Abstract
The genes from the oxygenase cluster nagAaGHAbAcAd of naphthalene-degrading Ralstonia sp. strain U2 were cloned and overexpressed. Salicylate 5-hydroxylase (S5H) activity, converting salicylate to gentisate, was present in vitro only in the single extract of cells with overexpressed nagAaGHAb or in a mixture of three cell extracts containing, respectively, NagGH (the oxygenase components), NagAa (ferredoxin reductase), and NagAb (ferredoxin). Each of the three extracts required for S5H activity was rate limiting in the presence of excess of the others but, when in excess, did not affect the rate of catalysis. S5H catalyzed the 5-hydroxylation of the aromatic rings of 3- and 4-substituted salicylates. However, the methyl group of 5-methylsalicylate was hydroxylated to produce the 5-hydroxymethyl derivative and the 6-position on the ring of 5-chlorosalicylate was hydroxylated, producing 5-chloro-2,6-dihydroxybenzoate. In an assay for the nag naphthalene dioxygenase (NDO) based on the indole-linked oxidation of NADH, three extracts were essential for activity (NagAcAd, NagAa, and NagAb). NDO and S5H were assayed in the presence of all possible combinations of the nag proteins and the corresponding nah NDO proteins from the “classical” naphthalene degrader P. putida NCIMB9816. All three oxygenase components functioned with mixed combinations of the electron transport proteins from either strain. The S5H from strain U2 is a unique monooxygenase which shares sequence similarity with dioxygenases such as NDO but is also sufficiently similar in structure to interact with the same electron transport chain and probably does so in vivo during naphthalene catabolism in strain U2.
The classical pathway for naphthalene catabolism in bacteria such as Pseudomonas putida strains NCIMB9816 and PpG7 (7, 52, 53) is via dihydroxylation and cleavage of the first ring and removal of the resulting aliphatic side chain to produce salicylate (2-hydroxybenzoate). This is then converted by the action of salicylate 1-hydroxylase to catechol (1,2-dihydroxybenzene), which undergoes extradiol cleavage via the same route used for a wide range of other aromatic compounds such as toluene and the xylenes (16) and phenol (38). The nah genes are located on two separate operons: the upper pathway operon encoding the conversion of naphthalene to salicylate and the lower (or meta) pathway operon encoding the conversion of salicylate to acetyl coenzyme A and pyruvate (5, 52). The induction of the two operons is linked by a common regulator protein, NahR (34).
Although a route for naphthalene catabolism involving the alternative conversion of salicylate to gentisate (2,5-dihydroxybenzoate) has been known for some time (14, 25, 30, 39, 41, 53), genetic analysis of such a pathway, in Ralstonia (formerly Pseudomonas) sp. strain U2, has only recently been reported (12, 54). There is similarity between the nag genes of strain U2 and the classical nah genes, but only in the conversion of naphthalene to salicylate, for which the genes are homologous and in the same order (nagAaAbAcAdBFCQED). There are, however, major differences. (i) Between nagAa and nagAb there are two genes (nagGH). The product of the larger one, NagG, has ∼18% sequence identity with the ISP α subunit of various naphthalene dioxygenases (NDOs) sharing both a Rieske (2Fe-2S) motif (CXHRGX8GNAKXFXCXYH), with conserved iron-binding His and Cys residues (shown in bold type), and a mononuclear Fe2+ motif (EX3DXYHXGX2H) with conserved iron-binding Asp, Glu, and Cys (shown in bold type) (18): it was proposed that nagGH encoded salicylate 5-hydroxylase (S5H), converting salicylate to gentisate (12). (ii) The genes for the complete pathway from naphthalene to central metabolites (in this case pyruvate and fumarate) appear to be on a single large operon spanning ca. 18 kbp, in which the genes for the further conversion of gentisate (nagJIKLMN) are directly downstream of the genes for conversion of naphthalene to salicylate.
In this study we report the cloning and expression in vitro of nagGH, together with the other components of the oxygenase gene cluster (nagAaGHAbAcAd). We have developed an assay procedure for S5H and, by using a modified in vitro assay for the NDO, have shown that both enzymes share the chain for the transport of electrons from NAD(P)H to substrate of ferredoxin reductase (NagAa) and ferredoxin (NagAb).
MATERIALS AND METHODS
Bacterial plasmids and strains.
The bacterial plasmids used and constructed in this study are listed in Table 1. Escherichia coli DH5α [φ80d lacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169] (Life Technologies) was used routinely as a host in cloning experiments. E. coli BL21(DE3)/pLysS [F− ompT hsdSB (rB− mB−) dcm gal λ(DE3) pLysS (Cmr)] (43) was purchased from Promega and used as a host for the overexpression of genes cloned in expression plasmid pET5a.
TABLE 1.
Plasmids used and constructed in this study
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| pUC18 | Vector: Apr; multiple cloning site in lacZα | 51 |
| pET5a | Expression vector: Apr; NdeI site overlapping the initiating start codon downstream of T7 promoter and multicloning site. | 43 |
| pDTG121 | A pT7-5 derivative overexpressing nahAcAd from strain NCIMB9816 | 45 |
| pDTG191 | A pT7-5 derivative overexpressing nahAa from strain NCIMB9816 | F. Lee and D. T. Gibson, unpublished |
| pWWF6 | 8.5-kb BamHI fragment from strain U2 inserted into pUC18 | 12 |
| pWWF19-25 | 1,286-bp NdeI, EcoRI-cut PCR fragment containing nagI inserted into pET5a | 54 |
| pWWF44 | EcoRI-cutPCR fragment containing nagAa inserted into pUC18 | This study |
| pWWF45 | EcoRI-cutPCR fragment containing nagAaGHAb inserted into pUC18 | This study |
| pWWF46 | EcoRI-cutPCR fragment containing nagAb inserted into pUC18 | This study |
| pWWF47 | EcoRI-cutPCR fragment containing nagGH inserted into pUC18 | This study |
| pWWF48 | EcoRI-cutPCR fragment containing nagG inserted into pUC18 | This study |
| pWWF49 | EcoRI-cutPCR fragment containing nagH inserted into pUC18 | This study |
| pWWF53 | 739-bp NdeI-EcoRI-cut PCR fragment containing nagL inserted into pET5a | 54 |
| pWWF54 | 1,407-bp NdeI-EcoRI-cut PCR fragment containing nagAa inserted into pET5a | This study |
| pWWF55 | 3,143-bp NdeI-EcoRI-cut PCR fragment containing nagAaGHAb inserted into pET5a | This study |
| pWWF56 | 341-bp NdeI-EcoRI-cut PCR fragment containing nagAb inserted into pET5a | This study |
| pWWF57 | 1,881-bp NdeI-EcoRI-cut PCR fragment containing nagGH inserted into pET5a | This study |
| pWWF58 | 1,346-bp NdeI-EcoRI-cut PCR fragment containing nagG inserted into pET5a | This study |
| pWWF59 | 528-bp NdeI-EcoRI-cut PCR fragment containing nagH inserted into pET5a | This study |
| pWWF96 | 0.8-kb MspA1 I fragment containing nagK from pWWF86 subcloned into pUC18 | 54 |
| pWWF111 | PCR fragment containing nagAcAd inserted into pUC18 | This study |
| pWWF112 | 2,153-bp NdeI-EcoRI-cut PCR fragment containing nagAcAd inserted into pET5a | This study |
Apr, ampicillin resistant.
Media and bacterial culture.
