Abstract
The initial binding of bacterial cells to a solid surface is a critical and essential step in biofilm formation. In this report we show that stationary-phase cultures of Escherichia coli W3100 (a K-12 strain) can efficiently attach to sand columns when they are grown in Luria broth medium at 28°C in fully aerobic conditions. In contrast, growth in oxygen-limited conditions results in a sharp decrease in adhesion to hydrophilic substrates. We show that the production of lipopolysaccharide (LPS) and of flagella, as well as the transcription of the fliC gene, encoding the major flagellar subunit, increases under oxygen-limited conditions. Inactivation of the global regulatory hns gene counteracts increased production of LPS and flagella in response to anoxia and allows E. coli W3100 to attach to sand columns even when it is grown under oxygen-limited conditions. We propose that increased production of the FliC protein and of LPS in response to oxygen limitation results in the loss of the ability of E. coli W3100 to adhere to hydrophilic surfaces. Indeed, overexpression of the fliC gene results in a decreased adhesion to sand even when W3100 is grown in fully aerobic conditions. Our observations strongly suggest that anoxia is a negative environmental signal for adhesion in E. coli.
In many natural environments, bacteria grow as a biofilm on solid surfaces rather than living in a free-swimming, planktonic state (4, 5, 32). Formation of a biofilm is a complex process (22, 23), whose initial step is the adhesion of microorganisms to a surface via chemical interactions that depend on both surface and bacterial hydrophobicity and charge (18, 28, 29). The initial adhesion step is a critical point in the process of biofilm formation and is particularly important in porous media, such as sediments and aquifers, where the opposing forces of convective transport and the attachment of bacteria to surfaces determine the movement of bacteria through soil (34). Bacteria generally have a negative electrical charge at physiological pH (13) and must overcome repulsion forces in order to adhere to solid surfaces, which are also negatively charged in most cases (8, 20, 28). Experimental evidence indicates that some proteins, such as outer membrane proteins, fimbriae, and flagella, are required for initial bacterial adhesion (6, 26). Extracellular polysaccharides, like lipopolysaccharide (LPS) in gram-negative bacteria, play an important role in this process (11, 30, 42). LPS can establish strong short-range interactions with solid surfaces via the formation of hydrogen bonds (11, 12). Outer membrane proteins and other extracellular structures such as fimbriae and flagella also appear to be important for initial adhesion. However, it is not yet clear if they play a direct role in the establishment of interactions with solid surfaces or if they affect adhesion indirectly by increasing motility toward solid surfaces or by strengthening cell-cell interactions in postadhesion events (26).
Interestingly, the expression of many extracellular proteins and polymers involved in adhesion of microorganisms to surfaces is genetically regulated in response to environmental stimuli. For example, flagella are growth phase-regulated (14), while curli, fimbria-like structures important for initial adhesion in some Escherichia coli strains (40), are subjected to a complex form of regulation, being expressed in response to low temperature and osmolarity (1). These observations strongly suggest that the adhesion properties of bacteria are affected by environmental and growth conditions. Indeed, growth medium composition, temperature, and ionic strength seem to be important for the adhesion properties in E. coli and Pseudomonas aeruginosa (27, 37). The mechanism of the regulation of genes involved in bacterial adhesion often involves specific factors, as well as global regulatory proteins that affect expression of a large number of genes (21, 27, 31).
In this report, we investigate the effects of different growth conditions on the ability of the E. coli W3100 strain to adhere to a sea sand-filled column, a model system mimicking an aquifer. We show that E. coli attaches more efficiently to sand when grown in fully aerobic conditions than when grown in anoxic conditions. The observed reduction of adhesion efficiency upon growth in oxygen-limited conditions is dependent on the global regulatory protein HNS via positive regulation of LPS and flagella biosynthesis. We show that high levels of flagellum expression result in a decrease in initial bacterial attachment to solid surfaces in a porous medium. Our results suggest that lack of oxygen is an important environmental signal that negatively affects E. coli adhesion.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
In this report we used for our investigation the E. coli W3100 strain (E. coli Genetic Stock Center). To obtain W3100-derivatives mutated in global regulatory genes, we used bacteriophage P1-mediated transduction. Strains carrying alleles of global regulatory genes inactivated by antibiotic resistance cassettes were used as donors. Transductants were selected by plating on medium supplemented with the required antibiotic, and the correct location of the antibiotic resistance gene was verified by PCR. When necessary, antibiotics were added at the following concentrations: tetracycline, 25 μg/ml; hygromycin, 25 μg/ml; kanamycin, 50 μg/ml; ampicillin, 80 μg/ml. To ensure fully aerated conditions during growth, W3100 and its derivatives were grown in flasks filled to one-fifth of their capacity, with constant shaking at 200 rpm. Bacteria were usually grown overnight at 28°C in Luria broth (LB); when indicated, the growth temperature was changed to 33 or 37°C. For growth in high osmolarity, NaCl was added to a final concentration of 0.6 M. Growth in defined medium was performed in M9 medium supplemented with 0.4% glucose and 0.2% Casamino Acids (weight/volume). Anoxic growth was achieved by incubating the strain in 10-ml capped tubes filled to the top with LB, with no shaking. These conditions are sufficient for full induction of anaerobiosis-dependent genes (16, 41, 43), suggesting that cells are growing anaerobically in the experimental conditions used. Little or no formation of biofilm was observed on glass tubes in any of the growth conditions used. Motility was determined after 5 h of growth in swarming agar plates as described previously (35). To measure in vivo transcription from the fliC promoter region, we introduced a fliC::lacZ [Mu d (bla lac)] chromosomal fusion (35) in W3100 (wild-type) and PL400 (hns) strains by P1 transduction, producing, respectively, strains PL403 and PL404. The β-galactosidase activity was determined as described elsewhere (19). For fliC overexpression the gene, including its promoter region, was amplified by PCR from W3100 DNA with primers carrying HindIII and EcoRI restriction sites (underlined in the sequence): FLIC1 (5′-ACAGAAAAGCTTACCGGGGTTATCGGTCTG), which anneals 279 bp upstream the fliC start codon, and FLIC3 (5′-GTGCGAGAATTCGGATGCGGCGTAAACGCC), which anneals 168 bp downstream of the fliC stop codon. The PCR-amplified DNA fragment was cloned into the HindIII and EcoRI restriction sites of plasmid pUC19, producing plasmid pJD102.
