Abstract
Integrons can insert and excise antibiotic resistance genes on plasmids in bacteria by site-specific recombination. Class 1 integrons code for an integrase, IntI1 (337 amino acids in length), and are generally borne on elements derived from Tn5090, such as that found in the central part of Tn21. A second class of integron is found on transposon Tn7 and its relatives. We have completed the sequence of the Tn7 integrase gene, intI2, which contains an internal stop codon. This codon was found to be conserved among intI2 genes on three other Tn7-like transposons harboring different cassettes. The predicted peptide sequence (IntI2*) is 325 amino acids long and is 46% identical to IntI1. In order to detect recombination activity, the internal stop codon at position 179 in the parental allele was changed to a triplet coding for glutamic acid. The sequences flanking the cassette arrays in the class 1 and 2 integrons are not closely related, but a common pool of mobile cassettes is used by the different integron classes; two of the three antibiotic resistance cassettes on Tn7 and its close relatives are also found in various class 1 integrons. We also observed a fourth excisable cassette downstream of those described previously in Tn7. The fourth cassette encodes a 165-amino-acid protein of unknown function with 6.5 contiguous repeats of a sequence coding for 7 amino acids. IntI2*179E promoted site-specific excision of each of the cassettes in Tn7 at different frequencies. The integrases from Tn21 and Tn7 showed limited cross-specificity in that IntI1 could excise all cassettes from both Tn21 and Tn7. However, we did not observe a corresponding excision of the aadA1 cassette from Tn21 by IntI2*179E.
Integrons are genetic elements that permit tandem integration and expression of mobile cassettes coding for antibiotic resistance genes (18, 31, 37). The gene cassettes move by a conservative site-specific recombination mechanism, catalyzed by an integrase encoded by the integron 5′-conserved segment (5′-CS) (29, 37). The integrase belongs to the phage integrase family of recombinases (3), now called tyrosine recombinases (13). Integration of cassettes generally occurs into a target site attI, at the junction between the 5′-CS and the first cassette (Fig. 1) (11, 32). A second type of cassette recombination sites are the attC sites (59-base elements) which occur either at the 3′ end of each inserted cassette (17, 18, 31, 38) or in a free cassette circle (10). The initially described type of integron (referred to as class 1) is generally borne on elements similar to Tn5090 (30) or on related defective transposons (8), which sometimes “piggyback” on competent transposons to form Tn21 and its relatives. In addition, a second class of integron occurs on the nonreplicative transposon Tn7. Tn7 preferentially inserts into a unique site in bacterial chromosomes (16, 21, 23) from which it is transferred to other bacterial cells by less-specific transposition onto a conjugative plasmid (46). The question of whether or not Tn7 carries a functional integron is interesting because it would add to the multiplicity of systems for gene exchange among microorganisms.
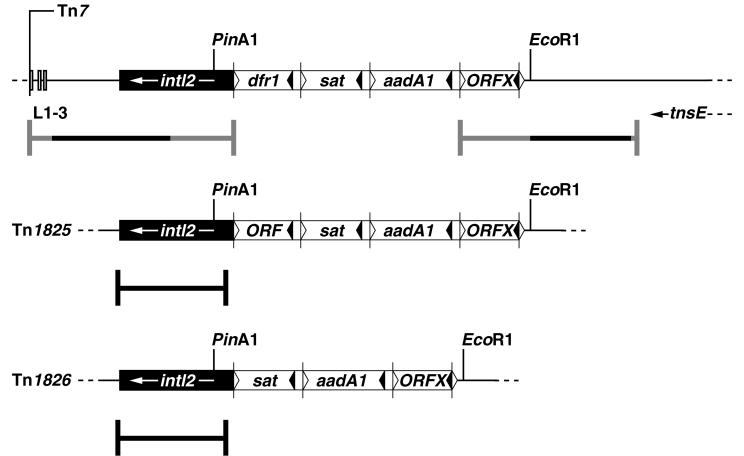
FIG. 1.
Maps of the integron regions in Tn7 and in the related transposons Tn1825 and Tn1826. Integron sequences are represented by boxes. Black boxes indicate the conserved sequence (5′-CS-2) with the integrase gene, intI2. Cassettes are shown by white boxes. Leftward-pointing filled arrowheads denote inverse core sequences inside the cassettes, and rightward-pointing open arrowheads denote core sequences where DNA strand transfer occurs. Vertical lines at cores indicate cassette borders. The PinAI and EcoRI restriction enzyme cut sites used for cloning the cassette regions are indicated. Nucleotide sequences determined in this study are underlined. Newly determined sequences are indicated with heavy black lines, while heavy gray lines indicate previously determined sequences that were checked in the present study. Vertical boxes denote the 22-bp repeats, L1-3, for transposase binding, which are clustered close to the left Tn7 end (2).
The integron of Tn7 (Fig. 1) has an organization similar to that of the class I integrons and carries three resistance gene cassettes—dfrA1, sat, and aadA1 (38, 39, 40)—close to an open reading frame, IntI2*, that is related to the class 1 integrase, IntI1, of Tn21 (Fig. 1) (19, 29). A variation of the cassette content in Tn7 was previously observed among related elements such as Tn1825 and Tn1826 (42) and Tn4132 (49). Since two cassettes in Tn7, dfrA1 and aadA1, have also been observed in class 1 integrons (14, 40), some cassettes appear to have been transferred among integron classes. Part of the integrase gene on Tn7 was included in a sequence by Simonsen et al. (36). The nucleotide sequence included an internal stop codon (TAA) in the intI2 gene. We reverified the presence of the internal stop codon which we also found to be present in the intI2 genes of Tn1825 and Tn1826 (Fig. 1). The sequence for intI2 in Tn7 was completed here by sequencing the previously unknown 3′ end of the gene, confirming that intI2 is closely related to intI1 of Tn21 and also resembles an integrase gene from a third integron class (intI3) (1).
We show here experimentally that the second class of integrase in Tn7 can promote recombination between cassette recombination sites present in the same plasmid (excision) or in different plasmids (cointegration), but only after changing the termination codon to a sense codon. Recombination activity was assayed in vivo by expressing the Tn7 integrase from a repaired form of intI2, intI2*179E, in which the internal stop codon has been altered into a triplet (GAG) encoding glutamic acid. Analysis of cassette recombination involving the class-specific attI sites revealed that the Tn7 integrase seems to be more restricted to recognition of its own attI site than the Tn21 integrase.