Liquid Luria-Bertani (LB) medium (24) containing the appropriate antibiotic was used for the cultivation of E. coli strains. Sensitivity test agar (LabM, Bury, United Kingdom) or LB agar was used with added ampicillin for the selection of strains carrying plasmids derived from pUC18 or pET5a. Minimal medium was prepared as described previously (49). LB medium and minimal medium plates contained 1.5% agar (LabM). For the culture of strain U2, 5 mM succinate, 2.5 mM salicylate, or 0.5% (wt/vol) powdered naphthalene was used, either added to liquid medium or, for naphthalene only, spread on the lids of inverted petri dishes. Ampicillin was used at 100 μg/ml where appropriate.
Plasmid extraction and DNA manipulation.
Restriction endonuclease digestions and ligations with T4 ligase were done in accordance with the manufacturer's instructions. E. coli DH5α was transformed by standard procedures (35). Plasmid DNA was purified by using a Concert rapid plasmid mini system (Life Technologies).
Expression of nagAa, nagAaGHAb, nagAb, nagGH, nagG, nagH, and nagAcAd.
Pairs of oligonucleotide primers were designed to produce PCR fragments containing the nagAa, nagAaGHAb, nagAb, nagGH, nagG, nagH, and nagAcAd genes. The upstream (forward) primers were designed with (i) a novel NdeI site introduced at the putative ATG start codon of each reading frame and (ii) a novel EcoRI restriction site upstream of the NdeI site. The downstream (reverse) primers either had a novel EcoRI restriction site downstream of the gene or was downstream of a native EcoRI restriction site. Initially, the amplified fragment was cut at the two EcoRI sites and inserted into pUC18. In this way, nagAa, nagAaGHAb, nagAb, nagGH, nagG, nagH, and nagAcAd genes were amplified from pWWF6 by using Pfu polymerase (Promega) to create pWWF44, pWWF45, pWWF46, pWWF47, pWWF48, pWWF49 and pWWF111, respectively (Table 1). The inserts of these clones were sequenced on a single strand to ensure that no mutation had been incorporated during the PCR. Each of these plasmids was then excised with NdeI and EcoRI, and the fragment carrying the gene was ligated into the expression vector pET5a to produce plasmids pWWF54 (nagAa), pWWF55 (nagAaGHAb), pWWF56 (nagAb), pWWF57 (nagGH), pWWF58 (nagG), pWWF59 (nagH), and pWWF112 (nagAcAd) (Fig. 1, Table 1), each of which was transformed into E. coli DH5α. All of the pET5a constructs were subsequently transformed into E.coli BL21(DE3)/pLysS. In the following list of the PCR primers used to make these plasmids, the NdeI site is italicized, the EcoRI site used for cloning into pUC18 is underlined, and the bases that differ from those in the wild-type sequence are in boldface: nagAa (forward), 5′-AAAAAGAATTCATATGGAACTGGTAGTAGAAC-3′; nagAa (reverse), 5′-CCTGGAATTCACCCTTGAGGCT-3′; nagAaGHAb (forward), 5′-AAAAAGAATTCATATGGAACTGGTAGTAGAAC-3′; nagAaGHAb (reverse), 5′-TGGATTGGATGGTTTGGTTG-3′; nagAb (forward), 5′-GAGGGAATTCATATGACTCAGAACTGG-3′; nagAb (reverse), 5′-TGGATTGGATGGTTTGGTTG-3′; nagGH (forward), 5′-CCAAGAATTCCAACATATGAGTGAACCCC-3′; nagGH (reverse), 5′-AATCTCGAATTCGACCTCGTAGAGCG-3′; nagG (forward), 5′-CCAAGAATTCCAACATATGAGTGAACCCC-3′; nagG (reverse), 5′-TTGGCGAATTCGCACACCATGGCGTA-3′; nagH (forward), 5′-TGGGGAATTCCATATGGTCGACTTCAAAAC-3′; nagH (reverse), 5′-TCAAGAGAATTCGCATCAATCCAGTTC-3′; nagAcAd (forward), 5′-GAGGAATTCCATATGATTTATGAAAATTTGGTG-3′; and nagAcAd (reverse), 5′-CTCGTCGACGCCCAGCATGTAATC-3′.
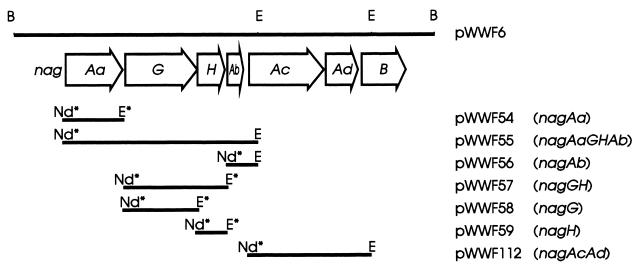
FIG. 1.
Physical map of the upstream region of the nag operon from the large catabolic plasmid of Ralstonia sp. strain U2 and of recombinant plasmids constructed from it. Open reading frames are marked by the open arrows, with the direction of the arrowheads indicating the direction of transcription. The restriction sites are indicated as follows: B, BamHI; E, EcoRI. Nd* represents an NdeI site and E* represents an EcoRI site which were engineered in the PCR primer used to generate the inserts of pET5a constructs. Only restriction sites relevant to the clones and constructs described in this figure are shown.
The Nag proteins encoded on the pET5a recombinants pWWF54 (nagAa), pWWF55 (nagAaGHAb), pWWF56 (nagAb), pWWF57 (nagGH), pWWF58 (nagG), pWWF59 (nagH) and pWWF112 (nagAcAd) were individually expressed in E. coli BL21(DE3)/pLysS by growth in LB medium at 37°C to an optical density at 600 nm (OD600) of 0.6 and then induced for 4 h by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 0.4 mM. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on a discontinuous gel in a Mini-Protean III electrophoresis cell (Bio-Rad) according to the manufacturer's instructions. The Nah proteins encoded on the plasmids pDTG121 (nahAcAd) and pDTG191 (nahAa) were also individually expressed in E. coli BL21(DE3)/pLysS by growth in LB medium at 30°C to an OD600 of 0.6 and then induced for 4 h by the addition of IPTG to 0.2 mM, the incubation temperature was lowered to 27°C after induction. Purified ferredoxinNAP (NahAb) was a kind gift from Rebecca Parales (University of Iowa).
Preparation of cell extract.
Cell extracts were prepared by resuspending the bacterial pellets in ice-cold 50 mM MES (2-N-morpholinoethanesulfonic acid) buffer (pH 6.4; ca. 0.1 g [wet weight]/ml) and disruption by sonication in an ice-water bath for three periods of 30 s with 30-s intervals, after which cell debris was removed by centrifugation at 100,000 × g for 1 h at 4°C.
Sequence determination and analysis.
Nucleotide sequences were determined by MWG-Biotech, Ltd. (Ebersberg, Germany). Sequences were analyzed with the Lasergene software package (DNAStar).
Enzyme assays.
All assays were performed in 50 mM MES buffer unless otherwise stated. S5H or NDO was assayed by measuring the decrease in absorbance at 340 nm due to the consumption of NADH or NADPH in the presence of substrates. The sample cuvette (1 ml) contained 50 mM MES buffer, 0.1 mM salicylate (for the S5H assay) or 0.1 mM indole (for the NDO assay), 0.3 mM NADH or NADPH, and 0.1 mM ferrous ammonium sulfate. The reference cuvette contained the same except the substrates. The assay was initiated by adding the components of S5H or NDO to both cuvettes. The specific activity of S5H was calculated from the initial ΔA340/min, taking the molar extinction coefficients at 340 nm as follows: for NADH or NADPH2, 6,220 M−1 cm−1, for salicylate, 120 M−1 cm−1, and for gentisate, 1,700 M−1 cm−1.
