer.
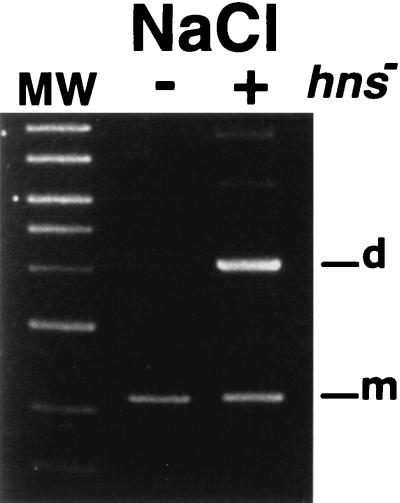
Resolution of pES dimers in an hns mutant is osmoregulated.
Since the H-NS protein has been implicated in regulation of osmotically controlled genes (25, 27, 28), we determined the efficiency of resolution of pES dimers in E. coli DS9012, an E. coli DS941 hns mutant derivative (21). Figure 3 shows that resolution was efficient in the mutant in the absence of NaCl while very poor in the presence of 0.5% NaCl. From these results, we concluded that H-NS is not directly involved in the osmoregulation of resolution of pES dimers.
FIG. 3.
pES dimer resolution in an H-NS-deficient mutant. E. coli DS941 and E. coli DS9012 (H-NS deficient) harboring dimers of pES were cultured in L broth containing 0.5% (+) or no (−) NaCl, and plasmid DNA was purified and analyzed by gel agarose electrophoresis. The left lane shows linear DNA molecular weight (MW) standards (10, 8, 6, 5, 4, 3, 2, and 1.5 kb). d, dimer; m, monomer.
Recombination at sites other than mwr is not highly osmoregulated.
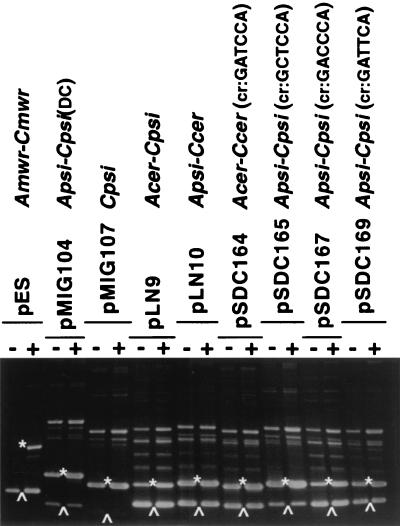
To find out if recombination at other sites was also osmoregulated, we performed studies on other derivatives. Since resolution of dimers harboring the sites A psi-C psi (psi accessory sequences-psi core recombination site) and A cer-C cer was highly efficient, irrespective of the presence or absence of NaCl, we studied osmoregulation of recombination at derivatives of these Xer recombination sites that were not as efficient. E. coli DS941was transformed with several plasmids carrying two directly repeated derivative sites described in Table 1. The transformed strains were cultured in medium with 0 or 0.5% NaCl, and plasmid DNA was extracted and analyzed by agarose gel electrophoresis. A plasmid carrying two directly repeated copies of dif, pSDC124, showed a small apparent difference in efficiency of resolution between the two conditions. A slightly larger percentage of resolved products was present when no NaCl was added to the growth medium (data not shown). However, the effect was much less than that seen when a dimer with two direct repeats of mwr was analyzed. Plasmids pMIG104 and pMIG107 have directly repeated modified psi sites and show a lower Xer site-specific recombination efficiency than that of the wild-type psi site. Plasmid pMIG104 contains two directly repeated psi site derivatives with a modified core site consisting of an inversion of the location of the XerC and XerD binding regions with respect to the accessory sequences [A psi-C psi(DC)]. Plasmid pMIG107 carries two directly repeated core psi sites and no accessory sequences (C psi). Figure 4 shows that the levels of resolution for both plasmids are identical in the presence or absence of NaCl in the medium. Plasmid pLN9, which has two directly repeated hybrid regions consisting of the cer accessory sequences and the psi core region, shows marginally better efficiency of recombination in the absence of NaCl (A cer-C psi, Fig. 4). Resolution assays using plasmids with directly repeated cer sites that have a modified central region (pSDC164), directly repeated psi sites with modified central regions (pSDC165, pSDC167, and pSDC169), or a directly repeated hybrid site consisting of psi accessory sequences and the cer core region (pLN10, A psi-C cer) showed a marginal difference in efficiency of resolution in the case of pSDC167 and no differences in all other plasmids. These results indicate that, under the assay conditions used, the efficiencies of Xer site-specific recombination at dif and derivatives of cer and psi were not substantially modified by the osmolarity of the external medium. This could reflect the fact that either recombination at wild-type cer or psi does not respond to a change in osmolarity or recombination at the impaired derivatives tested had lost the ability to be regulated by changes in the osmolarity of the growth medium.