The three previously defined cassettes borne on Tn7 mediate resistance to trimethoprim (dfrA1 [15, 40]), streptothricin (sat [39, 42]) and spectinomycin (aadA1 [14]). We checked for further excisable units and found a fourth cassette downstream of those described previously. This new cassette lacked the palindromic attC site that is generally found at the 3′ end of cassettes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids are listed in Table 1. Escherichia coli strains were cultured in twofold-concentrated YT broth (27), Luria broth (6), or IsoSensitest medium (Oxoid) supplemented with the appropriate antibiotic(s) at 37°C. The antibiotic concentrations used were as follows: ampicillin (50 μg ml−1), spectinomycin (50 μg ml−1), trimethoprim (50 μg ml−1), streptothricin (2 μg ml−1), chloramphenicol (40 μg ml−1), streptomycin (50 μg ml−1), sulfathiazole (0.5 mM), and nalidixic acid (25 μg ml−1).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44 ΔlacU169 (Φ80 lacZ′ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA | 47 |
| HB101 | F−hsdS20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 (Smr) xyl-5 mtl-1 supE44 λ− | 7, 34 |
| JM105 | thi rpsL endA sbcB15 hspR4 Δ(lac-proAB) F′ traD36 proAB lacIqZΔM15 | 48 |
| CU9276 | hsdR17 mcrAB recA1 Δ(lac-proAB) F′ traD36 proAB lacIqZΔM15 | 44 |
| IE972 | V601::thr leu thi Rifr (Tn1825) | 43 |
| IE970 | J53::met pro (Tn1826) | 43 |
| Plasmids | ||
| R388 | Tpr Sur Tra+ | 38, 41 |
| R388Δ1 | R388 deleted for the dfr2b and ORFA cassettes; Sur Tra+ | 20 |
| R483 | Tpr Strir Spr | 4 |
| pUC18/19 | Apr | 48 |
| pSU18/19 | Cmr | 5 |
| 636 | 1,620-bp SphI-HpaI fragment from R483 in pUC19; Apr | This study |
| 636∗179E | Plasmid 636 with a point mutation, changing the stop codon in the integrase gene at position 179 to a glutamic acid; Apr | This study |
| 670 | 2,919-bp PinA1-EcoRI fragment from R483 in pSU18; Cmr Tpr Strir Spr | This study |
| 671 | 2,919-bp PinA1-EcoRI fragment from R483 in pSU19; Cmr Tpr Strir Spr | This study |
| 674 | 352-bp SphI-PstI PCR fragment (Tn7-107, cup2) from plasmid 670 (deleted for the dfr1 and sat cassettes) in pSU19; Cmr | This study |
| 675 | 439-bp XbaI-PstI PCR fragment (Tn7-107, cup2) from plasmid 670 (deleted for the dfr1 and sat cassettes) in pSU18; Cmr | This study |
| 678 | Plasmid 670 deleted for the dfr1 and sat cassettes; Cmr Spr | This study |
| 699 | Plasmid 670 deleted for the dfr1 cassette; Cmr Strir Spr | This study |
| 2101 | 1,176-bp RsaI-BamHI of Tn21 in pUC18; Apr | 20 |
| 2123 | 1,618-bp SphI-HindIII PCR fragment (pSph, pHin) from Tn21 in pSU18; Cmr Spr | 20 |
| 2213 | 900-bp SphI-PstI PCR fragment (pSph, cup2) from Tn21 in pSU19; Cmr | 20 |
| 2280 | 1,618-bp SphI-HindIII PCR fragment (pSph, pHin) from Tn21 in pSU19; Cmr Spr | This study |
Smr, streptomycin resistance; Strir, streptothricin resistance; Apr, ampicillin resistance; Spr, spectinomycin resistance.
Standard DNA techniques.
Construction of recombinant DNAs, transformations, and restriction enzyme digestions were done according to standard procedures (34). Plasmid DNA preparations were performed with the Wizard Minipreps DNA Purification System (Promega) according to the manufacturer's instructions. Restriction enzymes, T4 DNA polymerase, polynucleotide kinase, and exonuclease III were from Boehringer Mannheim. T4 DNA ligase was from New England Biolabs. 5′-Methyl-dCTP and oligonucleotides (see Table 2) were purchased from Pharmacia LKB Biotechnology. For DNA sequencing, the dideoxyribonucleotide chain-terminating method developed by Sanger et al. (35) was used with synthetic primers listed in Table 2. The radioactively labeled compound [α-35S]dATP was from Amersham, and T7 DNA polymerase Sequenase version 2.0 was from U.S. Biochemical Corp., Cleveland, Ohio. Electrophoresis gels for sequencing contained 6% acrylamide-N,N′-methylenebisacrylamide (30:1) and 40% urea.
TABLE 2.
Oligonucleotides
| Oligo- nucleotide | Sequence | Positionsd |
|---|---|---|
| cdo2 | TATCTGCAGTTTGGAGAATGGCAGCGC | 465-488a |
| cup2 | TATCTGCAGTTOGCGCTTAGCTGG | 469-450a |
| cdo11 | TATCAAGCTTTACATCTGACAATGAG | 787-808b |
| cup11 | TATAAGCTTGAACGTGTTACGACCGC | 792-770b |
| Tn7int∗179E | CCCAGCAATAAAAGAGCTCATTGAGCAAGC | 1393-1364 |
| Tn7SRS-107 | TATCTAGAAACAGAGTGTCTTG | 2118-2131 |
| Tn7CRS+185 | TTAAAGCTTATGGAGTTGTCGTAGTTGC | 4424-4402 |
| Tn7 Fling | ATAAAGCTTACGACAATAGTCCATC | 4426-4444 |
| Tn7-int32 | ATTGGATCCTGATTGATAAGTAG | 940-954 |
| Tn7-int36 | AGTGACACGCTTGCTAAC | 1123-1140 |
| Tn7-int37 | AGTTGTCGTCTTGCTGAATAAG | 1338-1359 |
| Tn7-int38 | TAACTTGGTTGCGAGTATCC | 1542-1561 |
| Tn7-int53 | ATCATCTGCATGACTCCGTTG | 1202-1182 |
| Tn7-int55 | TATGCATGCTAGAATAGGCTGTATAG | 1962-1944 |
| Tn7left1 | TAGAATTCGTAGCGTCGTAAGCTAATAC | 95-115 |
| Tn7left2 | TTGGATCCTGCGGATGGACTGATG | 973-955 |
| Tn7left3 | TCCTATATCCAACTGATCAT | 204-223 |
| Tn7left4 | ATAAACAATCATAGAGGG | 875-858 |
| Tn7left5 | ACAGTAACAAGCATTACGAGG | 415-435 |
| Tn7left6 | TTGTTACTCCAAGAGCTTG | 585-603 |
| Tn7left7 | AATGGTCACTGTGATTGACAG | 787-807 |
| Tn7left8 | TCTGAAGCACATTTCTTATAC | 629-651 |
| Tn7left9 | ATGTTAATGGGCTATTACAATG | 391-369 |
| Tn7right1 | AAAGAAGATCAGACTCGTAG | 5278-5259 |
| Universal 17-nte | GTAAAACGACGGCCAGT | |
| Reverse 16-nt | AACAGCTATGACCATG | |
| sat1up2 | ATGGAACGATCTAGCCTCTAT | 249-269c |
| sat1do2 | TGTGCGCGACTCCTTTGCCTC | 319-299c |
| pHin | GGCAAGCTTAGTAAAGCCCTCGCTAG | 2055-2030a |
| pSph | TGATCCGCATGCCCGTTCCATACAG | 709-733a |
Published sequence (37).