Adhesion assays.
Attachment of bacterial cells to sand-filled columns was performed as described previously (34). Cells were grown overnight (14 to 18 h, optical density at 600 nm [OD600] of >2.0), washed with phosphate-buffered saline (pH 7.0; PBS) and resuspended in PBS (30 ml) at an OD280 of between 0.8 and 1.0 (corresponding to roughly OD600 = 0.2), corresponding to ca. 108 CFU/ml. The suspension was loaded at the flow rate of 0.5 ml/min onto a 12-cm column filled with 9 g of pure sea sand (Fluka, Geneva, Switzerland) preequilibrated in PBS. Similar conditions were used for glass columns, except that 12 g of glass beads was used. Observation of the column sand grains with an electron microscope shows that bacteria mostly attach as single cells in the conditions used in our experiments and that the sand surface is not fully occupied by the bacteria (data not shown), suggesting that cell-cell interaction does not play a major role in attachment to sand columns. Ten fractions (3 ml each) were collected, and the adhesion efficiency was calculated as the ratio between the OD280 of each fraction of the bacterial suspension collected at the column output (A) and the OD280 of the bacterial suspension loaded onto the column (Ao). For the experiments shown in Fig. 1 and 2, aliquots of randomly selected fractions were plated on L agar after opportune dilution; results of the plate counts showed good correlation with the spectrophotometric determination (data not shown). Each experiment was repeated at least three times unless otherwise stated.
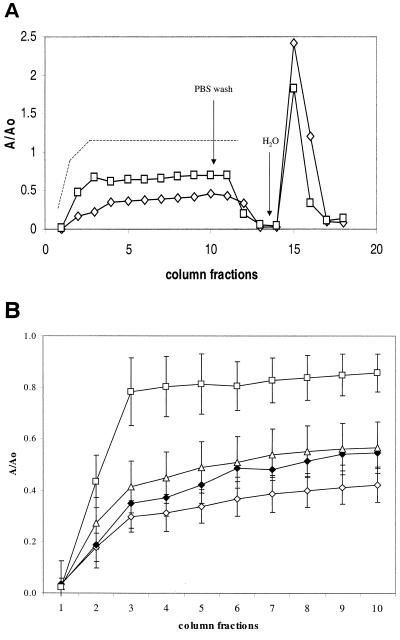
FIG. 1.
(A) Adhesion to a sand column by W3100 after overnight growth in different conditions. The A/Ao value was calculated as the ratio of bacteria recovered in the column flowthrough to bacteria loaded onto the column for each fraction, as described in Materials and Methods. Symbols: ◊, W3100 grown in LB at 28°C; □, W3100 grown in LB at 37°C. The dotted line represents the breakthrough curve of 1 mM KBr, used as a tracer in parallel experiments. (B) Effects of different growth conditions on W3100 adhesion. Symbols: ◊, growth in LB at 28°C in fully aerobic conditions; ▵, growth at 28°C in fully aerobic conditions in LB supplemented with NaCl to a 0.5 M final concentration; ⧫, growth in M9 medium (fully aerobic conditions); □, growth in LB in oxygen-limited conditions. The values shown are an average of four independent experiments.
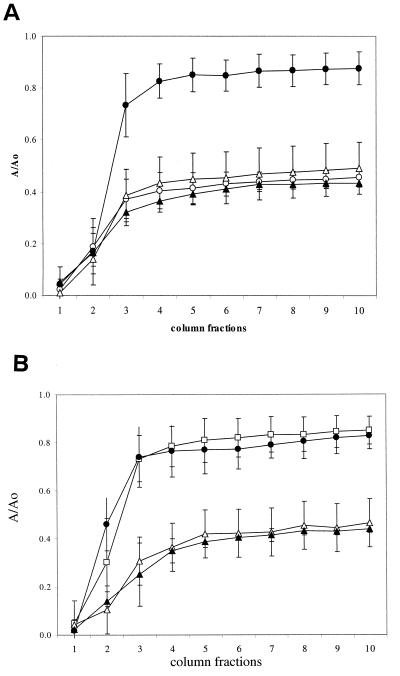
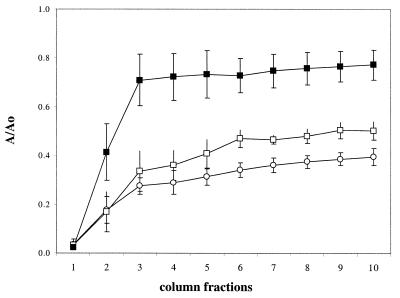
FIG. 2.
(A) Effect of the hns mutations on adhesion to sand. Open circles indicate W3100 grown in fully aerobic conditions (LB, 28°C). Open triangles indicate PL400 (hns) grown in fully aerobic conditions. Closed symbols indicate cells grown in oxygen-limited conditions. The values shown are an average of five independent experiments. (B) Adhesion to sand columns of W3100 (•), PL400 (hns ▴), PL400 transformed with pCI (▵), and PL400 transformed with plasmids pCI and pPLC2833 (□). Cells were grown overnight under oxygen-limited conditions at 33°C. Data are an average of three independent experiments.