Gentisate 1,2-dioxygenase (GDO), maleylpyruvate isomerase, and fumarylpyruvate hydrolase were assayed as described previously (54). Where substrates were generated in situ from salicylate or substituted salicylates by the action of S5H in BES buffer [N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid; pH 6.4], the pH of the buffer was adjusted to its pH optimum (7.4) before performing the GDO assays. Protein concentrations were determined by the biuret procedure. One unit of enzyme activity was defined as the amount required for the disappearance (or production) of 1 μmol of substrate (or product) per min at 30°C. Specific activities are expressed as units per milligram of protein.
Biotransformation of substituted salicylates and extraction of the products.
Biotransformations of substituted salicylates were performed by growing E. coli DH5α(pWWF45 [nagAaGHAb]) in 500 ml of LB medium (in 2-liter flasks) containing 2 mM substrates, 100 μg of ampicillin/ml, and 0.4 mM IPTG. The cultures were incubated with shaking at 200 rpm and 30°C. The cleared samples were checked periodically until the spectra of the sample had shown the characteristics of the expected products (i.e., by between 24 and 36 h). After centrifugation the culture supernatant was initially extracted with ethyl acetate. The aqueous phase was then acidified to pH 2.0 before extraction with diethyl ether to recover the acidic products.
Chemical and analytical procedures.
Reagents were obtained from commercial suppliers and used without further purification unless stated otherwise. Dichloromethane was distilled over calcium hydride and diethyl ether over Na wire. Organic solutions were dried over anhydrous MgSO4 and, unless stated otherwise, were evaporated at 14 mm Hg. Thin-layer chromatography was performed by using Aldrich Silica Gel 60 plates (F254). Compounds were visualized either by examination under UV or by exposure to I2 vapor. Column chromatography was conducted with a Merck 7736 silica gel under medium pressure. Methyl esters were synthesized by using diazomethane in diethyl ether solution.
Melting points were uncorrected. Infrared spectra were recorded on a Perkin-Elmer 1600 FTIR spectrometer as liquid films unless otherwise stated. Low-resolution nuclear magnetic resonance (NMR) spectra were measured by using a Finigan MAT 1020 spectrometer at 250 MHz (1H) and at 62.9 MHz (13C). 13C spectra were broad band decoupled and in most cases distortionless enhancement by polarization transfer (DEPT) spectra were also recorded.
RESULTS
Spectrophotometric assay of S5H activity.
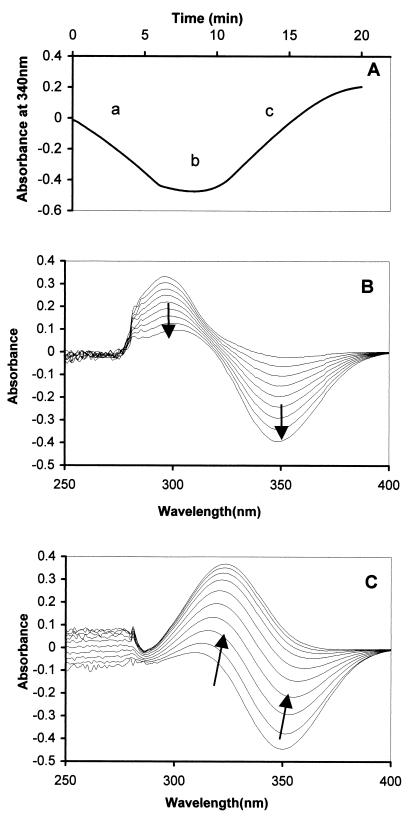
We developed an in vitro assay for S5H based on the salicylate-linked oxidation of NADH measured at 340 nm. In order to characterize the assay, we carried out repeat wavelength scans from 250 to 400 nm during the reaction. The problem of high endogeneous activity of NADH oxidase present in all of the extracts of the E. coli host was overcome by the blank cuvette containing all assay components except salicylate to compensate for the NADH oxidase. This meant that in order to get complete conversion of the salicylate, a considerable excess of NADH was required. Figure 2A shows the complete change of A340 from start to finish of a reaction. During the early part of the reaction (Fig. 2B) both NADH (λmax = 340 nm) and salicylate (λmax = 295 nm) diminished (Fig. 2A, stage a). When all of the salicylate was utilized, the rate of NADH disappearance in both experimental and blank (due only to NADH oxidase) was identical, so little net change in absorbance took place (Fig. 2A, stage b). However, as the NADH in the sample cuvette was finally exhausted, the spectrum of gentisate (λmax = 320 nm) (Fig. 2C), previously masked by the more strongly absorbing NADH, appeared, and the A340 also rose again (Fig. 2A, stage c).
FIG. 2.
Spectrophotometric changes during formation of gentisate from salicylate by cell extracts of E. coli BL21(DE3)/pLysS carrying pWWF54 (nagAa), pWWF56 (nagAb), or pWWF57 (nagGH) grown on LB medium containing 100 μg of ampicillin/ml and induced with 0.4 mM IPTG. Sample and reference cuvettes contained 50 mM MES buffer, 0.3 μmol of NADH, and 0.1 μmol of ferrous ammonium sulfate in 1-ml volumes. The sample cuvette also contained 0.1 μmol of salicylate. A mixture of cell extracts (70 μl containing 0.55 mg of protein [NagAa, NagAb, and NagGH]) was added to both sample and reference cuvettes in equivalent amounts. (A) Time course of absorbance change at 340 nm. (B and C) Scan of spectral changes from 0 to 5 min (B) and from 10 to 20 min (C).
NagAa, NagGH, and NagAb are required for salicylate transformation.
pWWF54, pWWF56, pWWF58, and pWWF59 carried the complete reading frames of nagAa, nagAb, nagG, and nagH individually in the expression vector pET5a (Table 1) whereas pWWF55 and pWWF57, respectively, carried the four complete reading frames of nagAaGHAb or the two complete reading frames of nagGH in the same vector. All six plasmids were transformed into E. coli BL21(DE3). A mixture of cell extracts of the IPTG-induced strains carrying pWWF54, pWWF56, and pWWF57 showed S5H activity which, at the optimum ratio of NagAa, NagAb, and NagGH (see below), had a specific activity of 0.051 U/mg of protein against salicylate. The cell extract of E. coli(pWWF55) alone also exhibited S5H activity but at a lower value (∼25% of the mixture of three extracts). No activity was detectable in controls where expression of the protein was not induced or where the expression vector contained no insert.
No S5H activity was detected in any single extract or pairs of the above three extracts. Furthermore, activity was lost when the extract containing NagGH was replaced by a mixture of extracts containing NagG (from pWWF58) and NagH (from pWWF59) even after the two extracts had been incubated together for 10 min at 30°C to allow for possible protein association.
Although S5H activity was measured as described, SDS-PAGE of the same extracts contained no obviously elevated levels of polypeptides of the expected sizes except in the case of E. coli(pWWF59), which showed an 18-kDa polypeptide band as expected from the calculated Mr for NagH (data not shown). This appears to be due to a high frequency of usage of codons rare for E. coli: nagG has 13 CCC (Pro) and 4 CGA (Arg) in 418 codons and nagAa has 18 rare codons in 329, respectively. Subsequent to the preparation of this paper, we transformed pWWF47 (nagGH) into the Rosetta strain of E. coli (Novagen), which enhances the expression of genes carrying codons rare for E. coli. Induction by IPTG produced heavy protein bands upon SDS-PAGE that corresponded to both subunits and were typical of overexpressed proteins (data not shown).
