Abstract
Plasmids of incompatibility group P (IncP) are capable of replication and stable inheritance in a wide variety of gram-negative bacteria. Three determinants of IncP plasmids are components of an active partition locus that is predicted to function in the segregation of plasmid copies to daughter cells. These determinants are incC, which codes for a member of the ParA family of partition ATPases; korB, which specifies a DNA-binding protein that also functions as a global transcriptional repressor; and OB, the DNA target for KorB, which occurs at multiple locations on IncP plasmids. To determine the importance and host range of the IncC/KorB partition system in the maintenance of IncP plasmids, we constructed an in-frame deletion of incC in the otherwise intact 60-kb IncPα plasmid R995. R995ΔincC was found to be highly unstable in Escherichia coli, Pseudomonas aeruginosa, Pseudomonas putida, Agrobacterium tumefaciens, and Acinetobacter calcoaceticus, whereas wild-type R995 is stable in all these hosts. In addition, R995ΔincC could not be established in Actinobacillus actinomycetemcomitans. trans-Complementation analysis showed that the coding region for IncC2 polypeptide, which is expressed from an internal translational start within the incC gene, was sufficient to restore stable maintenance to wild-type levels. The results show that the IncC/KorB active partition system of IncP plasmids is remarkably proficient for stable maintenance in diverse bacteria.
Active partition of bacterial plasmids and chromosomes into newly forming daughter cells is the functional equivalent of mitosis in eukaryotes (10, 14, 16, 19, 29). Early studies on plasmid maintenance and plasmid-plasmid incompatibility led to the identification of genetic loci for active partition (2, 27, 31-33). From sequence analysis, such loci are now known to be widespread among plasmids and bacterial chromosomes (12, 18, 51). The prototypical active partition locus consists of an autoregulated operon of two genes and a cis-acting DNA element functionally analogous to a chromosomal centromere. The genes specify an ATPase and a DNA-binding protein specific for the cis-acting element (3, 12). The partition locus constitutes a functional cassette sufficient to stabilize heterologous, unstable plasmids (1, 11, 27, 33). In a widely accepted model for active partition, replicated DNA molecules are paired in the form of nucleoprotein complexes containing the partition ATPase and the site-specific DNA-binding protein. The paired complexes are proposed to interact with a host DNA segregation apparatus at midcell, which facilitates the separation of the molecules and their translocation towards opposite poles of the cell (4, 21). The nucleoprotein partition complexes of plasmids P1, F, and R1 are understood in considerable molecular detail (8, 17, 21, 25, 37, 44). Additional critical support for the model has emerged from remarkable fluorescence microscopy studies of bacteria showing specific localization and migration of both plasmid and chromosomal DNA (14, 16, 19, 20, 36, 48). However, virtually nothing is known about the identity of the bacterial segregation apparatus or the specific events following the formation of nucleoprotein complexes.
The mechanism for ensuring the segregation of DNA in dividing bacterial cells promises to be especially interesting for the remarkable broad-host-range plasmids of incompatibility group P (IncP). These plasmids are stably maintained as autonomously replicating elements in a wide spectrum of gram-negative bacteria (9, 34). To understand how IncP plasmids are able to persist in markedly different bacterial hosts, it will be necessary to identify their strategies for stable inheritance and to learn how these systems interact productively with different hosts. From studies on the 60,099-bp IncPα plasmid RK2 (35), it is clear that stable plasmid maintenance in diverse hosts is not simply a function of replication control and that other plasmid determinants are required (42). Several stabilization loci have been identified thus far, including the parCBA operon, which specifies a multimer resolution system that maintains the normal plasmid copy number by reducing plasmid multimers to monomers (13, 38); the parDE operon, which encodes a plasmid addiction system that is toxic to plasmidless segregants (22, 39); and the kilE locus, which specifically stabilizes RK2 in Pseudomonas aeruginosa by an as yet unknown mechanism (50). In addition, RK2 encodes an active partition system with strong similarities to, but also notable differences from, partition systems common among other plasmids (30, 41). However, the significance of this partition system to the maintenance and host range of IncP plasmids has not been addressed.