Published sequence (39).
Published sequence (38).
Primer positions are as in Fig. 2A except as noted.
nt, nucleotide.
DNA sequencing strategies.
The integrase genes from Tn7, Tn1825, and Tn1826 were amplified with PCR primers Tn7-int32 and Tn7-int55 (Table 2) by using E. coli DH5α R483 (Tn7), IE972 (Tn1825), and IE970 (Tn1826) as templates. At least three independent amplification products of each integrase gene were sequenced on both strands by cloning in both orientations in M13mp18 and M13mp19. The DNA sequence was determined by using seven different oligonucleotides: universal 17-nucleotide primer specific for M13mp18 and M13mp19, Tn7-int32, Tn7-int36, Tn7-int37, Tn7-int38, Tn7-int53, and Tn7-int55 (Table 2). Three separately amplified products of the 860-bp DNA fragment obtained with primers Tn7left1 and Tn7left2 were cloned in both orientations in M13mp18 and M13mp19 and sequenced on both strands by using Tn7left1, Tn7left2, Tn7left3, Tn7left4, Tn7left5, Tn7left6, Tn7left7, Tn7left8, and Tn7left9 as primers (Table 2). The 797-bp EcoRI-HindIII fragment located between the aadA1 and tnsE genes in Tn7 was cloned in M13mp18 and M13mp19. Subclones were obtained by using restriction enzymes Sau3A and XbaI and the nucleotide sequence was determined on both strands by using Tn7right1 and the universal 17-nucleotide primer specific for the M13 vectors.
Mutagenesis of the internal stop codon in the intI2 gene.
Directed point mutations in the internal stop codon replacing amino acid 179 in intI2 of Tn7 were made according to the procedure described by Vandeyar et al. (44). A 1.6-kb SphI-HpaI fragment containing the Tn7 integrase gene from plasmid R483 was cloned in M13mp18. M13 clones were grown on E. coli strain JM105. Extracted single-stranded phage DNA was used as a template for the mutagenesis reaction, together with phosphorylated mutagenesis primer Tn7int*179E (Table 2). Mutant phage DNA was propagated in E. coli CU9276. Mutants were detected by restriction enzyme cleavage of a SacI site that was created by changing the stop codon (TAA) into a codon for glutamic acid (GAG). The mutants were further confirmed by DNA sequencing. Subcloning of the mutated gene into pUC19 resulted in plasmid 636*179E (T535G, A537G). A clone carrying the corresponding parental gene fragment was called plasmid 636.
PCR assay for detection of site-specific recombination.
Divergent primers specific for the cassettes of Tn7 or Tn21 (Table 2) were used for inverse PCR (28) to detect site-specific recombination between cassette ends. The cassette region from Tn7 was cloned on a 2.9-kb PinA1-EcoRI fragment into the XmaI and EcoRI sites in the polylinker of vector pSU18 and pSU19 to form plasmids 670 and 671. Plasmids 2123 and 2280 contained the SphI-HindIII cassette region from the integron of Tn21 cloned in pSU18 and pSU19, respectively (Table 1). The cassette clones, together with an integrase overproducing plasmid (636*179E or 2101, Table 1), were introduced into E. coli DH5α. Template DNA was prepared by boiling one loop of bacteria in 200 μl of water. Four sets of divergent PCR primers specific for the cassettes in Tn7 were used (cdo11+cup11, sat1up2+sat1do2, cdo2+cup2, and Tn7Fling+Tn7CRS+185, Table 2). PCR products arise when recombination correctly juxtaposes the two primers, either by circularization of an excised cassette or by tandem duplication of a cassette. The PCRs were run for 30 cycles at 94, 60, and 72°C by using a DNA thermal cycler 480 (Perkin-Elmer). Oligonucleotides specific for sequences flanking the mobile cassettes in the integrons of Tn7 and Tn21 (Table 2) were used for convergent PCR to detect cassette deletions. The PCR products obtained were cloned into the polylinker of pUC19, and the crossover points were confirmed by nucleotide sequencing.
Phenotypic test to detect cassette deletions.
Overnight cultures of DH5α harboring the clone overproducing integrase (plasmid 636*179E or plasmid 2101) and the plasmid to be tested were grown in IsoSensitest broth supplemented with ampicillin and under selection for the plasmid-borne cassettes and then diluted 1,000-fold in IsoSensitest broth without selection. DNA was prepared after cultivation at 37°C overnight, and 100 ng was used for the transformation of plasmid-free DH5α. Loss of resistance to antibiotics due to cassette deletions was checked among 600 colonies grown on chloramphenicol plates. Analogous experiments performed in the presence of plasmid 636wt (with the internal stop codon in the intI2 gene) did not result in any loss of antibiotic resistance cassettes.
Mating-out assay for site-specific recombination.
Donor E. coli cells were constructed by introducing R388 (trimethoprim resistant [Tpr], sulfonamide resistant [Sur]) or R388Δ1 (Sur) into HB101 by conjugation, followed by transformation of the nonconjugative pSU18-clone (chloramphenicol resistant [Cmr], Table 1) to be analyzed and the clone producing integrase (plasmid 636*179E or plasmid 2101). Exponentially growing cultures of donor cells (0.5 ml) were mixed with 0.5 ml of the recipient strain (DH5α, nalidixic acid resistant [Nalr]) in stationary phase. The mix was pelleted, and the cells were resuspended in 100 μl of Luria broth, spread on a Millipore 25-mm (pore-size) filter on a fresh IsoSensitest agar plate, and incubated at 37°C for 2 h. Filters were washed in 1 ml of 0.9% NaCl solution, and dilutions were plated on agar plates supplemented with nalidixic acid and either sulfathiazole or chloramphenicol to determine the recombination frequency. The recombination frequencies (incidence of cointegrate formation between either R388 or R388Δ1 and pSU18 derivatives) are given as the number of Cmr transconjugants per Sur transconjugant. Plasmid fusions in Nalr and Cmr transconjugants were analyzed by PCR by using either a cassette-specific primer (cup2 for the aadA1 cassette, sat1-down2 for the sat cassette, and cup11 for the dfrA1 cassette) and a primer hybridizing to the conserved sequence in R388 or R388Δ1 (pSph). To analyze the reciprocal crossover product in the cointegrates, a primer specific for qacEΔ1 (pHin), together with a vector-specific primer, was used. The PCR products were analyzed by nucleotide sequencing to confirm crossover points.
