Abstract
Most serine cycle methylotrophic bacteria lack isocitrate lyase and convert acetyl coenzyme A (acetyl-CoA) to glyoxylate via a novel pathway thought to involve butyryl-CoA and propionyl-CoA as intermediates. In this study we have used a genome analysis approach followed by mutation to test a number of genes for involvement in this novel pathway. We show that methylmalonyl-CoA mutase, an R-specific crotonase, isobutyryl-CoA dehydrogenase, and a GTPase are involved in glyoxylate regeneration. We also monitored the fate of 14C-labeled carbon originating from acetate, butyrate, or bicarbonate in mutants defective in glyoxylate regeneration and identified new potential intermediates in the pathway: ethylmalonyl-CoA, methylsuccinyl-CoA, isobutyryl-CoA, methacrylyl-CoA, and β-hydroxyisobutyryl-CoA. A new scheme for the pathway is proposed based on these data.
Methylobacterium extorquens AM1 is a facultative methylotroph that utilizes the serine cycle to assimilate formaldehyde and supply C3 units for cell biosynthesis (1, 30). While most of the reactions in this cycle have been characterized at the gene and/or enzyme level (6-12, 17), one part of the serine cycle that involves regeneration of a molecule of glyoxylate from a molecule of acetyl coenzyme A (acetyl-CoA) still remains unresolved. Earlier work in which the fate of carbon atoms originating from methanol, ethanol, or acetate was monitored indicated that this pathway is also essential for growth of M. extorquens AM1 on C2 compounds, and chemically induced mutants in this pathway had a characteristic phenotype: they were not able to grow on C1 or C2 compounds and were rescued on these compounds by the addition of glyoxylate or glycolate (15, 16, 30). Later, a region on the chromosome of M. extorquens AM1 containing three genes mutations in which produced this characteristic phenotype was identified. These were the genes for propionyl-CoA carboxylase (pccB), a putative alcohol dehydrogenase later identified as crotonyl-CoA dehydrogenase (ccr [19]), and a mutase similar to methylmalonyl-CoA mutase (MCM) but fulfilling a different and yet unknown function (meaA [12, 33]). Orthologs of meaA and crr were also found in Streptomyces species and were shown to be involved in C2 metabolism (19, 41), suggesting that this pathway might be found outside of serine cycle methylotrophs. In our more recent studies involving genes for poly-β-hydroxybutyrate (PHB) biosynthesis in M. extorquens AM1, we discovered that two genes essential for the pathway for PHB synthesis from acetyl-CoA, encoding β-ketothiolase (phaA) and acetoacetyl-CoA reductase (phaB), are also involved in the glyoxylate regeneration pathway (23). These data indicated that the first step of the pathway must be the condensation of two molecules of acetyl-CoA to form acetoacetyl-CoA (catalyzed by PhaA), which is later converted into (R)-β-hydroxybutyryl-CoA (catalyzed by PhaB), crotonyl-CoA (catalyzed by an R-specific crotonase), and butyryl-CoA (catalyzed by Ccr). Further steps of the pathway leading to propionyl-CoA and further to glyoxylate remained unknown, although the requirement for propionyl-CoA carboxylase suggested that propionyl-CoA must be an intermediate (12, 13). In this study we have identified additional genes involved in this pathway and have investigated mutants defective in these genes for accumulation of labeled compounds that may be the intermediates in the pathway. We have used sequences of the newly and previously identified enzymes for the novel glyoxylate regeneration pathway to search available microbial genome databases in order to assess the occurrence and distribution of this pathway in different groups of bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli DH5α (Bethesda Research Laboratories), JM109 (Promega), Top 10 (Invitrogen), and S17-1 (32) were used in the study. They were grown in Luria-Bertani (LB) medium in the presence of appropriate antibiotics as described by Maniatis et al. (26). M. extorquens AM1 was grown in the minimal medium described previously (20). Succinate (20 mM), methanol (100 mM), ethanol (50 mM), or ethylamine (20 mM) was used as the substrate. The following antibiotic concentrations were used for M. extorquens AM1: 10 μg of tetracycline ml−1, 100 μg of kanamycin ml−1, and 50 μg of rifamycin ml−1. The growth responses of mutants were tested on plates containing the substrates listed above in the presence or absence of supplements of glyoxylate (5 mM) or glycolate (20 mM). The following cloning vectors were used: pUC19 (Pharmacia) for cloning and subcloning, pAYC61 (5) as a suicide vector, pRK2013 (5) as a helper plasmid, pCR2.1 (Invitrogen) for cloning PCR-generated fragments, and pCM80 (27) for expression.
