Abstract
An extracellular matrix connects bacteria that live in organized assemblages called biofilms. While the role of the matrix in the regulation of cell behavior has not been extensively examined in bacteria, we suggest that, like mammalian cells, the matrix facilitates cell-cell interactions involved with regulation of cohesion, motility, and sensory transduction. The extracellular matrix of the soil bacterium Myxococcus xanthus is essential for biofilm formation and fruiting body development. The matrix material is extruded as long, thin fibrils that mediate adhesion to surfaces, cohesion to other cells, and excitation by the chemoattractant dilauroyl phosphatidylethanolamine. We report the identification of a putative matrix-associated zinc metalloprotease called FibA (fibril protein A). Western blotting with FibA-specific monoclonal antibody 2105 suggests extensive proteolytic processing of FibA during assembly into fibrils, consistent with the autoprocessing observed with other members of the M4 metalloprotease family. Disruption of fibA had no obvious effect on the structure of the fibrils and did not inhibit cell cohesion, excitation by dioleoyl phosphatidylethanolamine, or activity of the A- or S-motility motors. However, the cells lost the ability to respond to dilauroyl phosphatidylethanolamine and to form well-spaced fruiting bodies, though substantial aggregation was observed. Chemotactic excitation of the fibA mutant was restored by incubation with purified wild-type fibrils. The results suggest that this metalloprotease is involved in sensory transduction.
Mammalian cells arranged in organs are bound by a matrix of connective tissue that plays an active role in the maintenance of multicellularity. Similarly, a matrix interconnects bacteria existing in multicellular aggregates called biofilms. In spite of the obvious importance of the extracellular matrix to the organization of mammalian cells (44), there have been few studies on the structure and function of the extracellular matrix in prokaryotic biofilms. One of the best-defined biofilm systems is that of the soil bacterium Myxococcus xanthus, in which the matrix is essential for fruiting body development (37). The extracellular matrix of M. xanthus is extruded in long thin fibrils, about 30 to 40 nm in diameter, that emanate from many points on the cell surface (1). The fibrils consolidate with time to form a layer that surrounds colonies. Fibril synthesis is induced by starvation (1), stimulated by cell contact (4), and repressed by the stk locus (13). The fibrils are composed of a polysaccharide backbone to which many proteins are assembled (3). At least five of these polypeptides react with monoclonal antibody (MAb) 2105 (5).
M. xanthus mutants that lack the matrix, including dsp (1), which is also known as dif (26, 45), and mutants that fail to bind the textile dye calcofluor white (33) are defective in attachment, a prerequisite for biofilm formation. Though these mutants are defective in fruiting body development, attachment and development can be restored by the addition of wild-type fibrils (11, 45). The fibrils also modify cell behavior. Cells that fail to produce fibrils tend to move as individuals while those that overproduce fibrils tend to migrate in groups (13, 36). Fibrils are also involved in chemotaxis to some species of phosphatidylethanolamine (PE). Fibrils are essential for excitation by dilauroyl PE but not dioleoyl PE (22, 23). Addition of purified wild-type fibrils to a dsp mutant restored excitation by dilauroyl PE, suggesting that the extracellular matrix interacts with cells to complement the defect in the sensory pathway. The biochemical mechanisms of cell cohesion and dilauroyl PE excitation remain to be elucidated along with their role(s) in biofilm formation and development.
The purpose of this work is to identify proteins involved in matrix function. Behmlander and Dworkin (5) determined the N-terminal amino acid sequence of a fibril protein that reacts with MAb 2105. Cereon Genomics, LLC, which has sequenced about 95% of the M. xanthus genome, generously furnished the sequence of the only gene encoding this peptide. This paper describes the unusual features of this gene, which encodes a putative zinc metalloprotease, as well as the phenotype of the corresponding mutant cells.
MATERIALS AND METHODS
Strains and conditions.
M. xanthus strains DK1622 (wild type), DK2232 (aglB3), DK101 (pilQ1), and LS302 (dsp-1693) were grown in CYE broth [10 g of Difco Casitone per liter, 5 g of yeast extract per liter, 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS; pH 7.6), and 4 mM MgSO4] with vigorous shaking at 32°C.
DNA sequence analysis.
DNA and amino acid sequences were analyzed with the University of Wisconsin Genetics Computer Group software package and with the ExPASy (Expert Protein Analysis System) proteomics software from the Swiss Institute of Bioinformatics.
Mutagenesis.