Active partition in IncP plasmids was first indicated by Meyer and Hinds (28), who found an incompatibility determinant indicative of a maintenance function in the RK2 korA operon, which encodes the transcriptional repressors KorA and KorB. The gene responsible for incompatibility (incC) overlaps korA in a different reading frame and extends to the beginning of korB (Fig. 1) (45). incC expresses two polypeptides: full-length IncC1 (38.1 kDa) and the shorter IncC2 (27.5 kDa), which is initiated from an internal translation start site (24, 45). Based on their amino acid sequences, both polypeptides are members of the large family of predicted ATPases related to the ParA partition ATPase of plasmid P1 (12, 30). Fluorescence microscopy studies have shown that RK2 displays localization and migration behavior in Escherichia coli consistent with an active partition system (36). Recently, we reported that IncC protein interacts with KorB in vivo and in vitro and that IncC2, KorB, and one KorB-binding site (OB) are sufficient to confer the replication-independent incompatibility properties characteristic of an active partition system (41). These three elements are the RK2 counterparts to the partition ATPase, DNA-binding protein, and cis-acting DNA element, respectively, of the prototypical partition system. However, unlike in other systems, these RK2 partition elements are not sufficient for stabilization of a heterologous replicon in E. coli. A larger region of RK2 containing additional genes and OB sites is partially able to stabilize plasmids in E. coli in an incC-dependent manner (7, 41). Thus, while incC, korB, and an OB site are components of an active partition system, the genetic structure of the minimal IncP active partition system has not yet been determined.
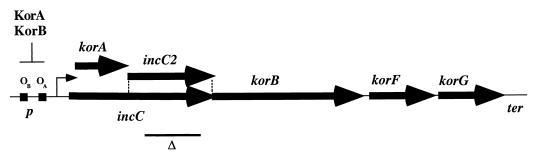
FIG. 1.
korA operon of IncPα plasmids. Genes are shown as bold arrows. The korA gene is within the incC coding sequence but in a different reading frame. incC2, the coding region for the IncC2 polypeptide translated from an internal initiation site in incC; line Δ, the 465-bp region deleted in R995ΔincC (pR9401); korF and korG, genes for basic proteins of unknown function (35); p, promoter; OA and OB, binding sites for the KorA and KorB repressors, respectively; angled arrow, transcriptional start site; ter, a putative transcriptional terminator.
In this study we sought to address a fundamental question concerning the IncP active partition system: is the system important for stable maintenance in a wide variety of bacteria or is it functional only in a subset of specific hosts? The answer has considerable significance. If the IncP system is host specific, then it will be critical to identify the appropriate host(s) before further analysis of the partition system can be undertaken. If the IncP active partition system is found to function in a broad range of hosts, then it will be important to understand how such a system can be expressed appropriately and function efficiently in very different hosts.
IncC is required for stable maintenance of R995 in E. coli.
We studied the function of the IncC/KorB partition system in the context of the natural ampicillin-sensitive IncPα plasmid R995, which is essentially equivalent to RK2 except for the absence of Tn1 in the kilC operon (Table 1) (47). The nucleotide sequence of the R995 incC gene is identical to that of RK2 (unpublished results). A derivative of R995 with a deletion of the incC gene (pR9401) was constructed by allelic exchange with a mutated incC subclone (Table 1). The deletion mutant is missing 465 bp of the 780-bp coding region of incC2, which lies between korA and korB (Fig. 1). The mutation is not expected to affect the regulation of the korA operon, whose transcription is controlled by the KorA and KorB repressors (Fig. 1). The in-frame deletion of incC2 leaves korA intact, and it is predicted to be nonpolar on expression of the downstream korB gene. This property was confirmed in E. coli by Western blot analysis using polyclonal antiserum specific for the KorB protein (data not shown).
TABLE 1.