Gene amplification and cloning.
Data from the M. extorquens AM1 genome project (http://vixen.microbiol.washington.edu/) were used for designing primers specific for putative genes of M. extorquens AM1 encoding the following enzymes: (R)-specific enoyl-CoA hydratase (rcro1, 5′-GAAGTCGAGGCTGTCGGCGATC-3′, and rcro2 reverse, 5′-CGAGCACCACCTCGTCGCCGAC-3′), first isobutyryl-CoA dehydrogenase (1adh1, 5′-CGTCACCCGCACCGCCATCTCG-3′, and 1adh2, 5′-CTCGCCGACGAACTCGACCCG-3′), second isobutyryl-CoA dehydrogenase (2adh1, 5′-CGATATCGAGAGCCGACTGCG-3′, and 2adh2, 5′-CATCCCGATGTCGATCATCG-3′), 3-hydroxyisobutyrate hydrolase (h1, 5′-GCCGTGCCCTAGCACATCCG-3′, and h2, 5′-CATCACGATCCCTTCGATCAG-3′), first 3-hydroxyisobutyrate dehydrogenase (1hibd1, 5′-CGCCGACCCTGTTCATTATG-3′, and 1hibd2, 5′-GACCAGCATGAAGTTGCCCGCG-3′), second 3-hydroxyisobutyrate dehydrogenase (2hibd1, 5′-GCATTTTGAATGTCGTGCGCGC-3′, and 2hibd2, 5′-GGGCGAGGATTGGCACTGGAC-3′), third (S)-specific crotonase (croB1, 5′-GCCGTTCGATGAACGTCATCAC-3′, and croB2, 5′-CCGCCTCGATCACGAGGTC-3′), fourth (S)-specific crotonase (croC1, 5′-CAGGCTGGCAGGGCTTTGGTG-3′, and croC2, 5′-CTAATTCTGCCCGTGATTCCG-3′), fifth (S)-specific crotonase (croD1, 5′-GTGGGGCGCGAGGGCGGATGAGC-3′, and croD2, 5′-GTGCACGAGTTCGTCCACCG-3′), succinate semialdehyde dehydrogenase (ssd1, 5′-GCTGATGCGCGAGCGCGTGGAG-3′, and ssd2, 5′-GGCGAAGACGGTCGGCTCGAAG-3′), the α subunit of MCM (mcm1, 5′-CAATATGCCGGCTTCTCGAC-3′, and mcm2, 5′-CCGTCATTCTCCTCGAAG-3′), the α subunit of propionyl-CoA carboxylase (pcca1, 5′-GGCAGGTCGGGACAGTCAC-3′, and pcca2, 5′-GCAGGCGCGTCCTGCGATC-3′), and methylmalonyl-CoA epimerase (ep1, 5′-GCCACCGTCGCGAACAAAGCG-3′, and ep2, 5′-GCGTGAATGCGCGACCTTTCC-3′).
DNA-DNA hybridization.
Preparation of nitrocellulose filters and DNA probes for DNA-DNA hybridization were carried out as described by Maniatis et al. (26).
DNA manipulations.
Plasmid isolation, E. coli transformation, restriction enzyme digestion, ligation, blunting of ends with T4 DNA polymerase, and filling in of ends with Klenow enzyme were carried out as described by Maniatis et al. (26). The chromosomal DNA of M. extorquens AM1 was isolated by the procedure of Saito and Miura (29).
Matings.
Triparental or biparental matings between E. coli and M. extorquens AM1 were performed overnight on nutrient agar at 30°C. Cells were then washed with sterile medium and plated on selective medium at appropriate dilutions. In triparental matings, pRK2013 (14) was used as a helper plasmid. Rifamycin was used for E. coli counterselection.
DNA sequencing.