Extracellular structures.
Outer membrane proteins were isolated with the Sarkosyl extraction method as described previously (6) and analyzed by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins of interest were extracted from the gel and identified by mass spectroscopy analysis of trypsin cleavage products as described earlier (3). For LPS isolation, cells were grown in a 2.5-ml overnight culture. After we adjusted the OD600 to 1, the cells were harvested by centrifugation, washed in H2O, and resuspended in PBS. The protein amount of the resuspended cells was measured and was found to be very similar for each sample. LPS was isolated with the hot phenol extraction method and analyzed by 18% PAGE as described previously (42). The gel was stained with silver staining.
Determination of physicochemical properties of bacterial strains.
The electrophoretic mobility of the bacterial cells was determined according to the method of Van Loosdrecht et al. (38), by using a Doppler electrophoretic light scattering analyzer (Zeta-Master; Malvern Instruments, Ltd.). For the measurements, cells grown in different conditions were harvested, washed, and resuspended in PBS (pH 7.0) at ca. 5 × 106 CFU/ml. Bacterial cell hydrophobicity was determined by using the contact angle measurement method as described previously (12).
RESULTS
Sand column assays.
E. coli W3100 strain was tested for adhesion to sand in a column system. As shown in Fig. 1, this experimental method allows the measurement of reversible initial adhesion by the bacteria. W3100 cells grown overnight in LB at 28°C with vigorous shaking (fully aerobic conditions) were able to attach rather efficiently to a sand column; >60% of the cells loaded in fractions 3 to 10 attached to the column (Fig. 1A, diamonds). The low concentration of bacteria in the first two fractions is a consequence of the void volume of the column, as can be seen from a curve obtained with a tracer dye (Fig. 1, dashed line), and does not indicate more efficient adhesion by bacteria in the initial fractions. Growth of W3100 at 37°C results in a decreased ability to adhere, with ca. 40% of bacterial cells attaching to the sand (Fig. 1A, squares). The initial bacterial adhesion to hydrophilic substrates is determined by the bacterial cell's surface charge (13) and can be reversed by lowering the ionic strength of the medium. Indeed, whereas washing the columns with PBS resulted in negligible elution of adhered bacteria from the sand column (fractions 11 to 13), a switch to sterile distilled water resulted in complete (>95%) elution of the attached bacteria. The different behavior of cells grown at 28 and 37°C suggests that reversible adhesion to hydrophilic surfaces might be affected by growth conditions. We further investigated the effects of different growth conditions on attachment to sand columns. As shown in Fig. 1B, growth in conditions of higher osmolarity (i.e., 0.6 M NaCl, 28°C in fully aerobic conditions) only had a small effect on adhesion to sand. Aerobic growth in defined M9 medium supplemented with glucose and Casamino Acids also resulted in a small reduction in adhesion. In contrast, growth in LB medium in anoxic conditions resulted in a dramatic loss of ability to attach to sand grains by W3100 cells. A comparable reduction in adhesion was observed when cells grown in anoxic conditions were loaded onto a column filled with glass beads (data not shown), suggesting that the lack of oxygen is a negative environmental signal for adhesion to hydrophilic surfaces. Interestingly, the growth rate does not play a major role in the ability of W3100 to attach to sand. Indeed, cells growing at the slowest rate (LB supplemented with 0.6 M NaCl, 0.64 doublings/h) and at the fastest rate (LB at 37°C, 2.05 doublings/h) displayed very similar adhesion properties. This observation suggests that the effect of anoxic growth on adhesion does not depend on variations in growth rate, but it is rather due to a specific switch in the expression of extracellular structures important for adhesion.
Regulation of adhesion properties in low-oxygen conditions.
To identify what regulatory genes might mediate the reduction in adhesion to sand in response to a lack of oxygen, we constructed a set of derivatives of W3100, in which genes encoding for gene expression regulators had been inactivated. The effects of their inactivation on adhesion were tested in the sand column assay. We targeted genes involved in regulation of anaerobic metabolism (fnr [33]), genes known to participate in the regulation of extracellular structures such as pili and outer membrane proteins (crp, lrp, and ompR [7, 40]), and genes involved in other global regulation processes (rpoS, hns, and fis [39]). Consistent with its role as principal regulator of anaerobic metabolism, the inactivation of the fnr gene resulted in extremely slow growth in low-oxygen conditions (<0.2 OD600 after 24 h of incubation at 28°C), making it impossible to test this mutant strain in the sand column assay. Inactivation of the crp, lrp, ompR, and fis genes had little or no effect on adhesion to sand by W3100 grown in oxygen-limited conditions (data not shown). In contrast, PL400, a W3100-derivative in which the hns genes had been disrupted by a hygromycin resistance cassette (44), was able to attach to the sand columns more efficiently than did the wild type (Fig. 2A). A similar effect, although to a lesser extent, was observed when the rpoS gene of W3100 was inactivated (data not shown). The hns gene encodes a DNA-binding protein involved in the negative regulation of a large number of genes (25); the rpoS gene encodes σS, an alternative σ factor of RNA polymerase mainly active in the stationary phase of bacterial growth (9). The effect of the hns mutation was also tested on adhesion of W3100 to glass bead columns; adhesion to glass by cells grown in anoxia were greatly increased by inactivation of the hns gene (data not shown).