Plasmids
| Plasmid | Marker(s)a | Relevant genotype | Description | Reference or source |
|---|---|---|---|---|
| pJAK16 | Cmr | lacIqtacp | IncQ replicon; expression vector | J. Kornacki |
| pRK21985 | Cmr | lacIq Φ(tacp-incC2) | pJAK16 with incC2 coding region expressed from tacp | 41 |
| pRK22324 | Apr | ΔincC | P15A replicon carrying the korA incC korB region of RK2 with 465-bp, in-frame deletion within incC | 41 |
| pR9401b | Kmr Tcr | ΔincC | R995 with 465-bp, in-frame deletion within incC | This work |
| R995 | Kmr Tcr | incC+ | Natural IncPα plasmid | 47 |
Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin; Tc, tetracycline; r, resistance.
A pRK22324/R995 cointegrate was selected by conjugation of E. coli donor cells containing both plasmids with a Nalr recipient. The cointegrate was transferred to pcnB mutant strain LS1443 (26), in which the P15A replicon of pRK22324 is unstable. Resolved cointegrates were screened for Aps, and the ΔincC mutation was detected by PCR. pR9401 was confirmed by restriction analysis.
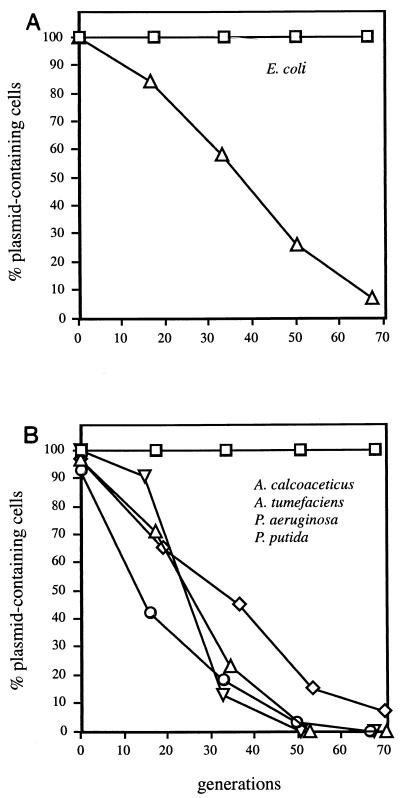
Wild-type R995 is completely stable in E. coli, with no detectable loss of the plasmid during 65 generations of unselected growth (Fig. 2A). In contrast, the R995ΔincC mutant was found to be highly unstable in E. coli, with 50% of the population having lost the plasmid in 35 to 40 generations of unselected growth (Fig. 2A). To confirm that the high loss rate of R995ΔincC resulted from the absence of the incC gene product, we tested the ability of incC2 to complement the ΔincC mutation in trans. The RK2 incC2 coding region was inserted downstream of the inducible tac promoter in a compatible IncQ plasmid vector, pJAK16, to generate pRK21985 (Table 1). The maintenance of R995ΔincC was then assayed in E. coli cells containing either the pJAK16 vector or the tacp-incC2 derivative (Fig. 3A). The pJAK16 vector showed no ability to stabilize R995ΔincC. In contrast, uninduced expression of incC2 from pRK21985 (without isopropyl-β-D-thiogalactopyranoside [IPTG]) allowed complete stabilization of R995ΔincC to wild-type levels. No plasmid loss was detected in 65 generations. With the pJAK16 vector in trans, 95% of the cells emerging over the same period lacked R995ΔincC. At 1 mM IPTG, R995ΔincC was destabilized, as expected from our previous results showing that elevated expression of incC2 causes rapid loss of IncP plasmid RK2 (41). Thus, low-level expression of incC2 from the leaky tacp promoter is sufficient to fully complement the ΔincC mutation. We conclude that incC is required for the stable maintenance of R995 in E. coli.
FIG. 2.