DNA sequencing from both strands was carried out with an Applied Biosystems automated sequencer by the Department of Biochemistry Sequencing Facility, University of Washington.
Computer analysis.
Translation and analyses of DNA and DNA-derived polypeptide sequences were carried out by using Genetics Computer Group (Madison, Wis.) and ORF Finder (National Center for Biotechnology Information [NCBI]) programs (http://www.ncbi.nlm.nih.gov).
Detection of intermediates of butyrate, propionate, and acetate metabolism.
Cells grown on succinate were harvested in the mid-exponential phase of growth by centrifugation at approximately 15,000 × g and suspended in 6 ml of fresh growth medium containing 25 mM ammonium sulfate and 25 mM phosphate buffer, to a concentration of 3 mg (dry weight)/ml. The suspension was incubated aerobically at 30°C in the presence of one the following carbon sources: methanol (0.2%), butyrate (0.04%), or propionate (0.1%). After 30 min, one of the following radioactive compounds was added: [1-14C]acetate (4 μCi), [1-14C]butyrate (4 μCi), or NaH14CO3 (10 μCi). Samples (1 ml) were withdrawn at suitable time intervals, immediately transferred to 3 ml of boiling ethanol, and incubated for 3 min. Then the ethanol suspensions were transferred to a water bath at 50°C for 10 min. Insoluble materials were removed by centrifugation for 10 min at approximately 15,000 × g, and the supernatants were evaporated to dryness under a vacuum. The pellets were resuspended in 20 mM NaON and placed in the water bath at 60°C to hydrolyze CoA esters to free acids. After 30 min the suspensions were neutralized by 20 mM HCl and evaporated to dryness. The residues were suspended in 0.02 ml of 20% (vol/vol) ethanol and spotted onto thin-layer chromatography (TLC) plates (Kieselgel 60; Merck). Chromatography was performed in an isopropylether-formic acid-water (90:7:3) solvent system. After the TLC plates were dried, radioactive compounds were visualized by autoradiography. Radioactive products were identified by cochromatography with either labeled or unlabeled standard compounds. These unlabeled standards were detected by staining with a bromocresol green solution. Radioactive compounds were also identified by gas chromatography-mass spectrometry (GC-MS). Radioactive compounds were eluted from their respective spots by aqueous 20% (vol/vol) ethanol, evaporated to dryness, suspended in a butanol-HCl mixture (4:1), heated to 70°C for 2 h, and evaporated to dryness. Butyl esters of carboxylic acids obtained from the procedure described above were injected into a DB1 capillary column (0.2 mm by 25 m; film thickness, 0.33 μm) (Hewlett-Packard, Palo Alto, Calif.) connected to a 5971A Mass Spectrometer/5890 Gas Chromatograph (Hewlett Packard). The flow rate of helium carrier gas was 2.7 ml/min. The initial column temperature of 50°C was held for 2 min, and then the temperature was raised by 5°C/min up to 280°C.
Determination of distribution of 14C from [1-14C]butyrate between CO2 and biomass.
Cells were harvested by centrifugation (for 5 min at 6,000 × g) and then washed and resuspended in a minimal medium containing 0.2% methanol. Assays were carried out in 2-ml glass screw-cap vials (Fisher) in a shaker at 25°C. Reaction mixtures contained 0.1 to 0.5 mg (dry weight) of cells in 0.3 ml of medium. [1-14C]butyrate (final concentration, 0.04%; 0.2 μCi per assay) was added, and cells were killed 0, 10, 20, 30, and 40 min after substrate addition, by injection of 0.1 N NaOH (0.3 ml). Vials were left overnight for complete trapping of CO2. After that, samples of cell suspensions (0.2 ml) were transferred into sealed 30-ml Wheaton glass bottles, and 0.2 N HCl (0.2 ml) was injected into the bottles. The evolved CO2 was trapped by phenylethylamine (0.2 ml) that was placed in the microtubes installed inside the bottles. The samples were left for 4 h for complete absorption of CO2. Phenylethylamine samples from the microtubes were mixed with Aquasol (5 ml) in scintillation vials, and the radioactivity was counted. Samples of cell suspensions (0.2 ml) were applied to nitrocellulose filters (pore size, 0.2 μm), dried, and then placed in Aquasol, and the radioactivity was counted. The percentage of total radioactivity in cell biomass versus CO2 was calculated at each time point.