Nucleotide sequence accession number.
The nucleotide sequences reported in this study have been assigned accession numbers AJ001816 and AJ002782 in the EMBL, GenBank, and DDBJ databases. The former sequence is a revision of the previously released sequence L10818.
RESULTS
Sequence characterization of the left end of Tn7.
In this work we have sequenced the complete intI2 gene, and all areas of Tn7 beyond those that have previously been analyzed, i.e., dfrA1 (15, 36), sat (42), aadA1 (14), and tnsA-tnsE (15a). The new data cover regions on either side of the cassettes, as indicated by horizontal lines in Fig. 1. The proteins coded by the new gene intI2 (325 codon triplets) and by intI1 (337 codon triplets) of Tn21 showed 46% amino acid identity, which indicates a closer relation than to other tyrosine recombinases and a possible role of IntI2* in incorporation of cassettes into Tn7. However, translation of intI2 is interrupted by an internal ochre codon (TAA) at amino acid position 179 (Fig. 2A and B), which must be suppressed or replaced in order to produce the full-length product. Unless read-through of TAA is allowed, a shorter and probably inactive polypeptide of 178 amino acids would be produced. The intI2 genes of both transposons Tn1825 and Tn1826 (Fig. 1) (42), which carry cassette arrangements different from those of Tn7, were sequenced (underlined areas in Fig. 1; positions 940 to 1970 in Fig. 2A). The sequences of the complete intI2 genes were identical, and all have a stop codon interrupting translation at position 179.
FIG. 2.
(A) The nucleotide sequence of the left end of transposon Tn7 containing intI2 and the new ORFX (translated) cassette. L1 to L3 indicate the multiple repeats clustered at the end of the transposon (2). Stop codons in the intI2 gene are in boldface and are marked with three dots below the sequence. The start codon of the gene is shown with an arrow. The G in the GTT sequences at the ORFX cassette borders are underlined by double dashed lines. Underlined sequences mark the core and inverse core in the ORFX cassette. The peculiar repetitions in ORFX are marked with horizontal lines above the sequence and are indexed by r1 to r7. The sequence at positions 4782 to 4856 is identical to a sequence in Arabidopsis (M55553). (B) Comparison of the amino acid sequences of the translated intI1, intI2, intI3, and intI4 (also called VchintIA). The well-conserved domains 1 and 2, with the four amino acids marked that are highly conserved among all members of the tyrosine integrase family, are underlined. (C) Nucleotide sequences and translated amino acid sequences of the 6.5 repetitions in ORFX. The sequence from positions 940 to 1970 was found to be identical in Tn1825 and Tn1826.
The amino acid identity between IntI1 and IntI3 from Serratia marcescens isolate AK9373 (1) is higher (59%) than between IntI2* and either IntI1 or IntI3 (46% in both instances) or IntI4 (48%) (Fig. 2B). The IntI1 (9, 26), IntI3 (F. Gagnon and P. H. Roy, unpublished results; M. Gullberg, K. Hansson, and L. Sundström, unpublished results), and IntI4 (33) proteins have been shown to be functional. We provide evidence below that the IntI2* protein is functional after mutagenesis of the internal stop codon to a glutamic acid codon (IntI2*179E). The sequence between the intI2 gene in Tn7 and the divergently transcribed cassettes showed no obvious resemblance to the corresponding regions in Tn21 or in the class 3 integron in S. marcescens.
The 936-bp sequence (position 1 to 936 in Fig. 2A) downstream of the integrase gene intI2 revealed no open reading frame of significant length. The beginning of this sequence contained the cluster of three transposase binding sites—L1, L2, and L3 (23)—with a defined role in transposase binding (2). It is an interesting coincidence that the 3′ end of the integrase gene intI1 in Tn5090 (30) is located even closer to similar binding sites that appear to be involved in transposon mobility.
Sequence of the region between aadA1 and tnsE.
The third cassette in Tn7 contains the aadA1 gene encoding an adenylyltransferase, which inactivates streptomycin and spectinomycin (14) (Fig. 1). The partially sequenced region downstream of aadA1 was checked and extended to fill in a previously unsequenced region. The beginning of the palindromic attC site (59-base element), occurring at the 3′ end of each cassette, generally is an inverse complement of the first nucleotides in the 5′ end of the same cassette (20). This complementarity between bases 4240 to 4248 and bases 4761 to 4769 (Fig. 2A) indicated the possibility of a fourth cassette downstream of aadA1 in Tn7. The cassette contains an open reading frame of 165 amino acids (named ORFX; Fig. 1 and 2A). ORFX was not followed by an attC site of the usual palindromic structure, such as those at the downstream ends of the dfrA1, sat, and aadA1 genes. Functional studies (see below) defined the terminal crossover points of the cassette. The function of the 165-amino-acid polypeptide remains unknown, but a series of 6.5 peculiar repetitions of a sequence coding for seven amino acids was observed (Fig. 2C). The amino acid sequence is almost completely conserved among the repeats, whereas the nucleotide sequence varied somewhat, hinting that ORFX might have a biological function.
The nucleotide sequence between ORFX and the HindIII site at position 5716 contained no open reading frame of known function. An 85-nucleotide region located just 3′ of the newly identified cassette showed full identity to a sequence downstream of a gene described in Arabidopsis thaliana (GenBank/EMBL file no. M55553 [24]), but this may be due to a cloning artifact in the latter.
Excision of a fourth cassette, ORFX, in Tn7.
The inverse PCR assay (see Materials and Methods and Fig. 3A) allowed detection of recombination products corresponding to excision of a unit containing the 165-amino-acid ORFX. The bands indicative of recombination were observed in the presence of overproduced IntI1 or IntI2*179E (see below). The crossover points were determined to be at positions 4239 to 4243 and positions 4776 to 4780 (Fig. 2A) by nucleotide sequencing of PCR products (data not shown). Although sequence identities at the ends of the cassette did not permit exact determination of the crossover point, it most likely is between G and TT in the recombination sites as shown previously (20, 32). The 3′ end of the cassette lacks a complete attC sequence but shows identity to 13 bases, CTAATAAAATGTT (nucleotides 2215 to 2227 in Fig. 1), of attI2 in Tn7.