To verify that increased adhesion was due to the lack of a functional HNS protein rather than to indirect effects of the hygromycin cassette that inactivates the gene, we performed a complementation experiment on the hns strain PL400. Since expression of the hns gene from multicopy plasmids is lethal for E. coli (36), we transformed the strain with plasmids pPLC2833 and pCI (a gift from C. Gualerzi, University of Camerino, Camerino, Italy). These plasmids carry, respectively, the hns gene under a cI-repressed promoter (pPLC2833) and the gene encoding λ cI857 protein (pCI); the λ cI857 protein is stable at 30°C but not at 37°C. Low-level expression of the HNS protein can be obtained by growing the bacteria at 33°C (M. Falconi and C. Gualerzi, unpublished data). The results of the complementation experiment in Fig. 2B indicate that low levels of hns expression restore the nonadherent phenotype of the wild type, showing that the effect on adhesion to sand in oxygen-limited conditions is indeed dependent on the hns gene.
Inactivation of the hns gene affects the cell's physicochemical properties.
Initial adhesion of bacteria to an abiotic surface depends primarily on nonspecific physicochemical interaction, such as the formation of hydrogen bonds and electric charge interaction (for hydrophilic surfaces) or of hydrophobic interaction (for hydrophobic surfaces). The interaction between cells and sand grains involves both kinds of interaction (34, 42). Thus, mutations in the hns genes might increase adhesion to sand by altering physicochemical properties, such as electrical charge, required for attachment to hydrophilic surfaces, or hydrophobicity.
Cell surface hydrophobicity was determined by using the contact angle measurement. This method basically consists in the measurement of the angle (θw) formed by a drop of water on a layer of bacteria obtained by filtration of a bacterial suspension on nitrocellulose filters (12). Low contact angle values correspond to low hydrophobicity of the cell surface. The values of [theta]w obtained for both W3100 and PL400 were close to 20°, suggesting that the highly hydrophilic nature of W3100 cell surface is not affected by the hns mutation.
Measurement of the zeta potential (ζ) and electrophoretic mobility (u) (Table 1) showed that the cell surface of W3100 is negatively charged at physiological pH, as is the case for most bacteria (13). Growth in oxygen-limited conditions resulted in an additional increase in the negative charge of W3100, which might negatively affect adhesion to sand. While the surface charge of PL400 was comparable to W3100 for aerobically grown cultures, the hns mutant strain displayed a less-negative charge when cells were grown in anoxic conditions, suggesting that the hns mutation might affect cell surface properties.
TABLE 1.
Zeta potential and electrophoretic mobility of bacterial strainsa
| Strain | Mean electrophoretic mobility (u) ± SD | Mean zeta potential (ζ [mV]) ± SD |
|---|---|---|
| W3100 (aerobic) | −1.869 ± 0.317 | −29.1 ± 4.35 |
| W3100 (anoxic) | −2.746 ± 0.368 | −38.7 ± 5.54 |
| PL400 (aerobic) | −1.981 ± 0.308 | −30.6 ± 4.01 |
| PL400 (anoxic) | −2.224 ± 0.234 | −32.1 ± 3.37 |
Determined in PBS (pH 7.0).
Effects of the hns mutation on outer membrane protein expression.
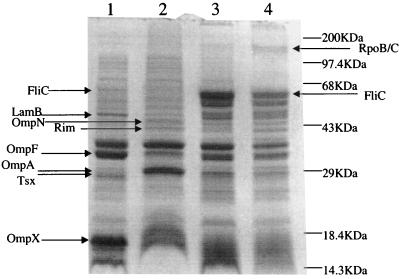
The sharp increase in adhesion to sand grains and the variation in the cell surface charge observed in the hns mutant are likely dependent upon altered expression of extracellular structures. Extracellular and outer membrane proteins can play an important role in initial bacterial adhesion due to their location on the cell envelope. Since the HNS protein is involved in the regulation of extracellular proteins such as flagella and curli (1, 7, 35), we isolated outer membrane proteins from strains W3100 (wild type) and PL400 (hns) grown in both oxygen-rich and oxygen-limited conditions (Fig. 3). Inactivation of the hns gene resulted in changes in the level of expression for several outer membrane proteins. These differences were detected mostly in aerobically growing strains (compare lanes 1 and 2) rather than in strains grown in anoxia (lanes 3 and 4). Interestingly, despite the fact that hns is mainly a negative regulator of gene expression (10), five of the eight proteins found to be expressed at different levels in strains W3100 and PL400 appeared to be positively regulated by hns, while the production of only three proteins was stimulated by hns inactivation. The outer membrane proteins whose expression was affected by the hns mutation were identified by mass spectrometry determination after gel extraction and trypsin digestion (3). Expression of the FliC protein, the major flagellar subunit, was reduced by the hns mutation, a finding consistent with results obtained by other investigators (10, 35); in addition, the expression of porins LamB, OmpF, Tsx, and OmpX was significantly decreased in the hns strain PL400. In contrast, expression of two porins, OmpN and OmpA, was stimulated by hns inactivation. Another protein found to be expressed at higher levels in the hns strain PL400 was Rim, the methylase subunit of the EcoRI restriction/methylation enzyme. The location of this protein is cytoplasmic, and its presence in the outer membrane protein fraction is likely to be an artifact, possibly due to nonspecific interaction of Rim with some membrane protein. Similarly, the β and β′ subunits of RNA polymerase are present as a contaminant in the outer membrane fraction of PL400 grown in oxygen-limiting conditions (lane 4). The pattern of outer membrane protein expression was much more homogeneous between the two strains when they were grown in oxygen-limited conditions (lanes 3 and 4). The most striking feature was the sharp increase of FliC production in both strains; however, the amount of the FliC protein was still four- to fivefold lower in PL400 than in the wild type. This ratio was determined by densitometric analysis of the gels, correcting for the apparent lower amount of total protein loaded in lane 4.