By using the in vitro assay we were unable to detect any S5H activity in the cell extract of strain U2 grown on naphthalene or salicylate, presumably because its specific activity was too low relative to the endogeneous NADH oxidation. However, GDO (gentisate dioxygenase, NagI) activity was evident in the same cell extract in both cases, at a level similar to that previously described (54).
Catabolism of salicylate to central metabolites by NagAaGHAb and NagIKL in vitro.
After the in vitro conversion of salicylate to gentisate by cell extracts in 50 mM BES buffer, the pH of the buffer was adjusted to 7.4. We demonstrated that the gentisate formed from salicylate could be converted directly to pyruvate (determined by using lactate dehydrogenase [54]) and fumarate by sequentially using cell extracts of E. coli (pWWF19-25) overexpressing NagI (GDO), E. coli(pWWF53) expressing NagL (maleylpyruvate isomerase) plus GSH, and then cell extracts of E. coli(pWWF96) containing NagK (fumarylpyruvate hydrolase). The sequence of the reaction happened as described in a previous study (54), demonstrating that salicylate can be converted to central metabolites solely by the above four enzymes in vitro.
Each component of S5H can be the limiting factor in the reaction.
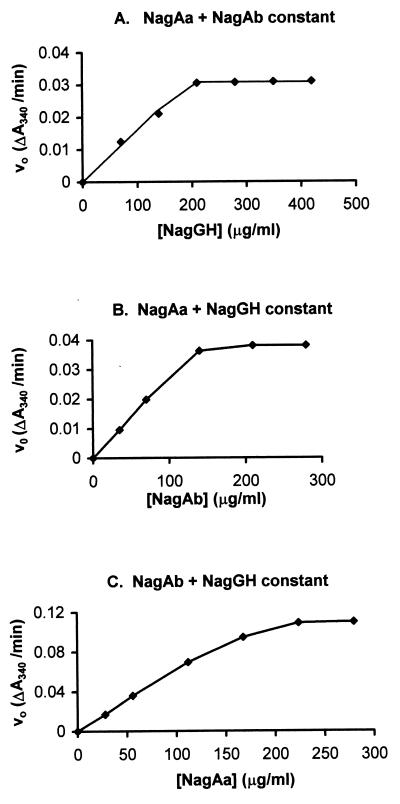
Our hypothesis has been that the hydroxylation of salicylate occurs through NagGH acting as a monooxygenase, with an associated electron transport chain consisting of a ferredoxin (NagAb) and a ferredoxin reductase (NagAa) supplying the electrons from a reduced nicotinamide cofactor. It would therefore be expected that each component of S5H will limit the rate of reaction when it becomes the “bottleneck.” To confirm this hypothesis, we increased from zero the amount of each component in the assay while keeping the other two constant. When NagGH was thus increased, with both NagAa and NagAb remaining constant, there was a clear linear dependence of activity upon the amount cell extract containing NagGH until it reached a point where the activity no longer increased and presumably either NagAa or NagAb (or both) became limiting (Fig. 3A). Similar results were also obtained by changing only the amount of NagAb (Fig. 3B) or the amount of NagAa (Fig. 3C). We used data from such experiments to calculate a mixture of all three extracts in which each was at a critical concentration below which it became limiting. At this point, this enzyme exhibited a maximum activity with a minimum amount of total extracts. In such a mixture, an increase of any one component should not change the activity because it would cause the other components to be limiting, but reduction of any component should proportionately reduce the activity since it would be the limiting component. The data in Table 2 confirm this expectation since reduction of any one component by 50% reduces the activity by ca. 50%, whereas an increase of any to 150% does not change the activity. However, a proportional increase or decrease in all three produces a proportional change in total activity. Such a mixture was used to determine the specific activity of the mixed extracts (see above).
FIG. 3.
Relationship between the enzyme activity and different concentrations of one component of the S5H, while the amounts of the other two components were kept constant. Sample and reference cuvettes contained 50 mM MES buffer, 0.3 μmol of NADH, and 0.1 μmol of ferrous ammonium sulfate in 1-ml volumes. The sample cuvette also contained 0.1 μmol of sodium salicylate. A mixture of cell extracts was added to both sample and reference cuvettes in equivalent amounts. Different amounts of cell extract of E. coli BL21(DE3)/pLysS carrying pWWF57 (nagGH) (A), pWWF56 (nagAb) (B), or pWWF54 (nagAa) (C) were added, respectively, while the amounts of the other two were kept constant.
TABLE 2.
Variation of S5H activity as a function of the ratio of the three cell extracts containing its components
| Ratio of each componenta | Relative activityb | ||
|---|---|---|---|
| NagAa | NagAb | NagGH | |
| 1 | 1 | 1 | 1.00 |
| 0.5 | 1 | 1 | 0.61 |
| 1 | 0.5 | 1 | 0.54 |
| 1 | 1 | 0.5 | 0.47 |
| 0.5 | 0.5 | 0.5 | 0.40 |
| 1.5 | 1 | 1 | 1.00 |
| 1 | 1.5 | 1 | 1.05 |
| 1 | 1 | 1.5 | 1.07 |
| 1.5 | 1.5 | 1.5 | 1.68 |
The amount of each extract in the assay was first adjusted so that each was at the position where it became limiting (see Fig. 3). This defined the initial ratio (1:1:1).
The specific activity of the 1:1:1 mixture was 0.051 U/mg, with a total activity of 0.028 U.
Substrate specificity of S5H in in vitro assay.
The active mixture of extracts (NagAa, NagGH, and NagAb) was tested against 14 substituted salicylates and other structurally similar compounds by using the spectrophotometric assay. No activity was detected against 2-hydroxycinnamate, 3-hydroxycinnamate, 2-hydroxyphenylacetate, 3-hydroxyphenylacetate, 2-hydroxybenzophenone, 1-hydroxy-2-naphthoate, 4-methoxysalicylate, and 2-hydroxyacetophenone. However, some substituted salicylates did stimulate NADH oxidation and a significant change in the absorption spectrum after the reaction was complete (see Table 3). It appears that essential requirements for a substrate are a ring-substituted −COO− group with an adjacent −OH: even the presence of a methylene group between the carboxylate and the ring, as in 2-hydroxyphenylacetate, is sufficient to stop catalysis. The relative rates of change of A340 between the different substrates at 100 mM varied by no more than a factor of 5 to 10 between the best (salicylate) and the worst (4-methylsalicylate).
TABLE 3.
Substrate specificity of S5H (NagAaGHAb)
| Substrate (λmax [nm]) | Product (λmax [nm]) | λmax (nm) of product formed by action of GDO |
|---|---|---|
| Salicylate (295) | Gentisate (320) | 330 (maleylpyruvate) |
| 3-Methylsalicylate (300) | 2,5-Dihydroxy-3-methylbenzoate (322) | 334 |
| 4-Methylsalicylate (296) | 2,5-Dihydroxy-4-methylbenzoate (315) | 324 |
| 5-Methylsalicylate (305) | 2-Hydroxy-5-hydroxymethylbenzoate (301) | NAa |
| 4-Chlorosalicylate (297) | 4-Chloro-2,5-dihydroxybenzoate (316) | 334 |
| 5-Chlorosalicylate (307) | 5-Chloro-2,6-dihydroxybenzoate (315) | NA |
| 2,4-Dihydroxybenzoate (291) | 2,4,5-Trihydroxybenzoateb (312) | 327 |
NA, no product was detected.