Stability of R995 and R995ΔincC in different hosts. (A) E. coli host. R995 (□) and R995ΔincC (Δ) in E. coli DF4063 (43). (B) Other gram-negative hosts. R995ΔincC in A. calcoaceticus BD413 (42) (◊), A. tumefaciens A136 (42) (▿), P. aeruginosa PAC452 (43) (Δ), and P. putida ATCC 12633 (6) (○). The results for all hosts with wild-type R995 were indistinguishable from the results for P. aeruginosa, which are displayed here (□). Strains were grown to saturation in the absence of selection and then diluted 105-fold in fresh broth. This procedure was repeated until the strains had grown for at least 65 generations. Growth conditions for all bacteria have been described previously (6, 42, 43). Colonies were screened for the presence of plasmids on medium containing kanamycin or tetracycline.
FIG. 3.
Complementation of R995ΔincC by incC2 in trans. (A) E. coli DF4063 host; (B) P. aeruginosa PAC452 host. Results for strains containing R995ΔincC and pJAK16 (tacp vector) without IPTG (□) and with 1 mM IPTG (◊), and containing R995ΔincC and pRK21985 (tacp-incC2) without IPTG (○) and with 1 mM IPTG (Δ) are shown. Assays were done as described in the legend to Fig. 2, except that selection for pJAK16 and pRK21985 was maintained.
Agarose gel electrophoresis revealed no differences in the amounts of R995 and R995ΔincC relative to that of a coresident IncQ plasmid (data not shown). Thus, the R995 copy number was not affected by the ΔincC mutation. Conjugative plasmids can promote their maintenance in a cell line by self-transfer to plasmidless segregants (43), raising the possibility that the ΔincC mutation affects conjugative transfer. However, the R995ΔincC mutant was found to transfer at the same frequency as that of the wild type, R995, in matings of E. coli with E. coli (data not shown). Thus, the properties of R995ΔincC are consistent with a defect in DNA segregation.
IncC is required for stable maintenance of R995 in diverse gram-negative hosts.
We compared the maintenance properties of R995 and R995ΔincC in broth cultures of P. aeruginosa, Pseudomonas putida, Acinetobacter calcoaceticus, and Agrobacterium tumefaciens (Fig. 2B). As expected for an IncP plasmid, R995 was stably maintained in all these hosts. In striking contrast, R995ΔincC was rapidly lost from growing populations of each of these hosts. To confirm trans complementation in a non-E. coli host, we placed the broad-host-range IncQ plasmid with tacp-incC2 (pRK21985) and the tacp vector control (pJAK16) in trans to R995ΔincC in P. aeruginosa. Similar to the results observed for E. coli, low-level expression of incC was sufficient to fully stabilize the R995ΔincC mutant in P. aeruginosa and high-level expression destabilized the plasmid (Fig. 3B). The results show that incC is required for stable maintenance of R995 in the four gram-negative hosts.
We encountered a severe phenotype expressed by R995ΔincC when we attempted to transfer the plasmid into Actinobacillus actinomycetemcomitans for stability assays. Wild-type R995 readily formed healthy transconjugants, but R995ΔincC produced transconjugant colonies at a frequency lower than 10−4 that of R995 (Table 2). The R995ΔincC transconjugant colonies that did appear were pinpoint and grew poorly, if at all, when restreaked on selective medium. Conjugative transfer of R995ΔincC was shown above to be normal with an E. coli recipient. To confirm that conjugative transfer to A. actinomycetemcomitans was also normal, we examined the ability of R995ΔincC to mobilize the IncQ plasmid pJAK16 to this host. IncQ plasmids are not able to self-transfer, but they are efficiently mobilized by the IncP plasmid transfer apparatus and they replicate in A. actinomycetemcomitans (15). We found that R995 and R995ΔincC mobilized pJAK16 from E. coli to A. actinomycetemcomitans at the same frequencies (Table 2). Thus, the failure to obtain R995ΔincC transconjugants of A. actinomycetemcomitans indicates a defect in plasmid establishment or maintenance in this host and leads to the prediction that incC in the recipient should complement the defect. This prediction was confirmed in matings with an A. actinomycetemcomitans recipient carrying the tacp-incC2 plasmid pRK21985. Healthy transconjugants of R995ΔincC appeared at the same frequency as those of wild-type R995 (Table 2). These results show that incC is essential for stable inheritance of R995 in A. actinomycetemcomitans.