FIG. 3.
SDS-PAGE analysis of outer membrane protein patterns of W3100 (wild type) and PL400 (hns) grown in fully aerobic (lanes 1 and 2) and oxygen-limited conditions (lanes 3 and 4). Proteins identified by mass spectrometry whose expression differs in the two strains are indicated.
The hns mutations affects LPS production.
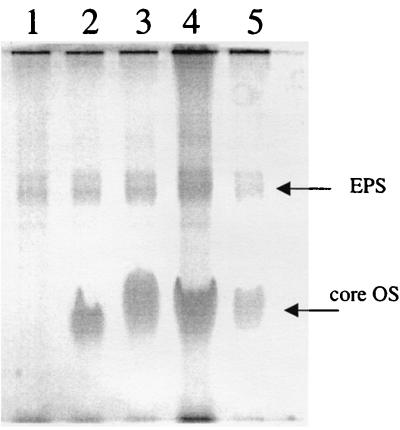
One of the main components of the cell envelope in gram-negative bacteria is LPS. LPS is a known determinant for initial bacterial adhesion to solid surfaces in several gram-negative bacteria (8, 11). To investigate its possible role in increased adhesion of the hns strain, we isolated and analyzed LPS from W3100 (wild type) and PL400 (hns) (Fig. 4). A crude LPS preparation was extracted by using the hot phenol method (42) and then analyzed on acrylamide gels. Two major bands were detected by silver staining: inactivation of the core LPS biosynthetic operon through insertion mutagenesis in the waaQ (previously rfaQ [24]) gene resulted in loss of the lower-molecular-weight band, suggesting that this band is a core oligosaccharide (lane 1). The high-molecular-weight band was readily stained by Alcian blue, indicating a stronger acidic nature, and its expression appears to be related to the core oligosaccharide. Inactivation of the hns gene did not result in any significant increase in LPS production under oxygen-saturating conditions (Fig. 4, lanes 2 and 3), whereas growth oxygen-limited conditions clearly stimulated LPS production in W3100 (compare lanes 2 and 4). In contrast, not only did LPS amount fail to increase in the hns strain, but it also decreased compared to PL400 grown in fully aerobic conditions (lanes 3 and 5). This suggests that the hns gene positively regulates LPS biosynthesis during anoxia.
FIG. 4.
PAGE analysis of LPS. Lane 1, PL519 (waaQ derivative of W3100) grown under oxygen-saturating conditions; lane 2, W3100 grown under fully aerobic conditions (LB at 28°C); lane 3, PL400 (hns) grown under fully aerobic conditions; lane 4, W3100 grown in oxygen-limiting conditions; lane 5, PL400 (hns) grown inder oxygen-limiting conditions. The two major bands revealed by silver staining are indicated.
Growth in oxygen-limiting conditions positively regulates flagellar expression at the transcription level.
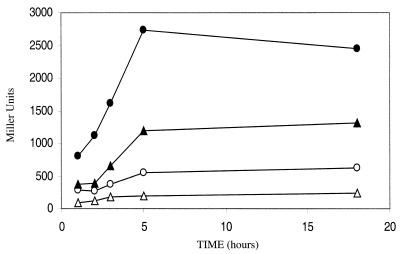
Growth in anoxic conditions dramatically increases the production of the FliC protein in W3100 (Fig. 3). We tested the possibility that this increased production is mediated by upregulation of the fliC promoter. We transduced a fliC::lacZ chromosomal fusion into either the W3100 (wild-type) or the PL400 (hns) strains, producing, respectively, strains PL403 and PL404. The strains carrying the fliC::lacZ fusions were grown overnight either in oxygen-rich or in oxygen-limited conditions, and the expression from the different promoters was determined as the β-galactosidase activity. These experiments (Fig. 5) showed that in the wild-type W3100 strain the levels of transcription from the fliC promoter increased when cells were grown in oxygen-limited conditions, a finding consistent with the results of direct outer membrane protein analysis (Fig. 3). Inactivation of the hns gene resulted in lower expression of fliC, a finding consistent with previous observations (35). Interestingly, growth in oxygen-limited conditions also stimulated fliC transcription in PL400, suggesting that this effect takes place independently of hns.
FIG. 5.
Expression of a fliC::lacZ chromosomal fusion in PL403 (wild type, circles) and in PL404 strain (hns, triangles). Open symbols indicate cells grown in fully aerobic conditions, and closed symbols indicate cells grown under oxygen-limited conditions. Overnight cultures (LB, 28°C, with shaking) were diluted 1:200, and samples were obtained after 1, 2, 3, 5, and 18 h. The β-galactosidase activity is expressed in Miller units (19). The data shown are the average of three independent experiments.
Overexpression of FliC negatively affects adhesion to sand.
The results of outer membrane protein and LPS analysis suggest that the increased production of FliC and/or of LPS might result in loss of W3100 adhesion abilities. In order to evaluate the possibility that an excess of FliC negatively affects adhesion, we transformed W3100 with a plasmid pJD102 carrying the fliC gene under the control of the lac promoter. pJD102-directed expression of fliC results in a strong reduction of attachment to sand columns by both W3100 (Fig. 6) and PL400 (data not shown), even though the strains were grown in fully aerobic conditions. No effects on adhesion were detected in either strain transformed with the pUC19 vector (Fig. 6) (data not shown). This result suggests that an excess of flagella production negatively affects the adhesion properties of the W3100 strain. Similar observations were reported for adhesion-deficient mutants of Pseudomonas fluorescens, which were found to express high levels of flagella. Despite the increased flagellar expression, the P. fluorescens mutants are nonmotile (6). Thus, we tested strains W3100 and PL400 for their motility in soft agar. Inactivation of the hns gene resulted in a reduction of motility, in agreement with previously published results and consistent with reduced production of flagella. However, anoxia-dependent increased expression of flagella did not result in loss of motility in either strain, suggesting that cell motility alone is not sufficient for efficient attachment to solid surfaces in our test system (data not shown).