Presumed product.
The products from 3-methylsalicylate, 4-methylsalicylate, and 4-chlorosalicylate were the corresponding substituted gentisates (2,5-dihydroxybenzoates) (see below): the product of 2,4-dihydroxybenzoate has not been identified but, by analogy, is tentatively assumed to be 2,4,5-trihydroxybenzoate. All of the products, formed by the action in vitro of S5H on 3- and 4-substituted salicylates, were substrates for GDO as measured by a change in λmax upon adding extracts containing GDO consistent with formation of the corresponding maleylpyruvates (Table 3). In contrast, the products from 5-methyl- and 5-chloro-salicylates were not substrates for GDO because they lacked the 2,5-dihydroxy substituents.
Identification of products of substituted salicylates.
Because of the difficulty of accumulating sufficient amounts of products for structural determinations from the conversion of salicylates by cell extracts, we chose to carry out biotransformations of the substrates with E. coli DH5α (pWWF45) carrying nagAaGHAb, as was originally used to demonstrate the conversion of salicylate to gentisate (11). Cultures were grown on rich medium in the presence of 2 mM substrates with IPTG as the inducer. The absorption spectra were measured after centrifugation to check that they were identical to those produced in vitro by cell extracts, after which the product was extracted into solvent.
In proton NMR, the product from 4-chlorosalicylic acid dissolved in CD3OD showed δH values of 7.4 (1H, s) and 6.9 (1H, s). Reaction with diazomethane gave the corresponding methyl ester which showed δH(CDCl3) values of 7.3 (1H, s), 7.0 (1H, s), and 3.9 (3H, s), characterized as methyl 2,5-dihydroxy-4-chlorobenzoate. To confirm the structure, an authentic sample of the acid was prepared from 4-chloro-2-hydroxybenzoic acid (2) and shown to have the same 1H NMR spectrum as the metabolic product; conversion to its ester again gave a compound that is identical, as determined by 1H NMR, to that described above.
The product from the metabolism of 4-methylsalicylic acid gave a 1H NMR spectrum in CD3OD which showed δH values of 7.23 (1H, br.s), 6.7 (1H, br.s), and 2.22 (3H, s). On the basis of the presence of just two singlets in the aromatic region of the spectrum, this compound was characterized as 2,5-dihydroxy-4-methylbenzoic acid (Fig. 4). Although this compound is known (28), no pertinent NMR data are available.
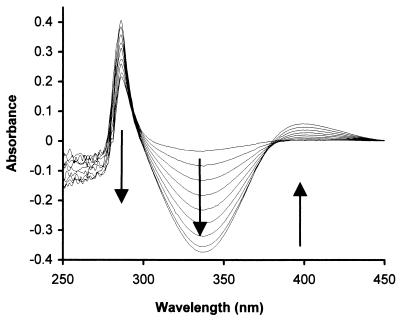
FIG. 4.
Spectrophotometric changes during formation of indoxyl from indole by cell extracts of E. coli BL21(DE3)/pLysS carrying pWWF54 (nagAa), pWWF56 (nagAb), or pWWF112 (nagAcAd) grown on LB medium containing 100 μg of ampicillin/ml and induced with 0.4 mM IPTG. Sample and reference cuvettes contained 50 mM MES buffer, 0.3 μmol of NADH, and 0.1 μmol of ferrous ammonium sulfate in 1-ml volumes. The sample cuvette also contained 0.1 μmol of indole. Spectra were recorded at 40-s intervals after the addition of a mixture of cell extracts (70 μl containing 0.45 mg of protein [NagAa, NagAb, and NagAcAd]). Cell extract was added to the reference cuvette in an equivalent amount.
The product from 3-methylsalicylic acid was soluble in CD3OD; it gave a 1H NMR spectrum which showed δH values of 7.10 (1H, d, J 3.1 Hz), 6.84 (1H, d, J 3.1 Hz), and 2.16 (3H, s). On the basis of a comparison with literature data (13, 42), it was characterized as 2,5-dihydroxy-3-methylbenzoic acid (Fig. 4). The 1H NMR spectrum in CD3OD was also identical to that of an authentic sample of this acid (provided by C. L. Poh).
The product from 5-methylsalicylic acid dissolved in CD3OD gave a 1H NMR spectrum which showed δH values of 7.86 (1H, s), 7.46 (1H, d, J 8.5 Hz), 6.91 (1H, d, J 8.5 Hz), and 4.53 (2H, s). The corresponding methyl ester was a yellow oil which showed δH(CDCl3) values of 10.6 (1H, s), 7.65 (1H, d, J 2.1 Hz), 7.31 (1H, dd, J 2.1, 8.5 Hz), 6.82 (1H, d, J 8.5 Hz), 4.45 (2H, br.s), 3.82 (3H, s), and 2.7 (1H, br.s). The 13C NMR spectrum showed δC values of 170.4(s), 160.9(q), 134.7(d), 131.8(s), 128.4(d), 117.6(d), 112.0(s), 64.3(t), and 52.2(q). The 1R spectrum had a νmax of 3,370 and 1,681 cm−1, and the mean spectrum contained two characteristic ions with an m/z of 182 (M+) of 150 (M+-CH3OH). It was thus characterized as 2-hydroxy-5-hydroxymethylbenzoic acid (Fig. 4). Although this compound is known (6), no NMR data are available for comparison. However, the coupling patterns in the 1H NMR spectra of both the acid and the ester are consistent with a 1,2,5-trisubstituted benzene, the presence of the singlets at δH 4.53 (4.45) are consistent with the CH2OH group, and the 13C spectrum of the ester shows both the correct numbers of carbons and the CH2OH and OCH3 groups at δC 64.1 and 52.2.
The product from 5-chlorosalicylic acid dissolved in CD3OD showed δH values of 7.3 (1H, d, J 8.8 Hz) and 6.41 (1H, d, J 8.8 Hz) and was characterized as 2,6-dihydroxy-3-chlorobenzoic acid (Fig. 4). An authentic sample was prepared from 2,6-dihydroxybenzoic acid as described in the literature (9) as a white solid that gave a 1H NMR spectrum identical to that described above.
NagAcAd exhibited naphthalene dioxygenase activity together with NagAa and NagAb.
Whereas NagG in particular show clears amino acid homologies to the α subunits of the NDO family, it is significantly divergent (12). NagAc and NagAd also represent a separate subgroup of the dioxygenase family but one which is sufficiently close to the classical NDOs for them to be reasonably assumed to be the genes in U2 for its NDO (12). In order to test this hypothesis, we modified an in vitro assay originally developed for an analogous toluene dioxygenase (19) which relies on the conversion by aromatic-dihydroxylating dioxygenases of indole to indigo (10) but with the intermediate formation of the yellow indoxyl (3-hydroxyindole) (λmax = 400 nm). By using protocols similar to those for the S5H assay we found that this approach was far more sensitive if NADH disappearance was measured at 340 nm rather than the indoxyl appearance at 400 nm (Fig. 5), as done by Jenkins and Dalton (19).
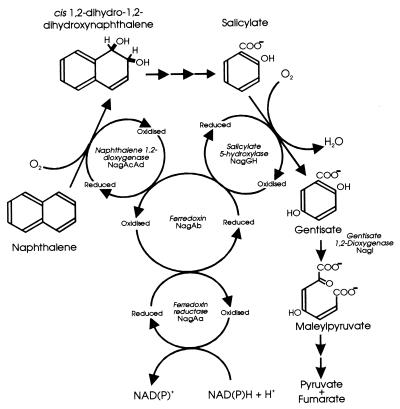
FIG. 5.