FIG. 4.
Xer site-specific recombination at several target sites. Dimers of pES and the different reporter plasmids were introduced by transformation into E. coli DS941. The cells were cultured in L medium containing 0.5% NaCl (+) or no NaCl (−). Plasmid DNA was isolated and subjected to agarose gel electrophoresis. The positions of unrecombined and recombined species are shown by asterisks and upward arrowheads, respectively.
The accessory sequences of mwr are responsible for its low dimer resolution efficiency in high-osmolarity medium.
To identify the region of mwr responsible for the low dimer resolution efficiency under conditions of high osmolarity, we generated hybrid sites containing the accessory sequences of mwr and the core region of cer (A mwr-C cer) and the accessory sequences of cer and the core region of mwr (A cer-C mwr) (Fig. 5a). Plasmids pMET mc and pMET cm contain the sites A mwr-C cer and A cer-C mwr, respectively. Plasmids pES and pKS492 include an intact copy of mwr (A mwr-C mwr) and cer (A cer-C cer), respectively (Fig. 5a). Dimers of all four plasmids were introduced in E. coli DS941 to determine their efficiency of resolution by Xer recombination. Figure 5b shows that efficient recombination was observed in the case of pKS492 (A cer-C cer) and pMET cm (A cer-C mwr). Conversely, the dimers were poorly resolved in both derivatives carrying the mwr accessory sequences (A mwr-C mwr and A mwr-C cer [see Fig. 5a]). These results indicate that the accessory sequences of mwr are responsible for the low efficiency of Xer recombination at this site in high-osmolarity medium. Furthermore, a comparison of the XerC and XerD binding capabilities of the core sites of cer and mwr showed that there are no substantial differences in their affinities for E. coli XerC and XerD (Fig. 5d). The Kd for XerD binding to mwr was 10.6 nM, a value similar to that of the Kd for XerD binding to cer (8). Addition of 0.28 μg of partially purified XerC to a reaction mixture containing radiolabeled mwr or cer DNA and XerD resulted in binding of ∼90% of the label in both cases. These results confirmed that the recombinases have similar affinities for the mwr and cer sites.
FIG. 5.
Resolution of dimers containing hybrid sites. (a) Recombination sites present in plasmids pKS492 (A cer-C cer), pES (A mwr-C mwr), pMET cm (A cer-C mwr), and pMET mc (A mwr-C cer). The bar in the ARG box in the mwr accessory sequences indicates that 1 nucleotide is different from the conserved consensus ARG box (ArgR binding site) sequence. (b and c) Dimers of pES, pMET mc, pMET cm, and pKS492 were introduced by transformation into E. coli DS941 and cultured in L medium containing 0.5% NaCl (b) or no NaCl (c). Plasmid DNA was isolated and subjected to agarose gel electrophoresis. Linear DNA molecular weight (MW) standards (10, 8, 6, 5, 4, 3, 2, and 1.5 kb) and the positions of dimer (asterisks) and monomer (arrowheads) species are shown. (d) In vitro protein-DNA binding. Oligonucleotides containing the mwr and cer core regions were end labeled and incubated in the presence of XerC, XerD, or both XerC and XerD. A control with no additions is shown. The products were separated by electrophoresis in 8% polyacrylamide gel.
A substitution of 1 nucleotide in the mwr ARG box results in a target site that recombines at high efficiency in high-osmolarity medium.
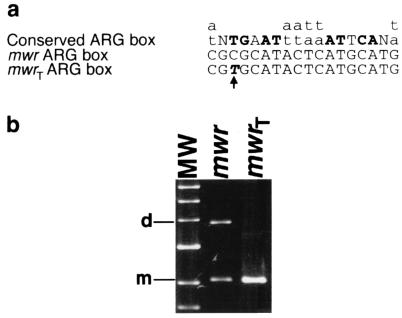
Unlike operator regions of ArgR-regulated genes, which have two adjacent ARG boxes (20), the cer and mwr accessory regions contain only one ARG box. In the synaptic complex formed between two recombination sites, one ArgR hexamer is believed to bind to both ARG boxes (1). To test if the apparent impairment of the mwr accessory sequences is due to its ARG box, we generated a mutation to create an ArgR binding site more similar to the E. coli consensus sequence as defined by Glansdorff (20). Plasmid pKD3 contains a substitution, C to T, at the third nucleotide of the mwr ARG box (mwrT). This is one of 8 nucleotides that are highly conserved in all ARG boxes (boldface in Fig. 6a). Figure 6b shows that dimers of pKD3, which contain two direct repeats of mwrT, were resolved more efficiently than dimers of pES in E. coli DS941. This result indicates that a nonoptimal ARG box may be responsible for the low efficiency of recombination at mwr in E. coli growing in high-osmolarity medium. It is possible that the mwr ARG box has evolved to efficiently bind the ArgR protein from K. pneumoniae. However, the E. coli ArgR protein shares 94% identity and 98% similarity with the ArgR from K. pneumoniae MGH78578, the strain for which the complete genome is being sequenced (http://genome.wustl.edu/gsc/Projects/K.pneumoniae/). Cloning of K. pneumoniae ArgR and complementation assays will permit us to determine if this is the case.