FIG. 6.
Effects of fliC overexpression on bacterial adhesion to sand. Symbols: ○, W3100 grown under fully aerobic conditions; □, W3100 transformed with pUC19 plasmid; ▪, W3100 transformed with pJD102 plasmid (carrying the fliC gene under the control of the lac promoter). The values shown are the average of three independent experiments.
Effects of fliC and waaQ inactivation on cell adhesion to sand.
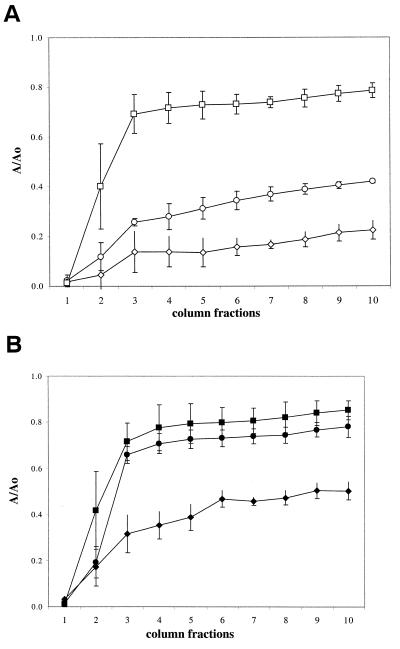
Results of the previous experiments suggest that increased expression of flagella and LPS negatively affects bacterial adhesion. Thus, we disrupted either the fliC (encoding the major flagellum subunit) or the waaQ gene (part of the LPS core biosynthetic operon) with a kanamycin resistance cassette, and we tested the ability of the deletion mutant strains to adhere to sand. The results of these experiments are shown in Fig. 7. Surprisingly, inactivation of fliC resulted in clear loss of adhesion to sand in aerobically growing cultures; in contrast, inactivation of the waaQ gene stimulated adhesion both in aerobic and anoxic conditions, suggesting a negative role of LPS in adhesion to sand.
FIG. 7.
Effects of inactivation of either the fliC or the waaQ genes on bacterial adhesion to sand. (A) Cells grown under fully aerobic conditions. Symbols: ○, W3100; ◊, PL519 (waaQ derivative of W3100); □, PL521 (fliC derivative of W3100). (B) Cells grown under oxygen-limited conditions. Symbols: •, W3100; ⧫, PL519 (waaQ derivative of W3100); ▪, PL521 (fliC derivative of W3100).
DISCUSSION
In this report we have shown that growth in oxygen-limited conditions results in loss of the ability of an E. coli laboratory strain (W3100) to adhere to hydrophilic porous media, such as sand grains and glass microbeads, in an experimental system mimicking an aquifer (Fig. 1B). Reduction in the attachment to solid substrates is not due solely to changes in growth rate but is specifically induced by oxygen deprivation (Fig. 1). This observation suggests that lack of oxygen is an environmental signal that reduces bacterial adhesion in E. coli. This effect could be mediated directly by oxygen starvation, or it might be due to the accumulation of some product of the anaerobic metabolism. Optimal anoxia-dependent negative regulation of adhesion requires a functional hns genes; indeed, a derivative of W3100 in which the hns gene has been inactivated (strain PL400) can efficiently attach to hydrophilic materials such as sand (Fig. 2) and glass (data not shown) independently of oxygen availability. The HNS protein is a global regulator of gene expression in E. coli, since it controls the transcription of a large number of genes, estimated to be ca. 5% of the E. coli genome (10). The expression of several outer membrane proteins is partly dependent on this global regulator in the E. coli MG1655 strain (10). In the W3100 strain used in our work, the HNS protein strongly affects the expression of several porins, outer membrane proteins involved in specific membrane transport (Fig. 3). The OmpF, Tsx, and OmpX porins were produced at higher levels in the wild type than in PL400, where the hns gene is not functional, suggesting that the HNS protein positively controls their regulation. In contrast, the expression of two other porins, OmpN and OmpA, appears to be downregulated by HNS. Thus, a functional hns gene determines the pattern of porin expression in cells growing with aerobic metabolism. Surprisingly, when cells are grown in oxygen-limited conditions, porin regulation by HNS is almost completely bypassed (Fig. 3). This observation strongly suggests that the effect of hns inactivation on adhesion is not directly mediated by the porin component of the outer membrane.
Analysis of the major components of the E. coli W3100 cell envelope suggests that anoxia-dependent reduction of its adhesion properties might depend on the concomitant increased expression of flagella and LPS (Fig. 3 and 4). Indeed, overexpression of fliC, encoding the major flagellar subunit, affects W3100 attachment to sand (Fig. 6), suggesting that high levels of flagellum production can be a negative determinant for bacterial adhesion in E. coli. Surprisingly, inactivation of fliC also resulted in an almost complete loss of adhesion to sand by W3100 (Fig. 7), a finding consistent with previous observations (26). The fact that both inactivation and overexpression of fliC negatively affect adhesion suggests a gene dosage effect: FliC, the major flagellum subunit, might be necessary for initial adhesion either in small amounts or only at a defined stage of the process. Excessive or untimely expression of the protein, as well as lack thereof, might affect adhesion properties of the bacterium. Interestingly, similar findings were already reported for Pseudomonas species: in a screening for adhesion-deficient mutants after transposon mutagenesis, both flagellum-deficient and flagellum-overexpressing mutants were found to be impaired in adhesion to solid surfaces (6). Unlike in Pseudomonas, high expression levels of flagella did not strongly affect E. coli cell motility (data not shown), suggesting that flagella might interfere with adhesion through steric hindrance; protruding flagella might not allow interactions between other components of the bacterial envelope and the solid substrate to take place.