TABLE 2.
Phenotype of R995ΔincC in A. actinomycetemcomitans
| Plasmid(s) in donora | Plasmid in recipientb | Relative no. of transconjugantsc |
|---|---|---|
| R995 | NA | 1.0 |
| R995ΔincC | NA | <10−4 |
| R995, pJAK16 | NA | 1.0, 0.9d |
| R995ΔincC, pJAK16 | NA | <10−4, 1.0d |
| R995 | tacp vector | 1.0 |
| R995ΔincC | tacp vector | <10−4 |
| R995ΔincC | tacp-incC2 | 1.0 |
E. coli LS1443 (26).
A. actinomycetemcomitans Y4Nal (46). NA, not applicable.
Four-hour matings at 37°C were done as described previously (46). Transconjugants were selected on media containing a combination of the following antibiotics: kanamycin (20 μg/ml), chloramphenicol (2 μg/ml), and nalidixic acid (20 μg/ml). The relative number of transconjugants was obtained by dividing the number of transconjugants per donor for the test plasmid by the number of transconjugants per donor for R995 for each experiment. Results are the average of two experiments.
Relative numbers of R995 and pJAK16 transconjugants, respectively.
Significance.
This study was undertaken to determine the importance and host range of the IncC/KorB active partition system of promiscuous IncP plasmids. The results show unequivocally that incC is required for stable maintenance of IncP plasmid R995 in E. coli and five other gram-negative hosts. In each case, the nonpolar ΔincC mutation of an otherwise wild-type IncP plasmid caused the plasmid to be lost rapidly from growing cultures. Furthermore, the shorter incC2 gene in trans was sufficient to restore a wild-type level of stability. The function of the full-length incC gene remains to be determined.
The R995ΔincC loss rates appear to be greater than those expected for a randomly diffusing plasmid with a copy number of 10 to 20 at cell division (5, 50). Recent fluorescence microscopy studies by Pogliano et al. (36) and Weitao et al. (49) may suggest an explanation. Their studies showed that multicopy plasmids are not randomly distributed in cells and that they may organize in clusters. Alternatively, abortive partition complexes may be formed in the absence of IncC, such that plasmids are localized but fail to segregate (7).
The significant rates of R995ΔincC loss observable for all hosts tested also underline the importance of the active partition system to the broad host range of IncP plasmids. R995ΔincC carries the determinants for multimer resolution (parCBA) and plasmid addiction (parDE), both of which function to stabilize IncP plasmids to varying degrees in these hosts (43). In addition, the kilE locus needed for maintenance in P. aeruginosa is intact. There is no reason to expect that multimer resolution and plasmid addiction are dependent upon the IncC protein, and indeed studies with a derivative of R995 with deletions of both the incC gene and the par region, which contains parCBA and parDE, show that the phenotypes are additive, as expected for independent pathways. At 33 generations, R995Δpar was present in 98% of cells, R995ΔincC was present in 75% of cells, and R995ΔparΔincC was present in 6% of cells. It is not yet known whether the kilE locus acts independently of incC.
Active partition systems are generally considered to be nonessential accessory functions that enable replicons to be stably maintained by their bacterial hosts. However, A. actinomycetemcomitans cells that have received R995ΔincC do not form colonies under selection. The parDE postsegregational toxicity system is not responsible for this phenotype, since the removal of parDE from the R995ΔincC mutant did not cause a change in phenotype (unpublished results). Clearly the absence of incC in A. actinomycetemcomitans has serious consequences, and there are several possible explanations for the phenotype. While the level of KorB is unchanged by the absence of IncC in E. coli, it remains possible that it is affected in A. actinomycetemcomitans. Increased levels of KorB could inhibit replication, whereas decreased levels of KorB may allow derepression of toxic plasmid genes (35). It has also been observed for plasmids F and P1 that, under certain circumstances, the DNA-binding protein causes the silencing of several kilobases of DNA surrounding the cis-acting DNA-binding site (17, 40). Perhaps a similar silencing of R995ΔincC is induced in A. actinomycetemcomitans by the absence of IncC. Another possibility is that the absence of IncC causes IncP plasmids to aggregate, thereby blocking their segregation to daughter cells. Such a phenotype has been suggested for partition mutants of P1 (5). The failure to form colonies might also be caused by inhibition of cell growth, which might, for example, be due to interference with the host DNA segregation apparatus caused by its interaction with an incomplete IncP partition system. The strong phenotype in A. actinomycetemcomitans may prove useful for selecting plasmid or host mutants that reveal other components or properties of the IncP partition system.