FIG. 6.
Mutagenesis of the mwr ARG box. (a) Nucleotide sequence of the mwr ARG box showing the substitution in mwrT and the consensus sequence of the ARG box according to Glansdorff (20). Boldface capital letters indicate the most important conserved nucleotides in the ARG box. (b) Dimers of pES (mwr) and pKD3 (mwrT) were introduced by transformation into E. coli DS941 and cultured in L broth containing 0.5% NaCl. Plasmid DNA was isolated and subjected to agarose gel electrophoresis. The left lane shows linear DNA molecular weight standards (6, 5, 4, 3, 2, and 1.5 kb). d, dimer; m, monomer.
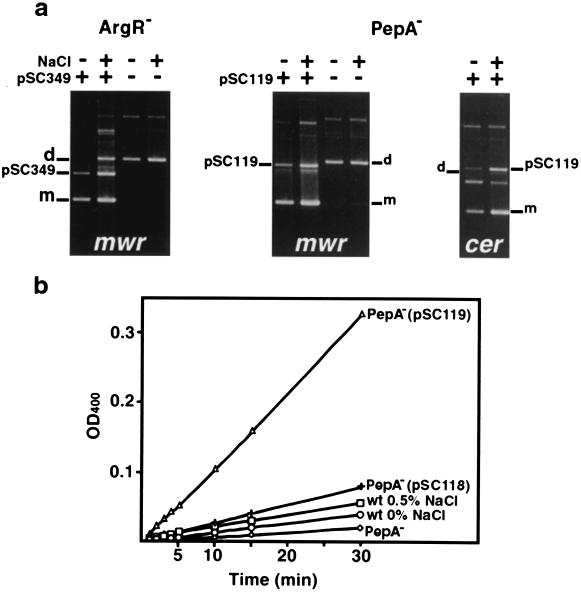
Increased levels of PepA suppress the poor recombination phenotype at high osmolarity.
To determine if higher levels of ArgR or PepA can overcome the low efficiency of recombination at high osmolarity, pES dimers were introduced into ArgR− and PepA− mutants of E. coli already harboring pCS349 and pSC119. These plasmids overexpress ArgR and PepA from plac, respectively. Figure 7a shows that in E. coli DS956(pCS349), which has a higher concentration of ArgR in the cell, the dimers were efficiently resolved in the absence of NaCl, while the efficiency was lower in the presence of 0.5% NaCl, as is the case with resolution in E. coli DS941. In contrast, in the PepA overproducer E. coli DS957(pCS119), the pES dimers were very efficiently resolved when the cells were cultured in medium containing either 0.5% or no NaCl (Fig. 7a). Control assays without the complementing plasmids pCS349 or pCS119 did not show any resolution (Fig. 7a). These experiments show that an excess of PepA can overcome the low efficiency of pES dimer resolution at high osmolarity in the growth medium. It has been shown before that at a high concentration of PepA, the in vitro recombination reaction at cer could occur in the absence of ArgR (15). In a control experiment, dimers of pKS492 were efficiently resolved in E. coli DS957(pCS119) in the presence or absence of NaCl (Fig. 7a). Figure 7b shows that the concentration of PepA in E. coli DS957(pCS119) is substantially higher than that in E. coli DS941. Conversely, the PepA concentration in E. coli DS957(pCS118), in which pepA was cloned in the orientation so that it could not be expressed from the lac promoter, was only slightly higher than that in E. coli DS941 (Fig. 7b). (In plasmid pCS118, pepA carries its natural promoter.) This result indicates that the high PepA expression from pCS119 is due to transcription from plac. Figure 7b also shows that PepA activity in the plasmidless E. coli strain DS957 is negligible.
FIG. 7.
Complementation of ArgR− and PepA− derivatives. (a) E. coli DS956 (argR) and DS957 (pepA) were transformed with dimers of pES (A mwr-C mwr) or dimers of pKS492 (A cer-C cer) and pCS349 (argR gene fusion) or pCS119 (pepA gene fusion), respectively. These derivatives, as well as control E. coli DS956 and DS957 harboring only pES dimers, were cultured in L broth containing 0.5% NaCl (+) or no NaCl (−). Plasmid DNA was isolated and subjected to agarose gel electrophoresis. d, dimer; m, monomer. (b) Aminopeptidase A was partially purified, and its activity was determined as described before (26) for E. coli DS957 (◊), E. coli DS957(pSC118) (+), E. coli DS957(pSC119) (Δ), and E. coli DS941 cultured in L broth with no NaCl (○) or 0.5% NaCl (□).