Chromosomal DNA was purified from M. xanthus DK1622 (Easy-DNA kit; Invitrogen) and an approximately 1-kb internal fragment of the M. xanthus fibA gene was PCR amplified with a hot start with a 5′ GCAACCTGGTCTTCAAGAAGGC 3′ forward primer and a 5′ GGAGATACCGGAGCTGTAGTG 3′ reverse primer (62°C annealing temperature) using Ready-To-Go PCR beads (Amersham Pharmacia Biotech, Inc.). The PCR product was cloned into pCR2.1 (Invitrogen) to create pDB23. pDB23 was electroporated (21) into M. xanthus DK1622 and plated on CYE medium containing 40 μg of kanamycin per ml. A single homologous crossover integrated pDB23 into the chromosome to disrupt the gene. This technique generates two defective alleles, one with a 5′ and one with a 3′ deletion in the fibA coding region. One kanamycin-resistant colony, LS2200 (fibA::pDB23), was selected for further study. The plasmid pDB23 was also electroporated into DK2232, LS302, and DK101 for genetic analysis of gliding motility. The structure of the cointegrate was confirmed by PCR analysis of genomic DNA.
Scanning electron microscopy.
M. xanthus cells were grown to 5 × 108 cells/ml in CYE, centrifuged, washed in 10 mM MOPS buffer, and resuspended in agglutination buffer (10 mM MOPS [pH 6.8], 10 mM MgCl2, 1 mM CaCl2) to 5 × 107 cells/ml. The suspension was incubated for 30 min at room temperature prior to collecting 200 μl of the cell suspension by vacuum filtration on Nuclepore filters (0.2-μm pore size). The cells were fixed in Parducz solution (3 parts 4% OsO4, 3 parts distilled H2O, and 1 part saturated HgCl2) (31) for 2 min, washed twice in cacodylate buffer (0.1 M cacodylate buffer [pH 7.2], 5% sucrose), and dehydrated with sequential washes in 50, 70, 90, 95, and 100% ethanol. The filters were dried in a Samdri-780A critical point drying apparatus with liquid CO2, coated in an Edwards vacuum evaporator with 25 nm of chromium, and examined with a Phillips 505 scanning electron microscope at 3.0 kV.
Fibril purification and Western blotting.
Fibrils were purified from DK1622 and LS2200 M. xanthus cells as described previously (22). Fibrils were quantified as a function of protein content according to the bicinchoninic acid method with bovine serum albumin as the standard (Pierce Chemical Co.) or carbohydrate content using the phenol-sulfuric acid assay with glucose as the standard (17). Cells (5 × 108) or 5 μg of protein from purified fibrils was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were electroblotted onto nitrocellulose and developed with a 1:500 dilution of primary antibody MAb 2105 and a 1:10,000 dilution of a secondary antibody composed of horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G. The immunoblot was developed with the ECL luminescence detection kit (Amersham Pharmacia, Inc.).
Cohesion assay.
The cohesion assay was similar to that described by Shimkets (36). M. xanthus cells (5 × 108) were resuspended in 1 ml of agglutination buffer in a spectrophotometer cuvette. The absorbance at 600 nm was recorded every 8 min.
Fruiting body formation and sporulation.
DK1622 and LS2200 cells were grown in CYE broth and resuspended to 5 × 109 cells/ml in MOPS buffer. Ten microliters of each suspension was spotted onto TPM (10 mM HCl [pH 7.6], 10 mM MgSO4, 1 mM K2HPO4, 1.5% agar [Difco]) plates and incubated at 32°C. Fruiting bodies were scored after 5 days by digital imaging. The plates were then incubated at 55°C for 4 h to kill vegetative cells. The fruiting bodies were harvested in 1 ml of MOPS buffer, sonicated, serially diluted in 3 ml of CYE containing 0.3% agar, and poured on CYE agar. Plates were incubated at 32°C for 5 days before counting of CFU.
Excitation assay.
The excitation assay measures changes in the reversal periods following exposure to a uniform concentration of PE and was performed as described previously (23). Rescue of excitation with purified fibrils was performed as described previously (22).
Nucleotide sequence accession number.
Sequences are available under GenBank accession number AF457462.