How is the IncP partition system able to function efficiently in a broad range of hosts? Recent experiments have shown that the Soj/Spo0J chromosomal partition system of Bacillus subtilis can function in E. coli, suggesting that DNA segregation machinery is highly conserved in bacteria. It is therefore possible that the broad-host-range IncP partition system has evolved to fully exploit the conserved features of bacterial systems for efficient function in multiple hosts. Alternatively, functioning of the basic IncC/KorB partition system of IncP plasmids in various bacteria may be facilitated by additional plasmid-encoded, host-specific adapters. For example, the kilE locus, which is required for stable maintenance in P. aeruginosa, may express partition adapters specific for this host. Such a collaboration might serve to explain why incC/korB and kilE are coregulated as part of the unique kor regulon of IncP plasmids (23). The answer awaits further investigation of the IncP plasmid partition system.
In summary, we have determined that the IncC partition protein of IncP plasmids is required for stable maintenance in diverse gram-negative bacterial hosts of the α and γ subdivisions of the Proteobacteria. The clear instability phenotype of the partition defect, as revealed by the deletion of incC, will allow the identification of other components of the IncP active partition system. In addition, the ability to fully complement the ΔincC mutation in trans has opened the door to a detailed molecular genetic analysis of the functions of the IncC protein. We anticipate that future studies on the IncP partition system will lead to a better understanding of plasmid host range and DNA segregation in bacteria.
Acknowledgments
We thank Tom Rosche for helpful discussions and Mrinal Bhattacharjee for advice on the growth of A. actinomycetemcomitans.
This work was supported by NIH grant R01-GM29085 to D.H.F.
REFERENCES
- 1.Austin, S., and A. Abeles. 1983. Partition of unit-copy miniplasmids to daughter cells. I. P1 and F miniplasmids contain discrete, interchangeable sequences sufficient to promote equipartition. J. Mol. Biol. 169:353-372. [DOI] [PubMed] [Google Scholar]
- 2.Austin, S., and A. Abeles. 1983. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J. Mol. Biol. 169:373-387. [DOI] [PubMed] [Google Scholar]
- 3.Austin, S., and K. Nordström. 1990. Partition-mediated incompatibility of bacterial plasmids. Cell 60:351-354. [DOI] [PubMed] [Google Scholar]
- 4.Austin, S. J. 1988. Plasmid partition. Plasmid 20:1-9. [DOI] [PubMed] [Google Scholar]
- 5.Austin, S. J., and B. G. Eichorn. 1992. Random diffusion can account for topA-dependent suppression of partition defects in low-copy-number plasmids. J. Bacteriol. 174:5190-5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayres, E. K., V. J. Thomson, G. Merino, D. Balderes, and D. H. Figurski. 1993. Precise deletions in large prokaryotic genomes by vector-mediated excision (VEX): the trfA gene of promiscuous plasmid RK2 is essential for replication in several gram-negative hosts. J. Mol. Biol. 230:174-185. [DOI] [PubMed] [Google Scholar]
- 7.Bignell, C. R., A. S. Haines, D. Khare, and C. M. Thomas. 1999. Effect of growth rate and incC mutation on symmetric plasmid distribution by the IncP-1 partitioning apparatus. Mol. Microbiol. 34:205-216. [DOI] [PubMed] [Google Scholar]
- 8.Bouet, J. Y., J. A. Surtees, and B. E. Funnell. 2000. Stoichiometry of P1 plasmid partition complexes. J. Biol. Chem. 275:8213-8219. [DOI] [PubMed] [Google Scholar]
- 9.Datta, N., and R. Hedges. 1972. Host ranges of R factors. J. Gen. Microbiol. 70:453-460. [DOI] [PubMed] [Google Scholar]
- 10.Firshein, W., and P. Kim. 1997. Plasmid replication and partition in Escherichia coli: is the cell membrane the key? Mol. Microbiol. 23:1-10. [DOI] [PubMed] [Google Scholar]