Since a higher concentration of PepA compensates for the lower efficiency of dimer resolution when the cells are cultured in the presence of 0.5% NaCl, we determined if the osmoregulation of pES dimer resolution is mediated by a higher expression of PepA. We measured aminopeptidase A activities in cells growing in L broth containing 0.5% or no NaCl. Figure 7b shows that the aminopeptidase A activity at low osmolarity is slightly lower than at high osmolarity, indicating that a higher concentration of PepA is not responsible for a higher-resolution efficiency at low osmolarity. Therefore, the effect of PepA observed in the experiments described in this section may be due to an ability of high levels of PepA to make the recombination reaction less dependent on ArgR.
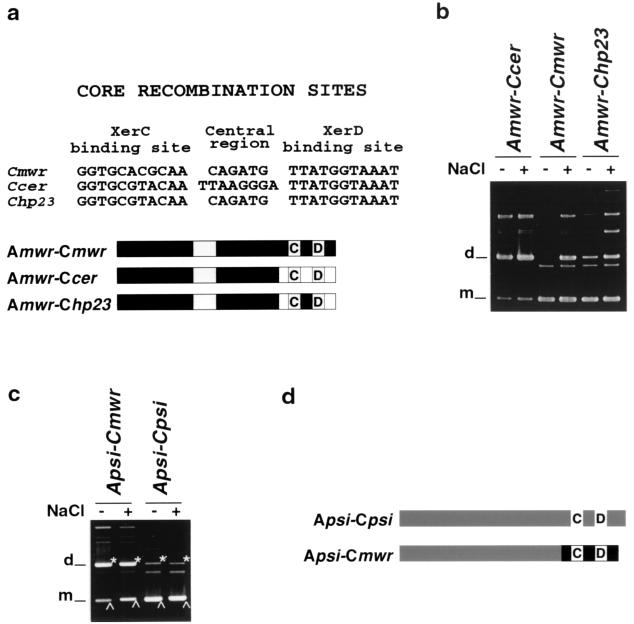
The central region of the mwr core recombination site is involved in osmoregulation.
The results described so far indicate that the mwr accessory sequences are responsible for the poor Xer recombination in high osmolarity. A high PepA concentration, a nucleotide substitution in the ARG box that improves ArgR binding, or low osmolarity in the growth medium can overcome this deficiency. To identify features of mwr involved in osmoregulation of Xer recombination, the efficiencies of resolution of dimers of plasmids containing intact cer, mwr, or the hybrid sites A cer-C mwr and A mwr-C cer were analyzed in the presence or absence of NaCl in the medium. Figure 5b and c show that while resolution of dimers of pES was substantially enhanced when the NaCl concentration in the medium was reduced, the level of resolution of dimers containing A mwr-C cer remained constant. Since the only difference between the derivatives A mwr-C mwr and A mwr-C cer is their core recombination sites and recombination at the latter is not regulated by the osmolarity of the medium, we generated a substitution in the hybrid A mwr-C cer derivative, A mwr-C hp23, in which the central region of the core recombination site has been modified to that of mwr (Fig. 8a). The efficiency of resolution of pHP2.3 dimers was increased in the absence of NaCl, although the effect was less than that observed for the wild-type A mwr-C mwr (Fig. 8b). This result strongly suggests that the core recombination site of mwr plays a role in osmoregulation through its central region. To determine if the presence of the mwr core recombination site was sufficient to confer the property of being osmoregulated, a site was constructed with the psi accessory sequences (A psi-C mwr, Fig. 8c). A dimer carrying this hybrid site had a low recombination frequency at high osmolarity (Fig. 8d), and only a small improvement in dimer resolution in the absence of NaCl was observed (Fig. 8d), suggesting that the presence of the mwr core recombination site is not sufficient for osmoregulation.
FIG. 8.
Resolution of dimers containing a modified core recombination site. (a) Comparison of the nucleotide sequence of core recombination sites of mwr, cer, and the mutant derivative called hp23 and diagram of the hybrid recombination site assayed. Gray boxes indicate the location of the XerC (C) and XerD (D) binding sites and the ARG box. (b and c) E. coli DS941 transformed with dimers harboring the indicated sites were cultured in L medium containing 0.5% NaCl (+) or no NaCl (−). Plasmid DNA was isolated and subjected to agarose gel electrophoresis. d, dimer; m, monomer.(d) Diagram representing A psi-C psi and the A psi-C mwr hybrid sites.