RESULTS
M. xanthus fibrils contain a zinc metalloprotease.
Many fibril proteins may be separated by SDS-PAGE of purified fibrils (4). This approach led to the discovery that a MAb raised against whole cells, MAb 2105, reacted with at least five fibril polypeptides ranging in size from 14 to 66 kDa. The 31 N-terminal amino acids from a 31-kDa integral fibril protein (IFP-31) that reacted with MAb 2105 were derived by Edman degradation (5). We submitted the N-terminal amino acid sequence to Cereon Genomics, LLC, for comparison with the unpublished M. xanthus genome sequence. The peptide coding region was identical to an internal sequence of a putative 79.8-kDa protein (Fig. 1). The coding region displayed a third-position mole percent G+C bias and a codon preference consistent with other known M. xanthus genes (38). Two potential translational start sites were identified but only one appeared to have a ribosome binding site. A BLAST search of the putative protein product revealed strong homology with the M4 family of zinc metalloproteases as follows: 39.5% identity (46.8% similarity) to thermolysin from Bacillus stearothermophilus and 34.1% identity (41.7% similarity) to elastase from Aeromonas hydrophila. The gene was named fibA (fibril protein A).
FIG. 1.
Amino acid sequence alignment of M. xanthus FibA with the AhpB elastase of A. hydrophila (accession number AAF07184) and LasB elastase of P. aeruginosa (accession number AAG07111). Open circles mark the FibA putative active site residues. The putative nucleotide binding P-loop is marked with closed circles. The putative type II secretion signal sequence is underlined with a solid black bar, while the Behmlander and Dworkin (5) amino acid sequence determined by N-terminal sequencing is underlined with a gray bar.
Zinc metalloproteases are widely distributed among prokaryotes and eukaryotes. They are usually secreted and are involved in a diverse range of activities, including bacterial pathogenesis as well as adhesion, cohesion, and movement of mammalian cells. In the active site a catalytic zinc atom is coordinated to three histidine residues and an activated water molecule is coordinated to a glutamate (9). The M. xanthus FibA amino acid sequence was compared with the LasB elastase from Pseudomonas aeruginosa (7) and the AhpB elastase from A. hydrophila (10), which revealed the conserved HExxH motif and a high level of similarity among the active site residues (Fig. 1). In particular, the LxYxxxxGxxNExxSD motif places FibA in the thermolysin/elastase family (20).
The comparison revealed three unusual features of FibA compared with other secreted zinc metalloproteases such as elastase. First, FibA is likely to be a lipoprotein. A putative lipoprotein secretion signal sequence is present at the FibA N terminus followed by the conserved cysteine (C21) that is presumably modified by the addition of diacylglycerol (Fig. 1). The second unusual feature of FibA is an insertion containing the consensus P-loop nucleotide binding motif (25). This domain typically forms a flexible loop between a β strand and an α helix and interacts with one of the phosphates of a nucleotide, typically ATP or GTP. The presence of this domain is unexpected, as FibA is an extracellular protein. However, this motif is sometimes found in proteins that do not bind nucleotides, such as chymotrypsin or human ferritin light chain. The third unusual feature of FibA is the presence of tandem copies of a highly conserved 107-amino-acid sequence at the C terminus which is found in a variety of unrelated proteases (data not shown). In the case of the A. hydrophila protease, the function of this domain is unknown and it is removed from the mature protein by autoprocessing (10). Proteolytic processing also appears to play a role in FibA structure, since the N-terminal sequence of IFP-31 determined by Behmlander and Dworkin (5) falls at the beginning of the first repeat.
FibA mutagenesis.
A 1-kb internal fragment of fibA was PCR amplified, cloned into pCR2.1 to generate pDB23, and electroporated into M. xanthus DK1622. A single homologous crossover between pDB23 and the chromosomal allele generated strain LS2200, a merodiploid containing two truncated copies of fibA (40). The adjacent genes are divergently transcribed, making it unlikely that the mutation is polar. The structure of the merodiploid was confirmed by PCR analysis (data not shown). The FibA mutant was compared with otherwise isogenic wild-type cells for the presence of fibrils and the phenotypic properties correlated with fibrils.
FibA is not essential for fibril biogenesis.
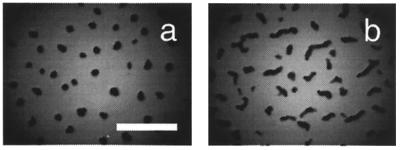
The presence of an extracellular matrix can be assayed in several ways. One assay involves binding of the dye Congo red to whole cells. When wild-type cells are placed on TPM agar containing 57 μM Congo red, the colony turns red after 6 h of incubation at 32°C (2). Matrix-deficient dsp colonies do not assimilate the dye (1). When LS2200 was spotted on TPM agar containing Congo red, the colony turned red, suggesting that fibrils are present in the mutant. Scanning electron microscopy revealed that LS2200 elaborated fibrils that are similar in structure and number to those of wild-type cells (Fig. 2). FibA does not appear to be essential for fibril assembly.