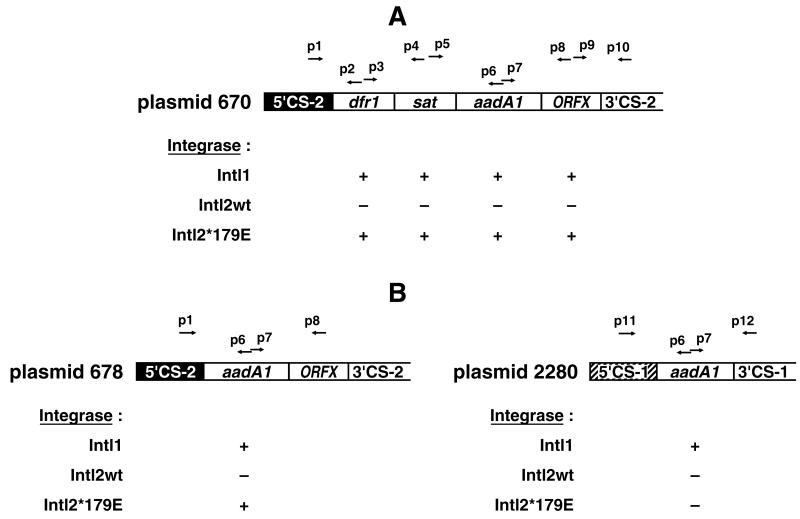
FIG. 3.
Site-specific recombination between cassette ends promoted by IntI1 or IntI2*179E from cloned integron fragments of class 1 and class 2 integrons. Recombination was detected by divergent and convergent PCR as described in Materials and Methods. The ability of the different integrases to promote excision of the tested cassettes is denoted by plus and minus signs. Identical results were obtained with divergent and convergent PCR assays. (A) Plasmid 670 carrying the cassette region from Tn7 (class 2 attI site). (B) Plasmid 678 with the cassette region from Tn7 deleted for the dfrA1 and sat cassettes and plasmid 2280 carrying the cassette region from Tn21 (Class 1 attI site). White boxes indicate cassettes, and arrows indicate the positions of the PCR primers used in this study. Black boxes indicate the conserved sequence (5′-CS-2) from the class 2 integron of Tn7, while the conserved sequence from the class 1 integron in Tn21 is hatched (5′-CS-1). The primers used are denoted as follows: p1, Tn7SRS-107; p2, cup11; p3, cdo11; p4, sat1do2; p5, sat1up2; p6, cup2; p7, cdo2; p8, Tn7CRS+185; p9, Tn7Fling; p10, reverse; p11, pSph; and p12, pHin.
Substitution of the internal stop codon in the Tn7 integrase gene with a sense triplet.
To test whether the IntI2* protein of Tn7 could be activated, we changed the internal ochre codon (TAA) into a triplet (GAG) coding for glutamic acid, the corresponding amino acid found in IntI1. After site-directed mutagenesis, we cloned the 1.6-kb SphI-HpaI fragment carrying the intI2*179E gene into pUC19 to form plasmid 636*179E. The ability of this clone to promote recombination between cassette sites was tested by using the inverse PCR assay (primers designed to give a product only if recombination has occurred) and the convergent PCR assay (primers arranged to give a smaller product if recombination has occurred) as described in Materials and Methods. As a recombination substrate, we used plasmid 670 containing the cassette region from the Tn7 integron (Fig. 3A). The inverse primers specific for the dfrA1, sat, aadA1, and ORFX cassettes revealed recombination products in the presence of plasmid 636*179E, from which the repaired Tn7 integrase (IntI2*179E) was overexpressed (Fig. 3A). These results were confirmed by PCR with convergent primers binding on either side of a given cassette (Fig. 3A). Sequencing of the PCR products revealed that DNA strand crossover promoted by IntI2*179E occurred between the GTT sequences at the ends of the cassettes (data not shown). Plasmid 636 encoding IntI2* failed to promote cassette recombination in any of the assays used here.
IntI1 and IntI2*179E show different substrate specificities.
We tested the ability of IntI1 and IntI2*179E to mediate crossovers at attC or attI sites in Tn7 and Tn21. All cassettes except the first cassette in an integron have attC sites at both ends, which should be recognized by all integron integrases. Both IntI1 and IntI2*179E mediated recombination between the attC sites at the ends of the aadA1 cassette in Tn7 (plasmid 670; Fig. 3A) (45). Qualitative detection of recombination was done by using the convergent and inverse PCR assays (Fig. 3A).
In contrast to the attC sites, the attI site includes the end of the class-specific 5′-CS and the beginning of the first cassette in an integron. Apart from an abundance of A, these conserved sequences are unique for each class (20) and recombination involving these attI sites could be class specific. Plasmid 2280 contains the aadA1 cassette adjacent to the 5′-CS-1 (attI1 of Tn21 [38]), whereas in plasmid 678 aadA1 has been inserted adjacent to the 5′-CS-2 (attI2 of Tn7) (Fig. 3B). IntI2*179E mediated recombination, as assayed by PCR with both divergent and convergent primers, between the attI2×attC sites of aadA1, excising this cassette when it was adjacent to 5′-CS-2 in plasmid 678 (Fig. 3B) but not when it was adjacent to 5′-CS-1 (attI1×attC; plasmid 2280). IntI1, however, mediated both attI1×attC and attI2×attC recombination, excising the aadA1 cassette when adjacent to either attI1 or attI2, and therefore seems to have a wider site specificity than IntI2*179E.
Which cassette is incorporated adjacent to the 5′-CS also affects the attI site (20). The attI2 sites including the first six bases of the dfrA1 or sat cassettes (plasmids 670 [Fig. 3A] and 699) were also shown by PCR detection to recombine in the presence of either IntI1 or IntI2*179E.
We conclude that IntI2*179E has a narrower specificity for attI sites compared to IntI1, which can mediate recombination involving either attI1 or attI2. The result was the same regardless of which cassette was inserted at attI, and the specificity therefore depends mainly on the conserved sequence 5′ of the crossover point.
Quantitation of excision frequency as measured by phenotypic tests.