DISCUSSION
In this report, we demonstrate that Xer recombination between the directly repeated mwr sites in a dimer is osmoregulated. Resolution experiments in vivo at various osmolarities obtained with several osmolites showed an inverse relationship between osmolarity and efficiency of resolution. To the best of our knowledge, osmoregulation of Xer site-specific recombination has not been previously observed. Experiments with hybrid sites showed that the mwr accessory sequences are responsible for the low efficiency of recombination in E. coli at high osmolarity, apparently because the ARG box in this site interacts poorly with ArgR. A substitution in the mwr ARG box that generates a more efficient E. coli ArgR binding region resulted in a site that recombines efficiently at high osmotic pressure. The mwr ARG box may have evolved to bind the K. pneumoniae ArgR more efficiently, thereby increasing the pJHCMW1 stability in this bacterium. However the amino acid sequences of ArgR in both E. coli and K. pneumoniae MGH78578 (http://genome.wustl.edu/gsc/Projects/K.pneumoniae/) are highly related (94% identity and 98% similarity), and experiments in the presence of the K. pneumoniae ArgR will have to be performed to determine if this is the case. The impairment in the pJHCMW1 ARG box could be overcome by increasing the levels of PepA in E. coli. Similarly, in vitro recombination assays showed that high levels of PepA can overcome the requirements for ArgR to form Holliday junctions (43). As a consequence of these observations, we considered the possibility that the levels of PepA in a wild-type E. coli strain increase at low osmolarity, thereby overcoming the defective interaction of the mwr accessory sequences with ArgR. However, our results indicated that this is not the case: the levels of PepA, as determined by aminopeptidase A activity, did not significantly change at high or low osmolarity. On the other hand, these results do not rule out the possibility that at lower osmotic pressure, there is an improvement in the interactions of the accessory proteins with the accessory sequences leading to a more efficient formation of the synaptic complex. Several DNA-binding proteins have been shown to change their interaction properties at different osmotic pressures (30, 31). In other cases, such as some transcriptional regulators, the binding affinity is not highly dependent on the intracellular ion concentration (11).
To cope with osmotic variations, bacteria respond by inducing several adaptation mechanisms. For some of them, such as the osmoprotectant proU uptake system, expression is kept at elevated levels as long as the osmotic stimulus persists (24), while for others, like the kdp operon, induction is transient (19). Since the higher efficiency of dimer resolution has been observed in several generations of cultures, it is most probable that the elements involved in Xer recombination at mwr undergo modifications that last for as long as the osmotic situation persists as is the case for induction of proU. Searches for regulators of proU failed to identify classical regulators. Instead, DNA-binding proteins such as H-NS, IHF, HU, TopA, or GyrAB were found to be associated with control of transcription of proU (10, 21, 29, 46). The involvement of these proteins together with the fact that the degree of DNA supercoiling is lower in cells cultured in low-osmolarity medium suggests that DNA structure may play a role in proU expression. Although our results indicate that H-NS seems not to be involved in the higher efficiency of recombination at mwr in low-osmolarity-grown cells, it is possible that one or more of these proteins can contribute to osmoregulation. Further analysis of the nature of the mwr DNA structure and the role of DNA-binding proteins will help to find out if changes in supercoiling and interactions with any of these DNA-binding proteins play a role in osmoregulation of recombination at mwr. Although we do not know at this time if Xer site-specific recombination at mwr is osmoregulated in K. pneumoniae, comparisons of the amino acid sequences of the recombinases and accessory proteins with those from E. coli showed that they are very closely related. Dimer resolution experiments with plasmids carrying mwr in K. pneumoniae will permit us to determine if the recombination reaction is osmoregulated and, if it is, what the significance of this property is.