FIG. 2.
The fibA mutant is proficient in fibril biosynthesis. Scanning electron micrographs of DK1622 wild type (a) and LS2200 fibA::pDB23 (b) are shown. Cells were incubated for 30 min in cohesion buffer prior to fixation. Bar = 1 μm.
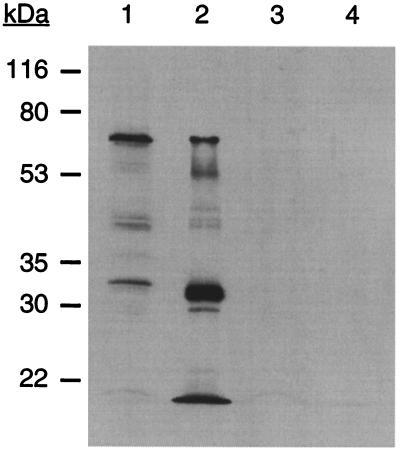
Members of the M4 metalloprotease family are typically self-processing and give protein profiles similar to those observed with MAb 2105. The N-terminal propeptide acts as an intramolecular chaperone that simultaneously inhibits protease activity in the cytoplasm and facilitates export (30). Both N-terminal and C-terminal processing sites have been observed in A. hydrophila elastase (10). In whole wild-type M. xanthus cells the most abundant protein that reacts with MAb 2105 has an apparent size of 66 kDa (Fig. 3, lane 1). Neither the prepropeptide (79.8 kDa) nor the form lacking the leader sequence (ca. 77 kDa) is observed. It is possible that processing occurs too rapidly to visualize the large forms or that the N-terminal lipid modification increases the electrophoretic mobility of the protein. In fibrils purified from wild-type cells, the 66-kDa form is visible, but the major form is about 31 kDa (Fig. 3, lane 2). Another abundant form is found at the dye front and has a size of <20.5 kDa.
FIG. 3.
The fibA mutant lacks all MAb 2105 reactive polypeptides. A Western blot with MAb 2105 as primary antibody is shown. Lane 1, lysate from 5 × 108 whole cells of wild-type DK1622; lane 2, 5 μg of protein from purified DK1622 fibrils; lane 3, lysate from 5 × 108 whole cells of LS2200 (fibA); lane 4, 5 μg of protein from purified LS2200 fibrils.
LS2200 (fibA::pDB23) does not produce any of the MAb 2105 antigens in whole cells (Fig. 3, lane 3) or purified fibrils (Fig. 3, lane 4), suggesting that the multiple protein forms are derived from FibA by posttranslational processing. It is unlikely that FibA positively regulates the production of a suite of fibril proteins, including those that react with MAb 2105. This point was examined by comparing wild-type and fibA fibrils on silver-stained SDS-PAGE gels. Except for small differences in the 66-, 52-, and 31-kDa regions of the gel, the vast majority of the fibril proteins were identical in size and quantity (data not shown). These data suggest that the MAb 2105 reactive proteins are derived from FibA.
fibA retains cohesion and motility.
One function of M. xanthus fibrils is cohesion, which is used to attach to other cells during biofilm formation (3). When wild-type M. xanthus cells are resuspended in starvation buffer containing Mg2+ and Ca2+, the cells agglutinate and fall out of suspension, a process that can be measured as a decrease in absorbance. A dsp mutant which is deficient in fibril production did not agglutinate, while LS2200 agglutinated at a rate similar to that of the wild type, demonstrating that FibA is not essential for cohesion (not shown).
M. xanthus gliding appears to be the product of two different motility motors (18, 19). One motor, which enables movement of isolated cells, powers the adventurous motility (A-motility) system; the mechanism of A-motility is not known. The other motor, which controls the movement of groups of cells, powers the social motility (S-motility) system. S-motility is analogous to twitching and uses retraction of polar type IV pili to power motility (29, 41, 43). Only when both systems are inactivated are the cells nonmotile. The dual propulsion systems provide a simple genetic assay for classifying motility mutants (18, 19). For example, if an A-motility mutation is transduced into an A− S+ strain, the double mutation in the A system has no further deleterious effect and the recipient exhibits S-motility. If, however, the same locus is transduced into an A+ S− strain, then the combination of the A-motility and S-motility defects renders the recipient nonmotile. M. xanthus cells remained motile when the fibA::pDB23 allele was placed in either an S-motile background (aglB3) or an A-motile background (dsp-1693 or pilQ1), suggesting that fibA is not essential for either motility system (data not shown).
fibA is defective in dilauroyl PE excitation.