The loss of antibiotic resistance markers borne on cassettes gives quantitative information of how efficient recombination is between cassette end sites. Cassette excision studied by the phenotypic tests for trimethoprim, streptothricin (39), and spectinomycin with plasmid 670 as substrate showed that the dfrA1, sat, and aadA1 cassettes were lost at frequencies of 1.1, 13, and 10%, respectively, in the presence of IntI2*179E (Table 3). Deletion rates were also measured for the aadA1 cassette located in the first position in class 1 and class 2 integrons (plasmids 2280 and 678 in Table 3). In the presence of IntI2*179E, the aadA1 cassette was lost at 0.33% when adjacent to 5′-CS-2, whereas no loss was detected when the same cassette was adjacent to 5′-CS-1. IntI1-mediated excision of the aadA1 cassette from both plasmids 2280 and 678 took place, but excision was much more efficient when the cassette was adjacent to 5′-CS-1 (59% from 2280) than when the cassette was adjacent to 5′-CS-2 (0.7% from 678). This demonstrates that, while IntI1 possesses some degree of specificity for its own attI site, IntI2*179E discriminates against a class 1 attI site. IntI1 also seems to be a more efficient enzyme than IntI2*179E since it can delete the aadA1 cassette when the latter is adjacent to 5′-CS-2 more efficiently than does IntI2*179E (Table 3). However, IntI2* may have undergone genetic drift, or glutamate may not be the optimal amino acid at position 179.
TABLE 3.
Deletion frequencies of cassettes borne on class 1 and 2 integrons promoted by class 1 and 2 integrases
| Plasmida | attI class | Integrase | Cassette deletion frequencies (%)b
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| dfr1 | sat | aadA1 | dfr1 aadA1 | sat dfr1 | aadA1 sat | aadA1 sat dfr1 | |||
| 670 | 2 | IntI2* | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | IntI2*179E | 1.1 | 13 | 10 | 0.17 | 1.5 | 0.8 | 0 | |
| 2 | IntI1 | 1.5 | 5.2 | 73 | 0.33 | 0.50 | 2.0 | 0.17 | |
| 671 | 2 | IntI2* | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | IntI2*179E | 0 | 0.67 | 0.67 | 0 | 0 | 0 | 0 | |
| 2 | IntI1 | 0 | 0 | 18 | 0 | 0 | 2.5 | 0 | |
| 2280 | 1 | IntI2* | -c | - | 0 | - | - | - | - |
| 1 | IntI2*179E | - | - | 0 | - | - | - | - | |
| 1 | IntI1 | - | - | 59 | - | - | - | - | |
| 678 | 2 | IntI2* | - | - | 0 | - | - | - | - |
| 2 | IntI2*179E | - | - | 0.33 | - | - | - | - | |
| 2 | IntI1 | - | - | 0.7 | - | - | - | - | |
See Fig. 3 for maps of cloned integron fragments in plasmids 670, 671, 2280, and 678.
Phenotypic tests were used as described in Materials and Methods to detect cassette excisions. DNA fragments were cloned in pSU18 or pSU19 to obtain substrates in either direction in respect to the lac promoter (in plasmid 671 the lac promoter is directed from the 5′ end of the cassettes, and in plasmids 670, 2280, and 678 the promoter is directed from the 3′ end of the cassettes). Excision rates are given as the percentage of sensitive colonies among 600 to relevant antibiotics.
-, Not done.
Cassette deletion is dependent on position and direction of transcription through the Tn7 integron.
Phenotypic tests were used to determine the frequencies of integrase-mediated excision of cassettes from the Tn7 integron cloned in plasmids 670 and 671 to yield both orientations of the cassette arrays in the polylinker sites of pSU18 (Table 3). In plasmid 671, the cassettes were transcribed from the integron promoter in 5′-CS-2 (positions 1930 to 1959, Fig. 2A), as well as from the lac promoter in the vector. In plasmid 670, the lac promoter was reversed compared to the cassettes such that transcription from the integron promoter alone ensured expression of the phenotype (resistance). In the presence of IntI1 integrase the dfrA1, sat, and aadA1 cassettes were lost at frequencies of 1.5, 5.2, and 73%, respectively. Placing the lac promoter in tandem with the cassette promoter (plasmid 671) significantly reduced the rate of cassette excision for all three cassettes tested (Table 3). Among 600 colonies checked the aadA1 cassette was lost from plasmid 671 at 18%, whereas no excision of either sat or dfrA1 cassettes was observed (Table 3).
The effect of the placing the lac promoter in tandem with the cassette promoter was similar with IntI2*179E. The excision rates for all cassettes were much lower with plasmid 671 than with plasmid 670 (Table 3). Our results thus suggest that the orientation of a cassette relative to the vector promoter influences the rate of excision. The effect of the vector promoter is noteworthy; antisense transcription or reduced forward transcription enhances loss of cassettes.
Site-specific recombination between integrons of classes 1 and 2.
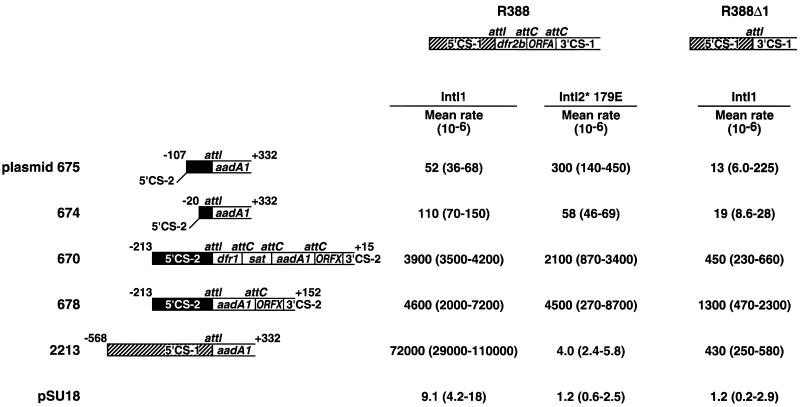
Plasmid clones of the Tn7 integron containing all cassettes (plasmid 670) or two of the cassettes (aadA1 and ORFX; plasmid 678) efficiently formed cointegrates with the conjugative plasmid, R388 (harboring a class 1 integron with two complete cassettes, dfrB2 and ORFA) in the presence of either IntI1 or IntI2*179E (Fig. 4). This mating-out assay for cointegration was first described by Martinez and de la Cruz (25; see also Materials and Methods). The most efficient recombination site of R388 is an attC downstream of ORFA (26), and most cointegrates result from crossovers at that site. Plasmid clones harboring an attI2 site (derivatives 674 and 675 in Fig. 4) were transferred with R388 at frequencies in the range of 100 per million matings. The rates were similar in the presence of IntI1 and IntI2*179E. For crossing over between attI1 (plasmid 2213) and R388, however, the IntI1 recombinase gave high rates (72,000 per million matings) of recombination, while IntI2*179E gave only background rates, thus confirming a specificity of the IntI2*179E protein for its own attI2 site.
FIG. 4.