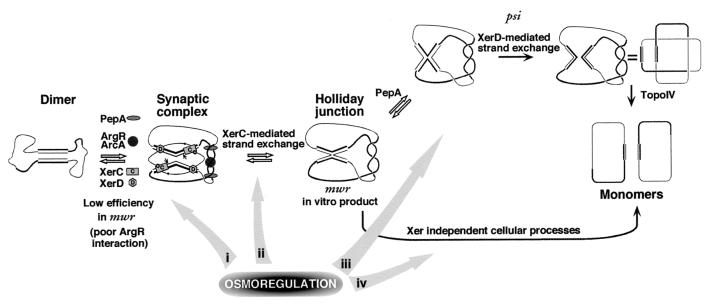
Replacement of the cer accessory sequences for those of mwr (hybrid site A mwr-C cer) resulted in a low efficiency of recombination comparable to that of the wild-type mwr at high osmolarity. However, unlike A mwr-C mwr, A mwr-C cer is not osmoregulated. Therefore, all or a portion of the core recombination site and its interaction with the recombinases may be responsible for this property. Replacement of the central region of the cer core recombination site (TTAAGGGA) for that from mwr (CAGATG) in the hybrid A mwr-C cer resulted in a site, A mwr-C hp23, with an intermediate level of osmoregulation compared to that of A mwr-C mwr (high osmoregulation) and A mwr-C cer (no osmoregulation). Therefore, we conclude that the mwr core recombination site and in particular the central region are involved in the regulation of recombination by osmotic pressure. The core central region plays an important role in determining the recombination properties of a site (2, 5). While in psi the central region favors a complete recombination reaction, in cer it favors only a strand exchange by XerC. These two central regions vary in length and purine content at the top strand: 6 nucleotides and 50% purine for psi and 8 nucleotides and 75% purine for cer. The central region of mwr is 6 nucleotides in length and has a 67% purine content. This intermediate purine content could be related to the ability to confer to a core recombination site the property of being osmoregulated. Figure 9 shows the steps for resolution of dimers harboring psi, cer, or mwr. While resolution of the Holliday junction at cer proceeds through a Xer-independent pathway, at psi, there is a change in conformation mediated by PepA followed by a XerD-mediated pair of strand exchanges. (It is possible that resolution at psi also proceeds through the Xer-independent pathway [2, 3, 15].) Although in vitro recombination at mwr yields Holliday junctions (43), we do not know whether they are resolved by Xer recombination (like psi), by a Xer-independent pathway (like cer), or both. The presence in a site of the accessory sequences of mwr results in an inefficient recombination site, probably by poor interaction with ArgR slowing the formation of the synaptic complex (Fig. 9). However, this reduction can be compensated for at low osmotic pressure by an increase in the efficiency of formation of the synaptic complex (by an increase in the rate of the forward reaction or a decrease in the rate of the backward reaction) or an increase in the efficiency of resolution of the Holliday junction (Fig. 9). We think it is likely that the osmoregulation is mediated through the XerCD-mwr core recombination site interaction. However, the stage at which this regulation acts is not known. It could act at the formation of the Xer-mediated synapsis (Fig. 9, arrow i) or at one of the catalytic steps (Fig. 9, arrows ii and iii). Alternatively, the osmoregulation might be mediated by unknown cellular processes, if resolution of the Holliday junction at mwr is Xer independent (Fig. 9, arrow iv). Experiments are being carried out to determine the step of the reaction that is regulated by the osmolarity of the growth medium. These possibilities do not rule out an additional contribution to osmoregulation mediated by a better interaction of PepA at a low salt concentration with the mwr accessory sequences. Another set of experiments suggests that the sole presence of the mwr core recombination site is not enough for osmoregulation of a site. The hybrid site A psi-C mwr, which recombines at low efficiency, was only barely osmoregulated (Fig. 8). Probably the interwrapping of the psi accessory sequences around the accessory proteins (ArcA and PepA) does not form the ideal topology for recombination at the mwr core recombination site. This deficiency is only minimally compensated for by osmoregulation mediated by the mwr core recombination site.
FIG. 9.
Osmoregulation of Xer recombination at mwr. The diagram shows the steps of the recombination process indicating proteins or features involved. For clarity, the proteins are shown only in the synaptic complex. The accessory proteins are PepA and ArgR (for cer and mwr) or PepA and ArcA (for psi). Bold lines represent accessory sequences. Small hexagons represent the C terminus of XerD activating XerC. The black circles represent the XerC tyrosine residue properly located to exchange the top strands. The Xer-mediated pathway of resolution of the Holliday junction at psi is shown at the top. The bottom shows the Xer-independent resolution pathway followed by cer (and probably by psi and mwr). For clarity, the supercoiling of the molecules is not shown. The ability to resolve through the Xer-dependent pathway is determined by the central region of the core recombination site (2). The gray arrows show possible levels of osmoregulation involving the central region.
Bacterial cells may encounter different environments through their life cycle. Upon entering a host, the pathogens' surroundings change dramatically, and they must have systems that enable them to grow or survive. Plasmids harbored by these bacterial cells carry genetic elements to be stably maintained in the various environments, and not all of them may be active under the same conditions. The plasmid pJHCMW1 has two systems for stability by multimer resolution: tnpR/res and xer/mwr (43). Although we do not know how by being osmotically regulated mwr helps the stability of pJHCMW1 in K. pneumoniae, one can envision that the mwr locus is not critical for stability under certain environments in which resolution occurs via the resolvase at res, but it is essential in others where the osmolarity must be lower. K. pneumoniae causes a substantial amount of hospital-acquired urinary tract infections, pneumonia, septicemias, meningitis, and soft tissue infections. One or more of the niches occupied in these diverse infections may present changes in osmolarity that influence the efficiency of Xer recombination at mwr.