We propose that, in addition to the negative effect of increased fliC expression, overproduction of LPS in the W3100 strain growing in oxygen-limiting conditions (Fig. 4) might affect cell-surface interactions. Consistent with this observation, inactivation of the waaQ gene, resulting in lack of LPS production (Fig. 4) clearly stimulated adhesion to sand by W3100. LPS can play both a positive and a negative role in initial adhesion to sand and other solid surfaces, as already shown in several reports (12, 17, 30, 42). The direct effect of LPS on initial cell-surface interactions is related to the formation of hydrogen bonds between bacterial cells and the solid surface (12). This hypothesis is supported by the fact that adhesion to sand in our system is strictly dependent on the ionic strength of the buffer (13) (Fig. 1A). The chemical composition, chain length, and overall charge of the LPS determine the nature of cell-surface interaction, allowing either attraction or repulsion forces to prevail (12, 13, 34). In strain W3100, anoxia-dependent LPS overproduction leads to a more negative electrical charge of the cell envelope (Table 1), which in turn would result in increased repulsion forces between the cell and negatively charged surfaces.
Inactivation of the hns gene counteracts anoxia-dependent increased expression of flagella and of LPS production (Fig. 3 and 4). Regulation of flagellar expression by hns takes place via indirect control of the main flagellar operon. hns negatively controls the hdfR gene, which in turn represses the flhDC operon, necessary for fliC expression (15). The presence of a functional hns gene results in higher levels of fliC transcription regardless of growth conditions, and oxygen starvation increases fliC expression in both W3100 and PL400 by roughly the same extent (Fig. 5). This shows that anoxia-dependent increase of fliC expression is independent of hns and hdfR. However, the lack of high-level expression of the FliC protein due to hns inactivation might positively affect bacterial adhesion.
In contrast to flagellar synthesis, the production of LPS depends on oxygen availability in an hns-dependent fashion (Fig. 4). While inactivation of the hns gene does not strongly affect LPS production in fully aerobic conditions, the amount of LPS is drastically reduced in the hns strain PL400 during anoxic growth, suggesting that hns might upregulate the LPS biosynthetic genes in response to oxygen starvation. To our knowledge, this is the first time that an effect of anoxia on LPS production is reported for E. coli. However, changes in the level of LPS gene expression were described for P. aeruginosa biofilms, in which bacteria grow in a lower oxygen concentration than as planktonic cells (2). Since the HNS protein is not directly involved in adaptation to anaerobiosis, anoxia-dependent LPS regulation is not likely to depend solely on HNS. However, hns takes part in the regulation of several genes that respond to oxygen starvation, such as the gad and glt regulons (10). We propose that LPS regulation involves the HNS protein, as well as at least another factor that responds specifically to lack of oxygen. Indeed, it has already been shown that HNS can regulate gene expression in concert with other factors at several promoters (1, 35, 39).
Acknowledgments
We thank Mike Volkert, Claudio Gualerzi, Roberto Kolter, Corinne Dorel, Steve Busby, Rod Welch, and Chris Whitfield for the gift of bacterial strains; Kirsten Lawlor for critical reading of the manuscript; and Teresa Colangelo for technical assistance.
This work was supported by the research grant 3100-058871 from the Swiss National Science Foundation.
REFERENCES
- 1.Arnqvist, A., A. Olsen, and S. Normark. 1994. σS-Dependent growth-phase induction of the agfBA promoter can be achieved in vivo by σ70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13:1021-1032. [DOI] [PubMed] [Google Scholar]
- 2.Beveridge, T. J., S. A. Makin, J. L. Kadurugamuwa, and Z. Li. 1997. Interactions between biofilms and the environment. FEMS Microbiol. Rev. 20:291-303. [DOI] [PubMed] [Google Scholar]
- 3.Chen, X., L. M. Smith, and E. M. Bradbury. 2000. Site-specific mass tagging with stable isotopes in proteins for accurate and efficient protein identification. Anal. Chem. 72:1134-1143. [DOI] [PubMed] [Google Scholar]
- 4.Costerton, W. J., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed]
- 5.Davey, M. E., and G. A. O' Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeFlaun, M. F., B. M. Marshall, E.-P. Kulle, and S. B. Levy. 1994. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl. Environ. Microbiol. 60:2637-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock, I. C. 1991. Microbial cell surface architecture, p. 21-59. In N. Moses, P. S. Handely, H. J. Busscher, and P. G. Rouxhet (ed.), Microbial cell surface analysis. VCH, Weinheim, Germany.