Structures of cloned integron fragments and recombination rates detected with the mating-out assay as described in Materials and Methods. Integron fragments from either Tn21 or Tn7 were tested for recombination with wild-type R388 and R388Δ1 in the presence of IntI1 or IntI2*179E. Conserved sequences from class 1 integrons are hatched (5′-CS-1) and from class 2 are in black boxes (5′-CS-2). Open white boxes indicate cassettes. The lengths of 5′ and 3′ flanks from the closest cassette border are given above the cloned integron fragments. Recombination frequencies are given as mean transfer frequency of million transferred R388 or R388Δ1, with ranges given within brackets. All experiments were repeated at least four times. Only background recombination rates were detected between the tested pSU18 clones and R388Δ1 with IntI2*179E.
Site-specific recombination has been demonstrated between two attI1 sites in the absence of the palindromic attC sites (20). To exclude the efficient attC sites from the recombination system a modified R388 (R388Δ1 [20]) harboring only an attI1 site was used. The frequencies of crossover between the attI1 in R388Δ1 and an attI2 (plasmids 674 and 675 in Fig. 4) were measured. In the presence of IntI1, the recombination rates for both plasmids were 13 to 19 × 10−6, whereas IntI2*179E gave insignificant recombination. In comparison, the recombination between two attI1 sites was previously shown to occur at 430 × 10−6 in the presence of IntI1 (20). These results again indicate a specificity of the IntI2*179E protein for its own attI2 site.
The crossover sequences in recombination between an attI1 and an attI2 were localized to within the core triplets GTT by nucleotide sequencing across crossover points in cointegrates formed by site-specific recombination. These results agree with previous data for the products of recombination between two attI sites of class 1 (20).
DISCUSSION
Tn7 is a remarkably widespread transposon which commonly accounts for trimethoprim resistance (Tpr) in gram-negative bacteria. Tn7 uses a nonreplicative transfer due to the production of a 5′ cleavage mechanism involving TnsA and shows a remarkable hierarchy of target specificities controlled by two proteins, TnsD and TnsE (12). In addition to its complex transposition mechanism, Tn7 has been postulated to carry an integron. Tn7 is known to carry two cassettes, dfrA1 and aadA1, identical to those in integrons as well as a third, sat (39). We have now completed the nucleotide sequence of Tn7. This analysis disclosed the presence of a fourth cassette downstream of those previously described, which all encode resistance to antibiotics used in human or veterinary medicine. The new cassette described here could not be assigned a resistance phenotype, but a biological function seems possible due to the conservation of internal 7-amino-acid repeats in the gene product. A particularly notable feature of the integrase gene in Tn7, intI2, is an internal stop codon at amino acid position 179. There are at least two possible explanations for the existence of an internal stop codon in the gene. Either the integrase gene is a pseudogene which is no longer active or the stop codon may permit a certain level of activity in ochre suppressor strains. Also, it is known that integrases often bind DNA through a helix-turn-helix DNA recognition motif in the N-terminal moiety of the protein. This makes it possible that a short, truncated form of the protein could bind to the same DNA sites as the complete integrase and have a regulatory function. Either the short form of the gene product could control the access to DNA-binding sites or it could autoregulate a promoter. The conservation of the stop codon throughout members of the Tn7 family carrying different cassettes in their integrons, found by the sequencing of the integrase genes of Tn1825 and Tn1826 described here, makes it possible that the short form of IntI2* could be an inhibitor polypeptide controlling the action of integrase.
The ability of Tn7 to use both site-specific and nonspecific modes of transposition (22, 46) could explain why it has become so widespread and persistent in bacterial populations. The integron of Tn7 has acquired a Tpr cassette which has probably contributed to its success. In contrast, there is a limited variety of cassette arrangements in the class 2 integrons, and this may be directly attributable to the internal stop codon of intI2. Again, the conservation of the internal stop codon makes it possible that the trimethoprim cassette in Tn7 and the open reading frame cassette in Tn1825 may have been inserted by trans action of IntI1 in cells where both classes of integron were present.
Mutagenesis of the stop codon to a glutamate codon restores activity to IntI2*. Both IntI1 and IntI2*179E efficiently excise cassettes (other than cassettes in first position) by attC×attC recombination. Excision of cassettes in the first position, however, occurs by attI×attC recombination, and the attI sites differ between class 1 and class 2 integrons. IntI1 and IntI2*179E differ in their ability to recognize these sites; while IntI1 recognizes both attI1 and attI2 (and thus can excise a cassette in first position in either class of integron), IntI2*179E recognizes only its own attI site and is unable to excise cassettes in first position in class 1 integrons. Cointegrate formation by attI×attC recombination confirms this specificity. Also, the low-level attI×attI recombination between class 1 and 2 forms of attI can be carried out by IntI1 but not by IntI2*179E. We cannot explain the observed effects of transcription on the frequencies of excision. Reduced excision frequency was observed in the presence of strong unidirectional transcription; it may be that RNA polymerase interferes with binding of the integrase.
In spite of the efficiency of Tn7 as a transposon, class 1 integrons have shown a far greater degree of cassette diversity than the Tn7-based class 2 integrons. Although the internal stop codon is an obvious explanation, the greater efficiency and looser specificity of IntI1 may also be important. Most integrons make up part of defective transposons lacking two of the four transposition genes borne on functional Tn5090 (30), which is a competent transposon. This disadvantage can be circumvented by the “piggybacking” of these defective transposons on an efficient transposon, as in the Tn21 family of transposons. Finally, the evolutionary success of an integron is no doubt determined by two other important factors: the resistance cassettes it carries and the host range of the plasmid on which it occurs.
Acknowledgments
This work was supported by Swedish Medical Research Council grants B96-16X-10849-03, B96-16X-00172-32C, and X-12638 to L.S. and Medical Research Council of Canada grant MT-13564 to P.H.R.