Pathway for naphthalene catabolism by Ralstonia sp. strain U2 highlighting the shared electron transport chain (NagAa and NagAb) by S5H (NagGH) and NDO (NagAcAd).
Plasmid pWWF112 was constructed to overexpress nagAcAd. After induction with IPTG, cell extracts of E. coli(pWWF112) were found to contain NDO activity (as measured by the indole-dependent oxidation of NADH) but only when mixed with the cell extracts of both E. coli(pWWF54) carrying nagAa and E. coli(pWWF56) carrying nagAb. The assay medium became yellow due to indoxyl formation, and, on being left to stand for ca. 30 min, a blue precipitate of indigo was formed, due to subsequent oxidation. There was no cross-reactivity between S5H activity and NDO: the former does not oxidize indole nor does the latter hydroxylate salicylate. NagAcAd was also active against styrene as a substrate in an NADH-linked assay (data not shown). NDO from Pseudomonas sp. strain NCIB 9816-4 reportedly transformed styrene (21) The NDO activity is also rate limited by each of the three components as described above for S5H (data not shown). The NDO (in the critical mixture of NagAa, NagAb, and NagAcAd) had a specific activity of 0.054 U/mg of protein against indole as the substrate.
The relationship of the two electron transport chains in nah (from P. putida NCIMB9816) and nag operons.
By using the two in vitro assays we have shown that the electron transport proteins from the nah system, namely, NahAa and NahAb, can be used by both the nag NDO and S5H enzymes. Furthermore, there is complete interchangeability in the protein interactions since the electron transport chain from the nag system (NagAa and NagAb) can also be used by NahAcAd for NDO activity. Interestingly, all three oxygenases were also functional with a mixed electron transport chain consisting of two components from different origin (NagAa + NahAb) or (NahAa + NagAb). Although it is difficult to compare absolute rates because the concentrations of the different components in the cell extracts could not be determined, the rates of the measured reactions with the various mixed systems did not appear to differ greatly from those with the single systems, indicating a fairly loose specificity in the various protein-protein electron transfers.
The electron transport chain from two systems and the mixed chains can also use both NADH and NADPH as electron donors at approximately similar rates (Table 4). Irrespective of the terminal oxygenases, each of which we tried to keep in excess, the ratio of the rate produced by NADH versus that produced by NADPH was remarkably consistent for each of the four possible combinations of electron transport proteins. Since the interaction with the nucleotide cofactor is solely with the ferredoxin reductase Aa, one might expect that the NADH/NADPH activity ratio would be determined exclusively by which Aa component was in the assay. We would clearly expect this to be true under assay conditions wherein the ferredoxin reductase is the rate-limiting component, but the complexity of ensuring this with all of the different extracts used in the experiment prevented us from doing this.
TABLE 4.
Relative activities of S5H and NDOs with NADH and NADPH as cofactors with different combinations of electron transport chainsa
| Oxygenasec | Source strain | Relative activitiesa with electron transport componentsb
|
|||
|---|---|---|---|---|---|
| NagAa + NagAb | NahAa + NahAb | NagAa + NahAb | NahAa + NagAb | ||
| S5H (NagGH) | U2 | 1.21 | 1.42 | 1.23 | 3.02 |
| NDO (NagAcAd) | U2 | 1.20 | 1.43 | 1.22 | 3.10 |
| NDO (NahAcAd) | NCIMB9816 | 1.19 | 1.42 | 1.22 | 3.12 |
The activity was measured with the same amount of each extract carrying the oxygenase present in all of the rows and the same amounts of each of the extracts containing the electron transport components in the columns. The activity with NADH as the cofactor was divided by the activity with NADPH as the cofactor.
The electron transport components were in extracts of E. coli carrying the following plasmids: for NagAa, pWWF54; for NagAb, pWWF56; and for NahAa, pDTG191. NahAb was added as a purified protein.
The oxygenase components were in extracts of E. coli carrying the following plasmids: for NagGH, pWWF57; for NagAcAd, pWWF112; and for NahAcAd, pDTG121.
DISCUSSION
In our original report of the nag genes of Ralstonia sp. strain U2 and based upon a limited amount of evidence, we proposed that they had two unique features (12). First, the products of two pairs of genes, each apparently α and β subunits of the dihydroxylating dioxygenase family, had separate functions, one being the NDO of the pathway (NagAcAd) and the other being a monooxygenase converting salicylate to gentisate. Second, they both shared the chain (NagAa and NagAb) transporting electrons from NAD(P)H through to the substrate (Fig. 5). In the present study we have added further evidence supporting these hypotheses. We have developed in vitro spectrophotometric assays for both activities, a direct assay for S5H and an indirect assay, involving the NAD(P)H-linked conversion of indole to 3-hydroxyindole for the NDO. Both assays only function if the pairs of genes for the α and β subunits are coexpressed in the same cell, but each reaction obligately requires the presence of the two electron transport proteins, which can be added as independent components from other extracts. All three components of the assay (oxygenase, ferredoxin, and ferredoxin reductase) are co-equal determinants of the reaction rate and, in diminishing quantities, each becomes rate limiting.
The nag oxygenase gene cluster represents an interesting example of genetic efficiency in which two electron transport proteins essential for activity are shared by two enzymes, one a dioxygenase and one a monooxygenase, that are separated by several steps within a metabolic pathway. There may be other examples of this. For example, in Sphingomonas spp. the organization of genes for aromatic catabolism appears to be less obviously structured than in other gram-negative organisms, and clusters of genes of uncertain metabolic function have been found to contain a multiplicity of genes for Rieske non-heme oxygenases with a relative paucity of genes for obvious associated electron transport proteins (34, 35). A converse situation has been found with a dioxin dioxygenase from Sphingomonas sp. strain RW1, in which two homologous ferredoxins are able to transfer electrons to the oxygenase (1).
In another way the NagGH is a further example of the diversity of oxygenase evolution. Oxygenases catalyzing hydroxylation reactions represent a diverse family of both catalytic proteins and electron transport components which have been reviewed and classified (3, 15, 23). Monohydroxylation reactions do not appear to require a complex enzymatic machinery, and at the simplest level they can be accomplished by simple single subunit flavoproteins, such as salicylate 1-hydroxylase NahG (50) or 4-hydroxybenzoate hydroxylase (37). However, the genetic complexity used for other reactions increases through to two genes, as with xylene monooxygenase XylMA (47), three genes with alkane hydroxylase AlkBGT (48), four genes with the methanesulfonic acid monooxygenase MsmABCD (8), through to five genes for soluble methane monooxygenase (40) and six genes for toluene 3-monooxygenase TbuA1UBVA2C (4) and phenol hydroxylase DmpKLMNOP (27, 32). Hydroxylating dioxygenases require from two genes, for 4-sulfobenzoate 3,4-dioxygenase (22), through three genes, for benzoate dioxygenase BenABC and the related toluate dioxygenase XylXYZ (26), to the NDO-like four-gene, three-protein systems (10). NagGH adds to the overall complexity of this diverse family by being the only reported example of a monooxygenase requiring four gene products, three of which are so closely similar to the NDO type III dioxygenases (3): the most similar monooxygenase to NagGH in its organization is the methanesulfonic acid monooxygenase, the oxygenase of which has a Rieske-type α-subunit, a smaller β-subunit plus an electron transport chain of a ferredoxin, and a ferredoxin reductase, but none show the same strong degree of similarity with the NDO-like class III dioxygenases (8). The similarities between the S5H of U2 and NDO further blurs the distinction between mono- and dioxygenases that is apparent from experimental results, particularly those from the laboratory of D. T. Gibson, that have shown that NDO from Pseudomonas strain NCIMB9816 can catalyze monooxygenation reactions, such as the conversion of 2-nitrotoluene to 2-nitrobenzyl alcohol (31), analogous to the reaction catalyzed by xylene (toluene) monooxygenase or the conversion of 5-methylsalicylate to 2-hydroxy-5-hydroxymethylbenzoate by S5H as shown in the present study (Table 3). A possible mechanism for S5H might be that it acts as a dioxygenase converting salicylate to a dihydrodiol which then loses water to form gentisate. However, such a mechanism is not easily compatible with the two unexpected reactions of S5H on the 5-substituted salicylates (Table 4), the products of both of which are difficult to reconcile with dihydrodiol intermediates.