In response to nutrient depletion, M. xanthus cells construct a fruiting body in which cells sporulate (Fig. 4a). The extracellular matrix is essential for fruiting body development as dsp mutants are blocked early in aggregation and fail to form appreciable levels of spores (39). The fibA mutation does not produce the serious developmental defect observed in a fibril-less dsp mutant. The fibA mutant produced elongated ridges that involved considerable aggregation but never consolidated into well-spaced fruiting bodies (Fig. 4b). The fibA mutant formed nearly normal levels of viable spores (95% of the wild type).
FIG. 4.
fibA produces elongated fruiting bodies. Wild-type (a) and fibA::pDB23 LS2200 (b) cells (5 × 107) were starved on TPM agar for 5 days to induce fruiting body formation. Bar = 2 mm.
The mechanism of M. xanthus chemotaxis relies on some features that have been described for flagellated organisms, though M. xanthus differs in the mechanism of motility. Escherichia coli swims through liquid media by means of rotary flagella and punctuates smooth swimming with tumbling to acquire new trajectories. M. xanthus cells glide along relatively straight paths on surfaces and reverse their direction of movement once every 6.8 min (8). Direction reversals are analogous to E. coli tumbles. Excitation by chemical attractants causes an increase in the length of time between E. coli tumbles (6) or M. xanthus reversals (23).
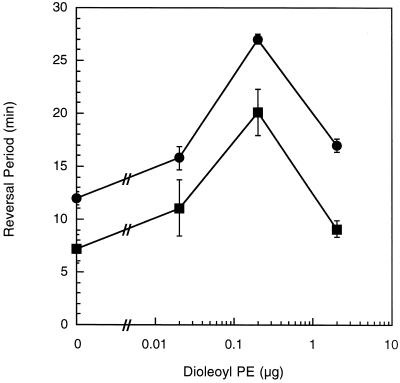
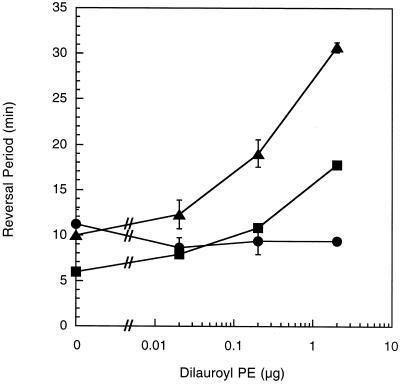
Excitation is measured by placing cells on a surface that has been coated by a uniform concentration of the stimulus and monitoring cell behavior. Changes in the duration of the reversal period are measured prior to the beginning of adaptation. The fibA mutant displayed an increased reversal period in the absence of either dioleoyl PE (Fig. 5) or dilauroyl PE (Fig. 6). The basal reversal period of 12 to 13 min is higher than the approximately 7-min reversal period of wild-type cells but lower than the 16- to 22-min reversal period of mutants lacking fibrils (22, 23). These results suggest that the fibA mutant has a phenotype similar to that of mutants lacking fibrils, though not as severe (22). While both dilauroyl PE and dioleoyl PE trigger excitation (23), the loss of fibrils eliminated excitation to dilauroyl but not dioleoyl PE (22). The fibA cells demonstrated excitation to dioleoyl PE (Fig. 5) but not dilauroyl PE (Fig. 6), similar to fibril-deficient mutants (22). These results suggest that FibA is part of the sensory pathway that regulates the activity of the motility motors in response to dilauroyl PE.
FIG. 5.
fibA has normal excitation to dioleoyl PE. The reversal periods of fibA::pDB23 LS2200 (circles) and the wild type (DK1622) (squares). Bars show the standard deviations of three replicates. The wild-type data are derived from that of Kearns et al. (22).
FIG. 6.
fibA is deficient in excitation to dilauroyl PE. The reversal periods of fibA::pDB23 LS2200 (circles), the wild type (DK1622) (squares), and LS2200 cells incubated with purified wild-type fibrils (triangles). Bars show the standard deviations of three replicates.