Enzyme assays.
Enzyme activities were determined in M. extorquens AM1 crude extracts obtained by passing cells through a French pressure cell at 1.2 × 108 Pa, followed by centrifugation for 10 min at approximately 15,000 × g. Propionyl-CoA carboxylase activity was determined by the method of Hunaiti and Kolattukudy (21). Two hundred nanomoles of butyryl-CoA in place of propionyl-CoA was used for the butyryl-CoA carboxylase activity assay. Spectrophotometric methods (22, 36) were used for protein determination.
Phosphorylation of proteins in crude extracts.
For protein phosphorylation in vivo, 4 μCi of [32P]orthophosphate was added to 2 ml of cell cultures grown on succinate. Suspensions were incubated aerobically at 30°C in the presence of either methanol or succinate. After 3 h of incubation, cells were harvested by centrifugation for 5 min at 15,000 × g and washed twice with 20 mM Tris-HCl (pH 7.0). Pellets were boiled in denaturing buffer (200 mM Tris-HCl [pH 6.8], 6% sodium dodecyl sulfate [SDS], 1% 2-mercaptoethanol, 4% glycerol, and 0.005% bromophenol blue) for 3 min. Samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) according to the method of Laemmli (24) by using 8-to-12% gradient acrylamide gels (Bio-Rad, Hercules, Calif.). After staining with Coomassie blue R-250, gels were dried and exposed to X-Omat film (Kodak) for autoradiography.
Nucleotide sequence accession numbers.
The sequences of nucleotides containing ibd2, meaB, and croR have been deposited with GenBank under accession numbers AY054980, AF416776, and AF416777, respectively.
RESULTS
Identification and analysis of genes involved in butyrate metabolism.
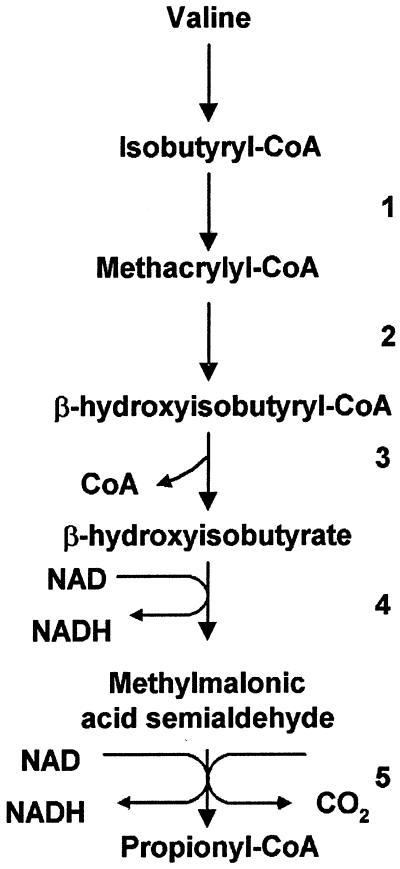
It is known that both bacteria and eucaryotes are capable of degrading valine to propionate, and isobutyryl-CoA, methacrylyl-CoA, β-hydroxyisobutyryl-CoA, β-hydroxyisobutyrate, and methylmalonic acid semialdehyde are intermediates in this pathway (34, 37, 40). The enzymes that carry out these steps are isobutyryl-CoA dehydrogenase (IBD), methacrylyl-CoA dehydratase [(S)-specific enoyl-CoA hydratase], β-hydroxyisobutyryl-CoA hydrolase, β-hydroxyisobutyrate dehydrogenase (HID), and methylmalonic acid semialdehyde dehydrogenase, respectively (34) (Fig. 1). In order to test the hypothesis that valine degradation enzymes might be involved in transformations of butyryl-CoA into propionyl-CoA in M. extorquens AM1, we attempted identification of these genes using the M. extorquens AM1 genome sequence database (http://vixen.microbiol.washington.edu/). Two putative genes were identified for IBD and were designated ibd1 and ibd2, respectively. Two putative genes were also identified for HID and were designated hid1 and hid2, respectively. One putative gene for β-hydroxyisobutyrate hydrolase (HIH), designated hih, was identified, but no genes potentially coding for methylmalonic acid semialdehyde dehydrogenase were identified. However, a putative gene encoding a related enzyme, succinate semialdehyde dehydrogenase (SSD), designated ssd, was found. Since these two enzymes carry out similar reactions, it was possible that ssd might encode an enzyme involved in this pathway.