- 11.Gerdes, K., J. E. L. Larsen, and S. Molin. 1985. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 161:292-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes, K., J. Moller-Jensen, and R. B. Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 13.Gerlitz, M., O. Hrabak, and H. Schwab. 1990. Partitioning of broad-host-range plasmid RP4 is a complex system involving site-specific recombination. J. Bacteriol. 172:6194-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser, P., M. E. Sharpe, B. Raether, M. Perego, K. Ohlsen, and J. Errington. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11:1160-1168. [DOI] [PubMed] [Google Scholar]
- 15.Goncharoff, P., J. K. K. Yip, H. Wang, H. C. Schreiner, J.-A. Pai, D. Furgang, R. H. Stevens, D. H. Figurski, and D. H. Fine. 1993. Conjugal transfer of broad-host-range incompatibility group P and Q plasmids from Escherichia coli to Actinobacillus actinomycetemcomitans. Infect. Immun. 61:3544-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681-708. [DOI] [PubMed] [Google Scholar]
- 17.Hanai, R., R. Liu, P. Benedetti, P. R. Caron, A. S. Lynch, and J. C. Wang. 1996. Molecular dissection of a protein SopB essential for Escherichia coli F plasmid partition. J. Biol. Chem. 271:17469-17475. [DOI] [PubMed] [Google Scholar]
- 18.Hayes, F. 2000. The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol. Microbiol. 37:528-541. [DOI] [PubMed] [Google Scholar]
- 19.Hiraga, S. 2000. Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Genet. 34:21-59. [DOI] [PubMed] [Google Scholar]
- 20.Jensen, R. B., and K. Gerdes. 1999. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J. 18:4076-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen, R. B., R. Lurz, and K. Gerdes. 1998. Mechanism of DNA segregation in prokaryotes: replicon pairing by parC of plasmid R1. Proc. Natl. Acad. Sci. USA 95:8550-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jovanovic, O. S., E. K. Ayres, and D. H. Figurski. 1994. Host-inhibitory functions encoded by promiscuous plasmids: transient arrest of Escherichia coli segregants that fail to inherit plasmid RK2. J. Mol. Biol. 237:52-64. [DOI] [PubMed] [Google Scholar]
- 23.Kornacki, J. A., C.-H. Chang, and D. H. Figurski. 1993. kil-kor regulon of promiscuous plasmid RK2: structure, products, and regulation of two operons that constitute the kilE locus. J. Bacteriol. 175:5078-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornacki, J. A., A. H. West, and W. Firshein. 1984. Proteins encoded by the trans-acting replication and maintenance regions of broad host range plasmid RK2. Plasmid 11:48-57. [DOI] [PubMed] [Google Scholar]
- 25.Lemonnier, M., J. Y. Bouet, V. Libante, and D. Lane. 2000. Disruption of the F plasmid partition complex in vivo by partition protein SopA. Mol. Microbiol. 38:493-505. [DOI] [PubMed] [Google Scholar]
- 26.Lopilato, J., S. Bortner, and J. Beckwith. 1986. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol. Gen. Genet. 205:285-290. [DOI] [PubMed] [Google Scholar]
- 27.Meacock, P. A., and S. N. Cohen. 1980. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell 20:529-542. [DOI] [PubMed] [Google Scholar]
- 28.Meyer, R., and M. Hinds. 1982. Multiple mechanisms for expression of incompatibility by broad-host-range plasmid RK2. J. Bacteriol. 152:1078-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moller-Jensen, J., R. B. Jensen, and K. Gerdes. 2000. Plasmid and chromosome segregation in prokaryotes. Trends Microbiol. 8:313-320. [DOI] [PubMed] [Google Scholar]
- 30.Motallebi-Veshareh, M., D. A. Rouch, and C. M. Thomas. 1990. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol. Microbiol. 4:1455-1463. [DOI] [PubMed] [Google Scholar]