- 9.Hengge-Aronis, R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72:165-168. [DOI] [PubMed] [Google Scholar]
- 10.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 11.Jucker, B. A., H. Harms, and A. J. B. Zehnder. 1998. Polymer interaction between five gram-negative bacteria and glass investigated using LPS micelles and vesicles as model system. Colloid Surfaces B Biointerfaces 11:33-45. [Google Scholar]
- 12.Jucker, B. A., H. Harms, S. J. Hug, and A. J. B. Zehnder. 1997. Adsorption of bacterial surface polysaccharides on mineral oxides is mediated by hydrogen bonds. Colloids Surfaces B Biointerfaces 9:331-343. [Google Scholar]
- 13.Jucker, B. A., Harms, H., and A. J. B. Zehnder. 1996. Adhesion of the positively charged bacterium Stenotrophomonas (Xanthomonas) maltophilia 70401 to glass and Teflon. J. Bacteriol. 178:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlinsey, J. E., S. Tanaka, V. Bettenworth, S. Yamaguchi, W. Boos, S. I. Aizawa, and K. T. Hughes. 2000. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 37:1220-1231. [DOI] [PubMed] [Google Scholar]
- 15.Ko, M., and C. Park. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J. Bacteriol. 182:4670-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landini, P., L. I. Hajec, and M. R. Volkert. 1994. Structure and transcriptional regulation of the Escherichia coli adaptive response gene aidB. J. Bacteriol. 176:6583-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makin, S. A., and T. J. Beveridge. 1996. The influence of A-band and B-band lipopolysaccharide on the surface characteristics and adhesion of Pseudomonas aeruginosa to surfaces. Microbiology 142:299-307. [DOI] [PubMed] [Google Scholar]
- 18.Marshall, K. C. 1986. Adsorption and adhesion process in microbial growth at interfaces. Adv. Colloid Interface Sci. 25:59-86. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 20.Norde, W., and J. Lyklema. 1989. Protein adsorption and bacterial adhesion to solid surfaces: a colloid chemical approach. Colloids Surfaces 38:1-13. [Google Scholar]
- 21.Nou, X., B. Braaten, L. Kaltenbach, and D. A. Low. 1995. Differential binding of Lrp to two sets of Pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 14:5785-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Toole, G. A., and R. Kolter. 1988. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 23.O'Toole, G. A., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-70. [DOI] [PubMed] [Google Scholar]
- 24.Parker, C. T., E. Pradel, and C. A. Schneitman. 1992. Identification and sequences of the lipopolysaccharide core biosynthetic genes rfaQ, rfaP, and rfaG of Escherichia coli K-12. J. Bacteriol. 174:930-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pon, C. L., Calogero, R. A., and C. O. Gualerzi. 1988. Identification, cloning, nucleotide sequence and chromosomal map location of hns, the structural gene for Escherichia coli DNA-binding protein H-NS. Mol. Gen. Genet. 212:199-202. [DOI] [PubMed] [Google Scholar]
- 26.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 27.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-464. [DOI] [PubMed] [Google Scholar]
- 28.Rijnaarts, H. H. M., W. Norde, E. J. Bower, J. Lyklema, and A. J. B. Zehnder. 1995. Reversibility and mechanism of bacterial adhesion. Colloids Surfaces B Biointerfaces 4:5-22. [Google Scholar]
- 29.Rijnaarts, H. H. M., W. Norde, E. J. Bower, J. Lyklema, and A. J. B. Zehnder. 1996. Bacterial deposition in porous media: effects of cell coating, substratum hydrophobicity, and electrolyte concentration. Environ. Sci. Technol. 30:2877-2883. [Google Scholar]
- 30.Rocchetta, H. L., L. L. Burrows, and J. S. Lam. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63:523-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Römling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 32.Shapiro, J. A. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81-104. [DOI] [PubMed] [Google Scholar]
- 33.Shaw, D. J., and J. R. Guest. 1981. Molecular cloning of the fnr gene of Escherichia coli K12. Mol. Gen. Genet. 181:95-100. [DOI] [PubMed] [Google Scholar]
- 34.Simoni, S. F., H. Harms, T. N. P. Bosma, and A. J. B. Zehnder. 1998. Population heterogeneity affects transport of bacteria through sand columns at low flow rates. Environ. Sci. Technol. 32:2100-2105. [Google Scholar]
- 35.Soutourina, O., A. Kolb, E. Krin, C. Laurent-Winter, S. Rimsky, A. Danchin, and P. Bertin. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spurio, R., M. Durrenberger, M. Falconi, A. La Teana, C. L. Pon, C. O. Gualerzi. 1992. Lethal overproduction of the Escherichia coli nucleoid protein H-NS: ultramicroscopic and molecular autopsy. Mol. Gen. Genet. 231:201-211. [DOI] [PubMed] [Google Scholar]
- 37.Stanley, P. M. 1983. Factors affecting the irreversible attachment of Pseudomonas aeruginosa to stainless steel. Can. J. Microbiol. 29:1493-1499. [DOI] [PubMed] [Google Scholar]
- 38.Van Loosdrecht, M. C. M., W. Norde, J. Lyklema, and A. J. B. Zehnder. 1990. Hydrophobic and electrostatic parameters in bacterial adhesion. Aquat. Sci. 52:103-114. [Google Scholar]
- 39.Vicente, M., K. F. Chater, and V. de Lorenzo. 1999. Bacterial transcription factors involved in global regulation. Mol. Microbiol. 33:8-17. [DOI] [PubMed] [Google Scholar]
- 40.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volkert, M. R., L. I. Hajec, Z. Matijasevic, F. C. Fang, and R. Prince. 1994. Induction of the Escherichia coli aidB gene under oxygen-limiting conditions requires a functional rpoS (katF) gene. J. Bacteriol. 176:7638-7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams, V., and M. Fletcher. 1996. Pseudomonas fluorescens adhesion and transport through porous media are affected by lipopolysaccharide composition. Appl. Environ. Microbiol. 62:100-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wing, H. J., S. M. Williams, and S. J. W. Busby. 1995. Spacing requirements for transcription activation by Escherichia coli FNR protein. J. Bacteriol. 177:6704-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada, H., T. Yoshida, K. Tanaka, C. Sasakawa, and T. Mizuno. 1991. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol. Gen. Genet. 230:332-336. [DOI] [PubMed] [Google Scholar]