FIG. 1.
Pathway of valine degradation in P. aeruginosa (34, 37). Enzymes: 1, IBD; 2, (S)-specific enoyl-CoA hydratase; 3, β-hydroxyisobutyryl-CoA hydrolase; 4, HID; 5, methylmalonic semialdehyde dehydrogenase.
Analysis of the M. extorquens AM1 genome database revealed that it contains five genes potentially encoding (S)-specific crotonases (enoyl-CoA hydratases). One of these, croA, has been mutated previously, and mutants with mutations in this gene had a wild-type phenotype (23). The second gene described above, hih, shows the highest identity to HIH from Caenorhabditis elegans. The amino acid sequence translated from the third gene showed the highest identity to a putative polypeptide of Rhodopseudomonas palustris in the fatty acid oxidation complex that includes enoyl-CoA hydratase, β-hydroxyacyl-CoA dehydrogenase, and β-hydroxybutyryl-CoA epimerase (http://jgi.doe.gov/JGI_microbial/html/index.html). We designated this gene croB. The products translated from the fourth and fifth genes showed the highest identities to enoyl-CoA hydratases of R. palustris and Pseudomonas aeruginosa, respectively. These genes were designated croC and croD. A gene predicted to encode the only (R)-specific crotonase (croR) was also identified as a result of BLASTP analysis, by using the amino acid sequence of an (R)-specific enoyl-CoA hydratase from Rhodobacter capsulatus (GenBank accession no. AF010496) as a query.
Another potential reaction involving isobutyryl-CoA that could play a role in this pathway is isobutyryl-CoA mutase, which converts isobutyryl-CoA to butyryl-CoA (35). However, no genes potentially coding for the subunits of isobutyryl-CoA mutase were identified in the M. extorquens AM1 genome.
Identification of meaB.
A cluster of genes in M. extorquens AM1 encoding the following three enzymes involved in the novel pathway for glyoxylate regeneration has been described before: the β subunit of propionyl-CoA carboxylase (pccB), an apparent B12-dependent mutase of unknown specificity (meaA), and crotonyl-CoA reductase (ccr, formerly adhA) (12). Mutations in these genes caused the characteristic C1- and C2-negative phenotype (12). In a more recent sequencing effort, by primer walking employing the cosmid containing the genes mentioned above (12), we identified meaB, which is separated by some 5 kb from meaA. MeaB is predicted to be a GTPase/ATPase, based on analysis with the COG program (NCBI). MeaB shows similarity to polypeptides translated from nearly all genomes whose sequences are available, including bacterial, archaeal, and eucaryotic genomes (3). Although in all but one case the polypeptides revealing identity with MeaB are not annotated, they seem to fulfill a function conserved in all life domains. In only one case was a function for the MeaB ortholog proposed: a similar polypeptide (ArgK) was ascribed a function in the transport of arginine, ornithine, and lysine in E. coli, by way of phosphorylation of specific periplasmic binding proteins (4). This functional assignment was challenged later, suggesting a function for ArgK/YgfD as a kinase/phosphatase involved in regulating activities of E. coli enzymes participating in the utilization of succinate and/or propionate as a substrate (18). This suggestion was based on the location of ygfD/argK in the cluster of genes involved in interconverting succinyl-CoA, methylmalonyl-CoA, and propionyl-CoA (3). Since these compounds are predicted intermediates in the novel acetyl-CoA oxidation pathway in M. extorquens AM1 (12), it seemed possible that MeaB might fulfill a function in the unknown pathway for glyoxylate regeneration. Orthologs of meaB are also found in close proximity to the genes encoding orthologs of propionyl-CoA carboxylase (PCC) and MCM in R. palustris, Caulobacter crescentus, Streptomyces cinnamonensis, Mycobacterium tuberculosis, and Bacillus halodurans (NCBI databases).
Identification of genes potentially involved in the conversion of propionyl-CoA to succinyl-CoA.