The NDO from P. putida NCIMB9816 has been shown by crystallography to be an α3β3 hexamer (20). It is our prediction that NagGH also has a similar structure. That it has a multimeric structure can be inferred from the fact that activity is only obtained when nagGH genes are coexpressed in the same host and not when they are expressed separately and the extracts are mixed. We have tried to show this on nondenaturing protein gels, but the level of expression was insufficient to visualize bands corresponding to a hypothetical multimer, as indeed it is on SDS-PAGE. However, the complete interchangeability of the electron transport chain proteins between the NDONAH, NDONAG, and S5HNAG activities suggests that they might have very similar surface structures to permit the necessary docking interactions between the oxygenase and the ferredoxins.
There are several previous reports of S5Hs. Where there has been characterization of the activity they appear to bear no structural or mechanistic relationship to the U2 enzyme. In Rhodococcus sp. strain B4, which, like strain U2, metabolizes naphthalene via salicylate and gentisate, the S5H activity required NADPH, ATP, and coenzyme A (14), and a purified enzyme from salicylate-grown R. erythropolis strain S1 was shown to be a homotetramer containing FAD and with a specific requirement for NADH (44). An uncharacterized activity was also reported in a salicylate-grown strain of Moraxella (33). However, very recently a four-gene cluster corresponding to and closely homologous to nagAaGHAb has recently been identified in close physical association to genes for chlorobenzoate catabolism (17). The gene products have S5H activity, but the surrounding DNA is completely devoid of any residues of associated NDO-like genes (17). Whereas we suggested that the nag oxygenase cluster arose by insertion of nagGH into a classical nah-like operon (12), this finding suggests that the four constituent S5H genes may also have an independent evolutionary history of their own.
It is not clear at present what is the relative frequency in the environment of nag-type genes compared with nah-type genes. To date, only one other strain has been identified which has a nag-like arrangement of genes, and it has not been definitively demonstrated that these show the same function (56). If both the significant number of sequenced genes in the data banks which have the nah-like oxygenase cluster and the relative rarity of reported strains degrading naphthalene through gentisate is added to this, it might suggest that the nag-like route is a relative rarity. However, nitrotoluene and dinitrotoluene degraders (30, 31, 45) have been reported that carry the four genes of the nag-NDO-like dioxygenase, but with nagGH-like pseudogenes intercalated between the Aa and Ab genes. These have a greater degree of sequence similarity with the nag genes than with the corresponding nah genes, suggesting that they have evolved from nag-like ancestors under relatively recent nitrotoluene selection, which might in turn point to their having an important evolutionary potential even if in low natural frequency.
Acknowledgments
This research was supported by the Biotechnology and Biological Sciences Research Council.
We thank Dave Gibson and Rebecca Parales for their gift of the plasmids and proteins from P. putida NCIMB9816 and Chit Laa Poh for providing gentisic acids.
REFERENCES
- 1.Armengaud, J., J. Gaillard, and K. N. Timmis. 2000. A second [2Fe-2S] ferredoxin from Sphingomonas sp. strain RW1 can function as an electron donor for the dioxin dioxygenase J. Bacteriol. 182:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharaya, S. C., and D. E. Seymour. 1950. 4-Aminosalicylic acid and its derivatives. Part II. The syntheses of 4-amino-2,5- and 4-amino-2,3-dihydroxybenzoic acid. J. Chem. Soc. 1950:1139-1140.
- 3.Butler, C. S., and J. R. Mason. 1997. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv. Microb. Physiol. 38:47-84. [DOI] [PubMed] [Google Scholar]
- 4.Byrne, A. M., J. J. Kukor, and R. H. Olsen. 1995. Sequence analysis of the gene cluster encoding toluene-3-monooxygenase from Pseudomonas pickettii. Gene 154:65-70. [DOI] [PubMed] [Google Scholar]
- 5.Cane, P. A., and P. A. Williams. 1986. A restriction map of the catabolic plasmid pWW60-1 and the location of some of its catabolic genes. J. Gen. Microbiol. 132:2919-2929. [Google Scholar]
- 6.Carpenter, A. T., and R. F. Hunter. 1953. Phenol-formaldehyde and allied resins. III. Further syntheses of resol molecules. J. Appl. Chem. 496:486-495. [Google Scholar]
- 7.Davies, J. I., and W. C. Evans. 1964. Oxidative metabolism of naphthalene by soil pseudomonads. Biochem. J. 91:251-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Marco, P., P. Moradas-Ferreira, T. P. Higgins, I. McDonald, E. M. Kenna, and J. C. Murrell. 1999. Molecular analysis of a novel methanesulfonic acid monooxygenase from the methylotroph Methylsulfomonas methylovora. J. Bacteriol. 181:2244-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyle, F. P., J. H. C. Nayler, H. R. J. Waddington, J. C. Honson, and G. R. Thomas. 1963. Derivatives of 6-aminopenicillanic acid. Part V. Analogues of 2,6-dimethoxyphenylpenicillin with enhanced stability towards acid. J. Chem. Soc. 1963i:497-506.