Since the fibrils are surface appendages that can contact other cells, we examined whether fibrils purified from wild-type cells would correct the excitation deficiency in fibA::pDB23. Wild-type fibrils were previously shown to restore dilauroyl PE excitation to mutants lacking fibrils. In this experiment fibrils are incubated with fibA cells for a period of time prior to exposure to PE. Wild-type fibrils complemented the mutational defect and restored excitation to dilauroyl PE (Fig. 6). In the absence of PE the fibrils did not increase the reversal period, so it is unlikely that the fibrils are contaminated with an endogenous attractant. Rather, the complementation suggests that wild-type fibrils associate with the fibA mutant in spite of the presence of fibA fibrils. Perhaps there are more fibril binding sites on the cells than are occupied by native fibrils. Fibrils prepared from the fibA mutant LS2200 did not restore excitation to the fibA mutant with 2 μg of dilauroyl PE, suggesting that the complementation is specific (reversal period = 9.00 ± 0.10). Unlike in fibril-less dsp mutants, the addition of wild-type fibrils did not decrease the fibA basal reversal period closer to that of the wild type (Fig. 6) (23).
DISCUSSION
The M. xanthus extracellular matrix is involved in a variety of processes, including adhesion, cohesion, motility, fruiting body formation, and excitation by dilauroyl PE (14, 24). An understanding of the molecular mechanisms by which the extracellular matrix facilitates these processes demands knowledge of the functions of the matrix proteins. We report the identification and functional analysis of the first matrix protein. FibA, which is homologous to members of the M4 family of zinc metalloproteases, is the target of MAb 2105. Previously, immunogold electron microscopy demonstrated localization of MAb 2105 to an extracellular matrix antigen (4). As the fibA mutant lacks all of the MAb 2105 reactive antigens observed with SDS-PAGE, it appears that the multiple sizes of polypeptides observed in wild-type cells are autoprocessed products of FibA. This hypothesis is consistent with observations of autoprocessing in other members of the elastase family (20, 27).
The fibA mutant displayed normal fibril formation, cohesion, excitation to dioleoyl PE, and A- and S-motility. The mutant demonstrated substantial developmental aggregation but failed to form well-spaced fruiting bodies. The mutant exhibited an unusually long reversal period that was not increased by dilauroyl PE. In previous work we have provided evidence that fibrils initiate a sensory cascade for the perception of dilauroyl PE (22). The presence of dilauroyl PE increases the reversal period, as might be expected of a chemical attractant, and excitation is lost in cells lacking fibrils (22). Fibrils purified from wild-type cells complemented the dilauroyl PE excitation deficiency in fibA mutants, confirming the location of this protein on fibrils and the involvement of this protein in chemotaxis.
Over 20 mammalian matrix metalloproteases are known and the selective degradation of matrix proteins by their cognate metalloproteases is essential for such diverse processes as embryogenesis, wound healing, connective tissue remodeling, and the inflammatory response (44). Many of the metalloprotease targets have been identified and include matrix collagens, elastin, fibronectin, and different metalloproteases (15). Excessive protease activity is responsible for notable pathologies, such as tumor metastasis (12), arthritis (35), multiple sclerosis (42), chronic ulcers (34), and Alzheimer's disease (32). Accordingly, the activity of each enzyme is regulated at the transcriptional and posttranslational levels. Matrix metalloprotease activity is inhibited in vivo by the propeptide during latency and through noncovalent interaction with a cognate tissue inhibitor of metalloprotease (TIMP) after activation (9, 15). In both cases inactivation of the protease occurs through interaction of the inhibitor with the catalytic zinc (9). The activation of each metalloprotease is thought to be mediated by distinct physiological cues, some of which activate the protease though hydrolysis of the propeptide and some of which inactivate TIMP. However, the physiological activators remain unknown.
The proteolytic product of one mammalian metalloprotease interfaces with a sensory pathway to modulate chemotaxis. Monocyte chemoattractant protein 3 is the physiological substrate of the metalloprotease gelatinase A (28). The hydrolytic product acts as a receptor antagonist to dampen the inflammatory response by binding to the chemokine receptors but fails to induce chemotaxis or the requisite calcium fluxes. Based upon the results with the mammalian systems, one can begin to devise a framework for studying the role of FibA in regulation of motility and sensory transduction. At the heart of the matter is the identity of the proteolytic substrate of FibA. Since the fibA mutant has an unstimulated reversal period about twice as long as that of wild-type cells, the proteolytic substrate may modulate reversal frequency. By analogy with the monocyte chemoattractant 3 story, there may be competition for binding at the gliding motor between the FibA substrate and its proteolytic product to regulate reversals.