Acknowledgments
This work was supported by Public Health grants AI47115-01 (M.E.T.) and LS Basin MIRT T37 TW00048-05 from the National Institutes of Health and a grant from Wellcome Trust (D.J.S.). H.P. and K.D. were supported by MSD grant R25 GM56820-03 and MIRT T37 TW00048-05 from the National Institutes of Health.
We thank Migena Bregu and Sean Colloms for generously providing plasmids and R. Allen for technical help.
REFERENCES
- 1.Alen, C., D. J. Sherratt, and S. D. Colloms. 1997. Direct interaction of aminopeptidase A with recombination site DNA in Xer site-specific recombination. EMBO J. 16:5188-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arciszewska, L. K., R. A. Baker, B. Hallet, and D. J. Sherratt. 2000. Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J. Mol. Biol. 299:391-403. [DOI] [PubMed] [Google Scholar]
- 3.Barre, F. X., M. Aroyo, S. D. Colloms, A. Helfrich, F. Cornet, and D. J. Sherratt. 2000. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 14:2976-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barre, F. X., B. Soballe, B. Michel, M. Aroyo, M. Robertson, and D. Sherratt. 2001. Circles: the replication-recombination-chromosome segregation connection. Proc. Natl. Acad. Sci. USA 98:8189-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blake, J. A., N. Ganguly, and D. J. Sherratt. 1997. DNA sequence of recombinase-binding sites can determine Xer site-specific recombination outcome. Mol. Microbiol. 23:387-398. [DOI] [PubMed] [Google Scholar]
- 6.Blakely, G., S. Colloms, G. May, M. Burke, and D. Sherratt. 1991. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 3:789-798. [PubMed] [Google Scholar]
- 7.Blakely, G., G. May, R. McCulloch, L. K. Arciszewska, M. Burke, S. T. Lovett, and D. J. Sherratt. 1993. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell 75:351-361. [DOI] [PubMed] [Google Scholar]
- 8.Blakely, G., and D. Sherratt. 1996. Determinants of selectivity in Xer site-specific recombination. Genes Dev. 10:762-773. [DOI] [PubMed] [Google Scholar]
- 9.Blakely, G. W., A. O. Davidson, and D. J. Sherratt. 2000. Sequential strand exchange by XerC and XerD during site-specific recombination at dif. J. Biol. Chem. 275:9930-9936. [DOI] [PubMed] [Google Scholar]
- 10.Bremer, E., and R. Kramer. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria, p. 79-97. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 11.Cayley, S., B. A. Lewis, H. J. Guttman, and M. T. Record, Jr. 1991. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J. Mol. Biol. 222:281-300. [DOI] [PubMed] [Google Scholar]
- 12.Clerget, M. 1991. Site-specific recombination promoted by a short DNA segment of plasmid R1 and by a homologous segment in the terminus region of the Escherichia coli chromosome. New Biol. 3:780-788. [PubMed] [Google Scholar]
- 13.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colloms, S. D., C. Alen, and D. J. Sherratt. 1998. The ArcA/ArcB two-component regulatory system of Escherichia coli is essential for Xer site-specific recombination at psi. Mol. Microbiol. 28:521-530. [DOI] [PubMed] [Google Scholar]
- 15.Colloms, S. D., R. McCulloch, K. Grant, L. Neilson, and D. J. Sherratt. 1996. Xer-mediated site-specific recombination in vitro. EMBO J. 15:1172-1181. [PMC free article] [PubMed] [Google Scholar]
- 16.Colloms, S. D., P. Sykora, G. Szatmari, and D. J. Sherratt. 1990. Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda integrase family of site-specific recombinases. J. Bacteriol. 172:6973-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornet, F., I. Mortier, J. Patte, and J.-M. Louarn. 1994. Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J. Bacteriol. 176:3188-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dery, K. J., R. Chavideh, V. Waters, R. Chamorro, L. S. Tolmasky, and M. E. Tolmasky. 1997. Characterization of the replication and mobilization regions of the multiresistance Klebsiella pneumoniae plasmid pJHCMW1. Plasmid 38:97-105. [DOI] [PubMed] [Google Scholar]
- 19.Epstein, W. 1986. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol. Rev. 39:73-78. [Google Scholar]
- 20.Glansdorff, B. 1996. Biosynthesis of arginine and polyamines, p. 408-433. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 21.Higgins, C. F., C. J. Dorman, D. A. Stirling, L. Waddell, I. R. Booth, G. May, and E. Bremer. 1988. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52:569-584. [DOI] [PubMed] [Google Scholar]
- 22.Kuempel, P., J. Henson, L. Dircks, M. Tecklenburg, and D. Lim. 1991. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 3:799-811. [PubMed] [Google Scholar]