REFERENCES
- 1.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcisczewska, L. K., R. L. McKown, and N. L. Craig. 1991. Purification of TnsB, a transposition protein that binds to the ends of Tn7. J. Biol. Chem. 266:21736-21744. [PubMed] [Google Scholar]
- 3.Argos, P., A. Landy, K. Abremski, J. B. Egan, E. Haggård-Ljungquist, R. H. Hoess, M. L. Kahn, B. Kalionis, S. V. L. Narayana, L. S. Pierson, N. Sternberg, and J. M. Leong. 1986. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 5:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barth, P. T., N. Datta, R. W. Hedges, and N. J. Grinter. 1976. Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. J. Bacteriol. 125:800-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartolomé, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 6.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 8.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collis, C. M., and R. M. Hall. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collis, C. M., and R. M. Hall. 1992. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol. Microbiol. 6:2875-2885. [DOI] [PubMed] [Google Scholar]
- 11.Collis, C. M., C. Grammaticopoulos, J. Briton, H. W. Stokes, and R. M. Hall. 1993. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 9:41-52. [DOI] [PubMed] [Google Scholar]
- 12.Craig, N. L. 1997. Target site selection in transposition. Annu. Rev. Biochem. 66:437-474. [DOI] [PubMed] [Google Scholar]
- 13.Esposito, D., and J. J. Scocca. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 25:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fling, M. E., J. Kopf, and C. Richards. 1985. Nucleotide sequence of the transposon Tn7 gene encoding an aminoglycoside-modifying enzyme, 3′(9)-O-nucleotidyltransferase. Nucleic Acids Res. 13:7095-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fling, M. E., and C. Richards. 1983. The nucleotide sequence of the trimethoprim-resistant dihydrofolate reductase gene harboured by Tn7. Nucleic Acids Res. 11:5147-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Flores, C., M. I. Qadri, and C. Liechtenstein. 1990. DNA sequence analysis of five genes; tnsA, B, C, D and E, required for Tn7 transposition. Nucleic Acids Res. 18:901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gary, P. A., M. C. Biery, R. J. Bainton, and N. L. Craig. 1996. Multiple DNA processing reactions underlie Tn7 transposition. J. Mol. Biol. 257:301-316. [DOI] [PubMed] [Google Scholar]
- 17.Hall, R. M., D. E. Brookes, and H. W. Stokes. 1991. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 5:1941-1959. [DOI] [PubMed] [Google Scholar]
- 18.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 19.Hall, R. M., and C. Vockler. 1987. The region of the IncN plasmid R46 coding for resistance to β-lactam antibiotics, streptomycin/spectinomycin and sulfonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 15:7491-7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansson, K., O. Sköld, and L. Sundström. 1997. Nonpalindromic attI sites of integrons are capable of site-specific recombination with one another and with secondary targets. Mol. Microbiol. 26:441-453. [DOI] [PubMed] [Google Scholar]
- 21.Heikkilä, E., L. Sundström, M. Skurnik, and P. Huovinen. 1991. Analysis of genetic localization of the type I trimethoprim resistance gene from Escherichia coli isolated in Finland. Antimicrob. Agents Chemother. 35:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lichtenstein, C., and S. Brenner. 1981. Site-specific properties of Tn7 transposition into the E. coli chromosome. Mol. Gen. Genet. 183:380-387. [DOI] [PubMed] [Google Scholar]
- 23.Lichtenstein, C., and S. Brenner. 1982. Unique insertion site of Tn7 in the E. coli chromosome. Nature 297:601-603. [DOI] [PubMed] [Google Scholar]
- 24.Ma, H., M. F. Yanofsky, and E. M. Meyerowitz. 1991. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 5:484-495. [DOI] [PubMed] [Google Scholar]
- 25.Martinez, E., and F. de la Cruz. 1988. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol. Gen. Genet. 211:320-325. [DOI] [PubMed] [Google Scholar]
- 26.Martinez, E., and F. de la Cruz. 1990. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 9:1275-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller-Hill, B., L. Crapo, and W. Gilbert. 1968. Mutants that make more lac repressor. Proc. Natl. Acad. Sci. USA 59:1259-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouellette, M., and P. H. Roy. 1987. Homology of ORFs from Tn2603 and from R46 to site-specific recombinases. Nucleic Acids Res. 15:10055. [DOI] [PMC free article] [PubMed]
- 30.Rådström, P., O. Sköld, G. Swedberg, J. Flensburg, P. H. Roy, and L. Sundström. 1994. Transposon Tn5090 of Plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J. Bacteriol. 176:3257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 32.Recchia, G. D., H. W. Stokes, and R. M. Hall. 1994. Characterization of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res. 22:2071-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowe-Magnus, D. A., A.-M. Guerout, P. Ploncard, B. Dychinco, J. Davies, and D. Mazel. 2001. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl. Acad. Sci. USA 98:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain termination inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simonsen, C. C., E. Y. Chen, and A. D. Levinson. 1983. Identification of the type I trimethoprim-resistant dihydrofolate reductase specified by the Escherichia coli R-plasmid R483: comparison with procaryotic and eucaryotic dihydrofolate reductases. J. Bacteriol. 155:1001-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 38.Sundström, L., P. Rådström, G. Swedberg, and O. Sköld. 1988. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol. Gen. Genet. 213:191-201. [DOI] [PubMed] [Google Scholar]
- 39.Sundström, L., Roy, P. H., and O. Sköld. 1991. Site-specific insertion of three structural gene cassettes in transposon Tn7. J. Bacteriol. 173:3025-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundström, L., and O. Sköld. 1990. The dhfrI trimethoprim resistance gene of Tn7 can be found at specific sites in other genetic surroundings. Antimicrob. Agents Chemother. 34:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swift, G., B. J. MacCarthy, and F. Heffron. 1981. DNA sequence of a plasmid-encoded dihydrofolate reductase. Mol. Gen. Genet. 181:441-447. [DOI] [PubMed] [Google Scholar]
- 42.Tietze, E., J. Brevet, and H. Tschäpe. 1987. Relationships among the streptothricin resistance transposons Tn1825 and Tn1826 and the trimethoprim resistance transposon Tn7. Plasmid 18:246-249. [DOI] [PubMed] [Google Scholar]
- 43.Tietze, E., and J. Brevet. 1990. Nucleotide sequence of the streptothricin-acetyltransferase gene sat-2. Nucleic Acids Res. 18:12783. [DOI] [PMC free article] [PubMed]
- 44.Vandeyar, M. A., M. P. Weiner, C. J. Hutton, and C. A. Batt. 1988. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene 65:129-133. [DOI] [PubMed] [Google Scholar]
- 45.Wiedemann, B., J. F. Meyer, and M. T. Zühlsdorf. 1986. Insertions of resistance genes into Tn21-like transposons. J. Antimicrob. Chemother. Suppl. C 18:85-92. [DOI] [PubMed] [Google Scholar]
- 46.Wolkow, C. A., R. T. DeBoy, and N. L. Craig. 1996. Conjugative plasmids are preferred targets for Tn7. Genes Dev. 10:2145-2157. [DOI] [PubMed] [Google Scholar]
- 47.Woodcock, D. M., D. M. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 49.Young, H.-K., M. J. Qumsieh, and M. L. McIntosh. 1994. Nucleotide sequence and genetic analysis of the type Ib trimethoprim-resistant, Tn4132-encoded dihydrofolate reductase. J. Antimicrob. Chemother. 34:715-725. [DOI] [PubMed] [Google Scholar]