Another question of great interest is the manner by which dilauroyl PE decreases reversal frequency. Perhaps the simplest possibility is that dilauroyl PE alters the activity of FibA or its cognate TIMP (if there is one). It would appear that this notion is too simple since the reversal period of about 10 min observed in the FibA mutant (Fig. 6) is less than the 14- to 16-min reversal period observed with wild-type cells in the presence of dilauroyl PE. Another possibility is suggested by the observation that a fibril protein similar in size to mature FibA is ADP ribosylated (16). As isolated fibrils carry out ADP ribosylation in vitro it appears that the ADP ribosyl transferase is also associated with the fibrils. In this case the lipid could modulate the activity of the transferase and regulate proteolytic activity indirectly. These and other intriguing aspects of this system will likely be resolved with the purification of FibA and the identification of the FibA substrate.
Acknowledgments
We are grateful to M. Dworkin for the gift of MAb 2105. We are grateful to Bill Timberlake and Steve Slater of Cereon Genomics, LLC, for generously providing the sequence of fibA prior to public release. We thank John Shields and Mark Farmer for their assistance with the electron microscopy.
This material is based on work supported by the National Science Foundation under grant no. MCB0090946.
REFERENCES
- 1.Arnold, J. W., and L. J. Shimkets. 1988. Cell surface properties correlated with cohesion in Myxococcus xanthus. J. Bacteriol. 170:5771-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, J. W., and L. J. Shimkets. 1988. Inhibition of cell-cell interactions in Myxococcus xanthus by Congo red. J. Bacteriol. 170:5765-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behmlander, R., and M. Dworkin. 1994. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. J. Bacteriol. 176:6295-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behmlander, R. M., and M. Dworkin. 1991. Extracellular fibrils and contact-mediated cell interactions in Myxococcus xanthus. J. Bacteriol. 173:7810-7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behmlander, R. M., and M. Dworkin. 1994. Integral proteins of the extracellular matrix fibrils of Myxococcus xanthus. J. Bacteriol. 176:6304-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, H. C., and D. A. Brown. 1972. Chemotaxis in Escherichia coli analyzed by three-dimensional tracking. Nature 239:500-504. [DOI] [PubMed] [Google Scholar]
- 7.Bever, R. A., and B. H. Iglewski. 1988. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J. Bacteriol. 170:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackhart, B. D., and D. R. Zusman. 1985. Frizzy genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc. Natl. Acad. Sci. USA 82:8767-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borkakoti, N. 2000. Structural studies of zinc metalloproteinases. J. Mol. Med. 78:261-268. [DOI] [PubMed] [Google Scholar]
- 10.Cascon, A., J. Yugueros, A. Temprano, M. Sanchez, C. Hernanz, J. M. Luego, and G. Naharro. 2000. A major secreted elastase is essential for pathogenicity of Aeromonas hydrophila. Infect. Immun. 68:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, B.-Y., and M. Dworkin. 1994. Isolated fibrils rescue cohesion and development in the dsp mutant of Myxococcus xanthus. J. Bacteriol. 176:7190-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curran, S., and G. I. Murray. 2000. Matrix metalloproteinases: molecular aspects of their roles in tumour invasion and metastasis. Eur. J. Cancer 36:1621-1630. [DOI] [PubMed] [Google Scholar]
- 13.Dana, J. R., and L. J. Shimkets. 1993. Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. J. Bacteriol. 175:3636-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dworkin, M. 1999. Fibrils as extracellular appendages of bacteria: their role in contact-mediated cell-cell interactions in Myxococcus xanthus. BioEssays 21:590-595. [DOI] [PubMed] [Google Scholar]
- 15.Ellerbroek, S. M., and M. S. Stack. 1999. Membrane associated matrix metalloproteinases in metastasis. Bioessays 21:940-949. [DOI] [PubMed] [Google Scholar]
- 16.Hildebrandt, K., D. Eastman, and M. Dworkin. 1997. ADP-ribosylation by the extracellular fibrils of Myxococcus xanthus. Mol. Microbiol. 23:231-235. [DOI] [PubMed] [Google Scholar]
- 17.Hodge, J. E., and B. T. Hofreiter. 1962. Determination of reducing sugar and carbohydrates, p. 380-394. In R. L. Whistler and L. Wolfrom (ed.), Methods in carbohydrate chemistry, vol. 1. Academic Press, New York, N.Y. [Google Scholar]