- 23.Leslie, N. R., and D. J. Sherratt. 1995. Site-specific recombination in the replication terminus region of Escherichia coli: functional replacement of dif. EMBO J. 14:1561-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucht, J., and E. Bremer. 1994. Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system ProU. FEMS Microbiol. Rev. 14:3-20. [DOI] [PubMed] [Google Scholar]
- 25.Lucht, J. M., P. Dersch, B. Kempf, and E. Bremer. 1994. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J. Biol. Chem. 269:6578-6588. [PubMed] [Google Scholar]
- 26.McCulloch, R., M. E. Burke, and D. J. Sherratt. 1994. Peptidase activity of Escherichia coli aminopeptidase A is not required for its role in Xer site-specific recombination. Mol. Microbiol. 12:241-251. [DOI] [PubMed] [Google Scholar]
- 27.Mourino, M., C. Balsalobre, C. Madrid, J. M. Nieto, A. Prenafeta, F. J. Munoa, and A. Juarez. 1998. Osmolarity modulates the expression of the Hha protein from Escherichia coli. FEMS Microbiol. Lett. 160:225-229. [DOI] [PubMed] [Google Scholar]
- 28.Nieto, J. M., C. Madrid, A. Prenafeta, E. Miquelay, C. Balsalobre, M. Carrascal, and A. Juarez. 2000. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet. 263:349-358. [DOI] [PubMed] [Google Scholar]
- 29.Owen-Hughes, T., G. Pavitt, D. Santos, J. Sidebotham, C. Hulton, J. Hinton, and C. Higgins. 1992. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 71:255-265. [DOI] [PubMed] [Google Scholar]
- 30.Robinson, C., and S. Sligar. 1998. Changes in solvation during DNA binding and cleavage are critical to altered specificity of the EcoRI endonuclease. Proc. Natl. Acad. Sci. USA 95:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson, C., and S. Sligar. 1996. Participation of water in Hin recombinase-DNA recognition. Protein Sci. 5:2119-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sherratt, D. J., L. K. Arciszewska, G. Blakely, S. Colloms, K. Grant, N. Leslie, and R. McCulloch. 1995. Site-specific recombination and circular chromosome segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 347:37-42. [DOI] [PubMed] [Google Scholar]
- 34.Stirling, C. 1987. Cloning and characterization of E. coli xer genes. Ph.D. thesis. Institute of Genetics, University of Glasgow, Glasgow, Scotland.
- 35.Stirling, C. J., S. D. Colloms, J. F. Collins, G. Szatmari, and D. J. Sherratt. 1989. xerB, an Escherichia coli gene required for plasmid ColE1 site-specific recombination, is identical to pepA, encoding aminopeptidase A, a protein with substantial similarity to bovine lens leucine aminopeptidase. EMBO J. 8:1623-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stirling, C. J., G. Szatmari, G. Stewart, M. C. Smith, and D. J. Sherratt. 1988. The arginine repressor is essential for plasmid-stabilizing site-specific recombination at the ColE1 cer locus. EMBO J. 7:4389-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Summers, D. 1998. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol. Microbiol. 29:1137-1145. [DOI] [PubMed] [Google Scholar]
- 38.Summers, D. K. 1989. Derivatives of ColE1 cer show altered topological specificity in site-specific recombination. EMBO J. 8:309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Summers, D. K., C. W. Beton, and H. L. Withers. 1993. Multicopy plasmid instability: the dimer catastrophe hypothesis. Mol. Microbiol. 8:1031-1038. [DOI] [PubMed] [Google Scholar]
- 40.Summers, D. K., and D. J. Sherratt. 1984. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell 36:1097-1103. [DOI] [PubMed] [Google Scholar]
- 41.Summers, D. K., and D. J. Sherratt. 1988. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 7:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tecklenburg, M., A. Naumer, A., O. Nagappan, and P. Kuempel. 1995. The dif resolvase locus of the Eschrichia coli chromosome can be replaced by a 33-bp sequence, but function depends on location. Proc. Natl. Acad. Sci. USA 92:1352-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tolmasky, M. E., S. Colloms, G. Blakely, and D. J. Sherratt. 2000. Stability by multimer resolution of pJHCMW1 is due to the Tn1331 resolvase and not to the Escherichia coli Xer system. Microbiology 146:581-589. [DOI] [PubMed] [Google Scholar]
- 44.Tolmasky, M. E., and J. H. Crosa. 1987. Tn 1331, a novel multiresistance transposon encoding resistance to amikacin and ampicillin in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 31:1955-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tolmasky, M. E., M. Roberts, M. Woloj, and J. H. Crosa. 1986. Molecular cloning of amikacin resistance determinants from a Klebsiella pneumoniae plasmid. Antimicrob. Agents Chemother. 30:315-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood, J. M. 1999. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63:230-262. [DOI] [PMC free article] [PubMed] [Google Scholar]