- 10.Ensley, B. D., D. T. Gibson, and A. L. Laborde. 1982. Oxidation of naphthalene by a multicomponent enzyme-system from Pseudomonas sp. strain NCIB9816. J. Bacteriol. 149:948-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ensley, B. D., B. J. Ratzkin, T. D. Osslund, M. J. Simon, L. P. Wackett, and D. T. Gibson. 1983. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science 222:167-169. [DOI] [PubMed] [Google Scholar]
- 12.Fuenmayor, S. L., M. Wild, A. L. Boyes, and P. A. Williams. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giles, R. G. F., P. R. K. Mitchell, and M. V. Sargent. 1983. Approaches to the synthesis of 4-hydroxypiloquinone. J. Chem. Soc. Perkin Trans. 1:2147-2152. [Google Scholar]
- 14.Grund, E., B. Denecke, and R. Eichenlaub. 1992. Naphthalene degradation via salicylate by Rhodococcus sp. strain B4. Appl. Environ. Microbiol. 58:1874-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harayama, S., M. Kok, and E. L. Neidle. 1992. Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 46:565-601. [DOI] [PubMed] [Google Scholar]
- 16.Harayama, S., M. Rekik, A. Wasserfallen, and A. Bairoch. 1987. Evolutionary relationships between catabolic pathways for aromatics: conservation of gene order and nucleotide sequences of catechol oxidation genes of pWW0 and NAH7. Mol. Gen. Genet. 210:241-247. [DOI] [PubMed] [Google Scholar]
- 17.Hickey, W. J., G. Sabat, A. S. Yuroff, A. R. Arment, and J. Pérez-Lesher. 2001. Cloning, nucleotide sequencing and functional analysis of a novel, mobile cluster of biodegradation genes from Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 67:4603-4609. [DOI] [PMC free article] [PubMed]
- 18.Jain, H., R. E. Parales, N. A. Lynch, and D. T. Gibson. 1996. Site-directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: potential mononuclear non-heme iron coordination sites. J. Bacteriol. 178:3133-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins, R. O., and H. Dalton. 1985. The use of indole as a spectrophotometric assay substrate for toluene dioxygenase. FEMS Microbiol. Lett. 30:227-231. [Google Scholar]
- 20.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 21.Lee, K., and D. T. Gibson. 1996. Stereospecific dihydroxylation of the styrene vinyl group by purified naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. J. Bacteriol. 178:3353-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Locher, H. H., T. Leisinger, and A. M. Cook. 1991. 4-Sulfobenzoate 3,4-dioxygenase: purification and properties of a desulfonative two-component system from Comomonas testosteroni T2. Biochem. J. 274:833-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason, J. R., and R. Cammack. 1992. The electron transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277-305. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Monticello, D. J., D. Bakker, M. Schell, and W. R. Finnerty. 1985. Plasmid-borne Tn 5 insertion mutation resulting in the accumulation of gentisate from salicylate. Appl. Environ. Microbiol. 49:761-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neidle, E. L., C. Hartnett, L. N. Ornston, A. Bairoch, M. Rekik, and S. Harayama. 1991. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J. Bacteriol. 173:5385-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordlund, I., J. Powlowski, and V. Shingler. 1990. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp strain CF600. J. Bacteriol. 172:6826-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nudenberg, W., A. M. Gaddis, and L. W. Butz. 1943. The synthesis of condensed ring compounds. XIII. The preparation of 5- and 6-carbalkoxy-1,4-toluquinones, addition of 5-carbomethoxy-1,4-toluquinone and 6-carbomethoxy-1,4-toluquinone to butadiene. J. Org. Chem. 500:500-508. [Google Scholar]
- 29.Ohmoto, T., K. Sakai, N. Hamada, and T. Ohe. 1991. Salicylic acid metabolism through a gentisate pathway by Pseudomonas sp. TA-2. Agric. Biol. Chem. 55:1733-1737. [Google Scholar]
- 30.Parales, J. V., A. Kumar, R. E. Parales, and D. T. Gibson. 1996. Cloning and sequencing of the genes for 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene 181:57-61. [DOI] [PubMed] [Google Scholar]
- 31.Parales, J. V., R. E. Parales, S. M. Resnick, and D. T. Gibson. 1998. Enzyme specificity of 2-nitrotoluene 2,3-dioxygenase from Pseudomonas sp. strain JS42 is determined by the C-terminal region of the α-subunit of the oxygenase component. J. Bacteriol. 180:1194-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powlowski, J., and V. Shingler. In vitro analysis of polypeptide requirements of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J. Bacteriol. 172:6834-6840. [DOI] [PMC free article] [PubMed]
- 33.Rani, M., D. Prakash, R. C. Sobti, and R. K. Jain. 1996. Plasmid-mediated degradation of o-phthalate and salicylate by a Moraxella sp. Biochem. Biophys. Res. Commun. 220:377-381. [DOI] [PubMed] [Google Scholar]
- 34.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schell, M. A. 1990. Regulation of the naphthalene degradative genes of plasmid NAH7: example of a generalized positive control system in Pseudomonas and related bacteria, p. 165-176. In S. Silver, A. M. Chakrabarty, B. Iglewski, and S. Kaplan (ed.), Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. American Society for Microbiology, Washington, D.C.
- 37.Schreuder, H. A., W. G. J. Hol, and J. Drenth. 1990. Analysis of the active-site of the flavoprotein p-hydroxybenzoate hydroxylase and some ideas with respect to its reaction mechanism. Biochemistry 29:3101-3108. [DOI] [PubMed] [Google Scholar]
- 38.Shingler, V., J. Powlowski, and U. Marklund. 1992. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J. Bacteriol. 174:711-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skryabin, G. K., V. V. Kochetkov, A. A. Ereyomin, A. N. Perebityuk, I. I. Starovoytov, and A. M. Boronin. 1980. pBS4: a new naphthalene-degradative plasmid. Dokl. Akad. Nauk. SSSR 250:212-215. [Google Scholar]
- 40.Stainthorpe, A. C., V. Lees, G. P. C. Salmond, H. Dalton, and J. C. Murrell. 1990. The methane monooxygenase gene cluster of Methylococcus capsulatus (Bath). Gene 91:27-34. [DOI] [PubMed] [Google Scholar]
- 41.Starovoitov, I. I., M. Y. Nefedova, G. I. Yakovlev, A. M. Zyakun, and V. M. Adanin. 1975. Gentisic acid as a microbial oxidation product of naphthalene. Izv. Akad. Nauk. SSSR Ser. Khim. 9:2091-2092. [Google Scholar]
- 42.Still, I. W. J., and D. J. Snodin. 1972. The structures of the trihydroxyflavans from the acid-catalysed rearrangement and dimerization of 3-carene-2,5-dione. Can. J. Chem. 50:1276-1282. [Google Scholar]
- 43.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage-T7 RNA-polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 44.Suemori, A., R. Kurani, and N. Tomizuka. 1993. Purification and properties of 3 types of monohydroxybenzoate oxygenase from Rhodococcus erythropolis S1. Biosci. Biotechnol. Biochem. 57:1487-1491. [Google Scholar]
- 45.Suen, W.-C., B. Haigler, and J. C. Spain. 1996. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenases. J. Bacteriol. 178:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suen, W. C., and D. T. Gibson. 1994. Recombinant Escherichia coli strains synthesize active forms of naphthalene dioxygenase and its individual alpha-subunit and beta-subunit. Gene 143:67-71. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, M., T. Hayakawa, J. P. Shaw, M. Rekik, and S. Harayama. 1991. Primary structure of xylene monooxygenase: similarities to and differences from alkane hydroxylation system. J. Bacteriol. 173:1690-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witholt, B., J. Sijtsema, M. Kok, and G. Eggink. 1990. Oxidation of alkanes by Pseudomonas oleovorans, p. 141-150. In S. Silver, A. M. Chakrabarty, B. Iglewski, and S. Kaplan (ed.), Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. American Society for Microbiology, Washington, D.C.
- 49.Worsey, M. J., and P. A. Williams. 1975. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J. Bacteriol. 124:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto, S., M. Katagiri, H. Maeno, and O. Hayaishi. 1965. Salicylate hydroxylase, a monooxygenase requiring flavin adenine dinucleotide. I. Purification and general properties. J. Biol. Chem. 240:3408-3413. [PubMed] [Google Scholar]
- 51.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 52.Yen, K.-M., and I. C. Gunsalus. 1982. Plasmid gene organization: naphthalene/salicylate oxidation. Proc. Natl. Acad. Sci. USA 79:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yen, K.-M., and C. M. Serdar. 1988. Genetics of naphthalene catabolism in Pseudomonas. CRC Crit. Rev. Microbiol. 15:247-268. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, N.-Y., S. L. Fuenmayor, and P. A. Williams. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zylstra, G. J., and E. Kim. 1997. Aromatic hydrocarbon degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 19:408-414. [DOI] [PubMed] [Google Scholar]
- 56.Zylstra, G. J., E. Kim, and A. K. Goyal. 1997. Comparative molecular analysis of genes for polyaromatic hydrocarbon degradation. Genet. Eng. 19:257-269. [DOI] [PubMed] [Google Scholar]