- 31.Nordstrom, K., S. Molin, and H. Aagaard-Hansen. 1980. Partitioning of plasmid R1 in Escherichia coli. I. Kinetics of loss of plasmid derivatives deleted of the par region. Plasmid 4:215-227. [DOI] [PubMed] [Google Scholar]
- 32.Novick, R. P., and F. C. Hoppensteadt. 1978. On plasmid incompatibility. Plasmid 1:421-434. [DOI] [PubMed] [Google Scholar]
- 33.Ogura, T., and S. Hiraga. 1983. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell 32:351-360. [DOI] [PubMed] [Google Scholar]
- 34.Olsen, R. H., and P. Shipley. 1973. Host range and properties of the Pseudomonas aeruginosa R factor R1822. J. Bacteriol. 113:772-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 36.Pogliano, J., T. Q. Ho, Z. Zhong, and D. R. Helinski. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radnedge, L., B. Youngren, M. Davis, and S. Austin. 1998. Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partition systems. EMBO J. 17:6076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts, R. C., R. Burioni, and D. R. Helinski. 1990. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. J. Bacteriol. 172:6204-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, R. C., A. R. Ström, and D. R. Helinski. 1994. The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J. Mol. Biol. 237:35-51. [DOI] [PubMed] [Google Scholar]
- 40.Rodionov, O., M. Lobocka, and M. Yarmolinsky. 1999. Silencing of genes flanking the P1 plasmid centromere. Science 283:546-549. [DOI] [PubMed] [Google Scholar]
- 41.Rosche, T. M., A. Siddique, M. H. Larsen, and D. H. Figurski. 2000. Incompatibility protein IncC and global regulator KorB interact in active partition of promiscuous plasmid RK2. J. Bacteriol. 182:6014-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidhauser, T. J., and D. R. Helinski. 1985. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J. Bacteriol. 164:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sia, E. A., R. C. Roberts, C. Easter, D. R. Helinski, and D. H. Figurski. 1995. Different relative importances of the par operons and the effect of conjugal transfer on the maintenance of intact promiscuous plasmid RK2. J. Bacteriol. 177:2789-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Surtees, J. A., and B. E. Funnell. 2001. The DNA binding domains of P1 ParB and the architecture of the P1 plasmid partition complex. J. Biol. Chem. 276:12385-12394. [DOI] [PubMed] [Google Scholar]
- 45.Thomas, C. M., and C. A. Smith. 1986. The trfB region of broad host range plasmid RK2: the nucleotide sequence reveals incC and key regulatory gene trfB/korA/korD as overlapping genes. Nucleic Acids Res. 14:4453-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS 903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 181:7298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villarroel, R., R. W. Hedges, R. Maenhaut, J. Leemans, G. Engler, M. Van Montagu, and J. Schell. 1983. Heteroduplex analysis of P-plasmid evolution: the role of insertion and deletion of transposable elements. Mol. Gen. Genet. 189:390-399. [DOI] [PubMed] [Google Scholar]
- 48.Webb, C. D., A. Teleman, S. Gordon, A. Straight, A. Belmont, D. C. Lin, A. D. Grossman, A. Wright, and R. Losick. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]
- 49.Weitao, T., S. Dasgupta, and K. Nordstrom. 2000. Plasmid R1 is present as clusters in the cells of Escherichia coli. Plasmid 43:200-204. [DOI] [PubMed] [Google Scholar]
- 50.Wilson, J. W., E. A. Sia, and D. H. Figurski. 1997. The kilE locus of promiscuous IncPα plasmid RK2 is required for stable maintenance in Pseudomonas aeruginosa. J. Bacteriol. 179:2339-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaichi, Y., and H. Niki. 2000. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:14656-14661. [DOI] [PMC free article] [PubMed] [Google Scholar]