- 18.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167-176. [Google Scholar]
- 19.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 20.Jiang, W., and J. S. Bond. 1992. Families of metalloendopeptidases and their relationships. FEBS Lett. 312:110-114. [DOI] [PubMed] [Google Scholar]
- 21.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 22.Kearns, D. B., B. D. Campbell, and L. J. Shimkets. 2000. Myxococcus xanthus fibril appendages are essential for excitation by a phospholipid attractant. Proc. Natl. Acad. Sci. USA 97:11505-11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearns, D. B., and L. J. Shimkets. 1998. Chemotaxis in a gliding bacterium. Proc. Natl. Acad. Sci. USA 95:11957-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kearns, D. B., and L. J. Shimkets. 2001. Lipid chemotaxis and signal transduction in Myxococcus xanthus. Trends Microbiol. 9:126-129. [DOI] [PubMed] [Google Scholar]
- 25.Koonin, E. V. 1993. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 229:1165-1174. [DOI] [PubMed] [Google Scholar]
- 26.Lancero, H., J. E. Brofft, J. Downard, B. W. Birren, C. Nusbaum, J. Naylor, W. Shi, and L. J. Shimkets. 2002. Mapping of Myxococcus xanthus social motility dsp mutations to the dif genes. J. Bacteriol. 184:1462-1465. [DOI] [PMC free article] [PubMed]
- 27.McIver, K. S., E. Kessler, J. C. Olson, and D. E. Ohman. 1995. The elastase propeptide functions as an intramolecular chaperone required for elastase activity in Pseudomonas aeruginosa. Mol. Microbiol. 18:877-889. [DOI] [PubMed] [Google Scholar]
- 28.McQuibban, G. A., J.-H. Gong, E. M. Tam, C. A. G. McCulloch, I. Clark-Lewis, and C. M. Overall. 2000. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science 289:1202-1206. [DOI] [PubMed] [Google Scholar]
- 29.Merz, A., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-101. [DOI] [PubMed] [Google Scholar]
- 30.O'Donohue, M. J., and A. Beaumont. 1996. The roles of the prosequence of thermolysin in enzyme inhibition and folding in vitro. J. Biol. Chem. 271:26477-26481. [DOI] [PubMed] [Google Scholar]
- 31.Parducz, B. 1967. Ciliary movement and coordination in ciliates. Int. Rev. Cytol. 21:91-128. [DOI] [PubMed] [Google Scholar]
- 32.Peress, N., E. Perillo, and S. Zucker. 1995. Localization of tissue inhibitor of matrix metalloproteinases in Alzheimer's disease and normal brain. J. Neuropathol. Exp. Neurol. 54:16-22. [DOI] [PubMed] [Google Scholar]
- 33.Ramaswamy, S., M. Dworkin, and J. Downard. 1997. Identification and characterization of Myxococcus xanthus mutants deficient in calcofluor white binding. J. Bacteriol. 179:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravanti, L., and V. M. Kahari. 2000. Matrix metalloproteinases in wound repair. Int. J. Mol. Med. 6:391-407. [PubMed] [Google Scholar]
- 35.Shaw, T., J. S. Nixon, and K. M. Bottomley. 2000. Metalloproteinase inhibitors: new opportunities for treatment of rheumatoid arthritis and osteoarthritis. Expert Opin. Investig. Drugs 9:1469-1478. [DOI] [PubMed] [Google Scholar]
- 36.Shimkets, L. J. 1986. Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus. J. Bacteriol. 166:837-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimkets, L. J. 1999. Intercellular signaling during fruiting body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 38.Shimkets, L. J. 1993. The myxobacterial genome, p. 85-107. In M. Dworkin and D. Kaiser (ed.), Myxobacteria II. American Society for Microbiology, Washington, D.C.
- 39.Shimkets, L. J. 1986. Role of cell cohesion in Myxococcus xanthus fruiting body formation. J. Bacteriol. 166:842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimkets, L. J., and S. J. Asher. 1988. Use of recombination techniques to examine the structure of the csg locus of Myxococcus xanthus. Mol. Gen. Genet. 211:63-71. [DOI] [PubMed] [Google Scholar]
- 41.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinman, L. 1996. Multiple sclerosis: a coordinated immunological attack against myelin in the central nervous system. Cell 85:299-302. [DOI] [PubMed] [Google Scholar]
- 43.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143-1146. [DOI] [PubMed] [Google Scholar]
- 44.Vu, T. H., and Z. Werb. 2000. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 14:2123-2133. [DOI] [PubMed] [Google Scholar]
- 45.Yang, Z., X. Ma, T. Leming, H. B. Kaplan, L. J. Shimkets, and W. Shi. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182:5793-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]