The gene for the second subunit of PCC (pccA) has been identified in the genome sequence by high similarity of the translated polypeptide with known PCC α subunits. pccA and pccB are not linked on the M. extorquens AM1 chromosome. We were interested in testing MCM for participation in the pathway, since MCM and PCC usually catalyze sequential reactions. The genes predicted to encode the MCM subunits (mcmA and mcmB) were identified by the similarities of the translated polypeptides to known MCM subunits, and these are also not linked on the M. extorquens AM1 chromosome. A gene (epm) encoding a putative methylmalonyl-CoA epimerase was identified by the similarity of the translated polypeptide to the sequence of epimerase from Bacillus subtilis (YqjC, GenBank accession number D84432).
Construction of insertion mutations.
Insertion mutations in croB, croC, croD, croR, ibd1, ibd2, hih, hid1, hid2, ssd, pccA, mcmA, epm, and meaB were constructed in vitro using the kanamycin resistance gene cartridge as described earlier (5). Mutants were selected in the presence of kanamycin, and kanamycin-resistant colonies were checked for their resistance to tetracycline. Tcs colonies were chosen as potential double-crossover recombinants. The identities of these double-crossover recombinants were confirmed by diagnostic PCR with primers specific to the insertion sites. Mutations in all the genes produced double-crossover recombinants (null mutants) on succinate plates with a frequency of 10 to 50% relative to the total number of recombinants. These results demonstrated that none of these genes was required for growth on succinate.
Mutant phenotypes.
The newly generated mutants fell into two groups, based on their growth responses: the croB, croC, croD, ihd1, hih, hid1, hid2, ssd, and epm mutants grew normally on multicarbon compounds (succinate and pyruvate), C2 compounds (ethylamine and ethanol), and C1 compounds (methanol and methylamine), while croR, ihd2, pccA, mcmA, and meaB mutants lost the ability to grow on C1 (methanol and methylamine) and C2 (ethanol and ethylamine) compounds. Growth of these mutants on C1 and C2 compounds was restored by the addition of glyoxylate or glycolate; thus, the mutants showed a phenotype characteristic of mutants defective in glyoxylate regeneration (15, 16).
Detection of intermediates of butyrate metabolism.
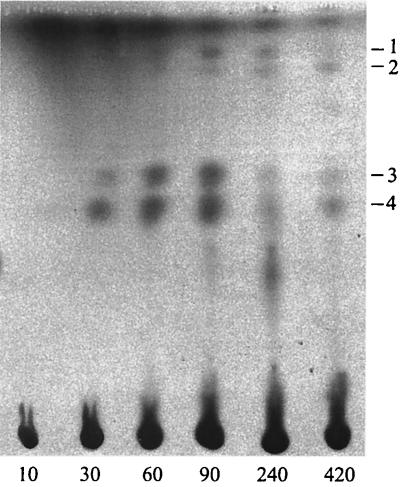
Our previous results have suggested that butyryl-CoA must be an intermediate in the pathway for glyoxylate regeneration in M. extorquens AM1 (23). We used 14C-labeled butyrate in order to detect potential intermediates in this pathway downstream of butyryl-CoA. Cell suspensions of wild-type M. extorquens AM1 and appropriate mutants were incubated in the presence of [1-14C]butyrate and unlabeled methanol. Identification of the labeled products (after removal of CoA moieties) was based on cochromatography with appropriate standards, followed by GC-MS monitoring (see Materials and Methods). Cells of wild-type M. extorquens AM1 incorporated isotope weakly, and mainly accumulated β-hydroxybutyrate. However, more labeled intermediates could be detected in the mutants (Table 1). We were able to detect β-hydroxybutyrate, methacrylate, methylsuccinate, and β-hydroxyisobutyrate in the ccr mutant. While methylsuccinate and methacrylate were early-labeled compounds, β-hydroxybutyrate and β-hydroxyisobutyrate were later intermediates. β-Hydroxybutyrate, methylsuccinate, β-hydroxyisobutyrate, and an unknown compound were detected in the meaA mutant (Fig. 2 shows a representative chromatogram). In the ihb2 mutant, β-hydroxybutyrate was the main 14C-labeled product and no β-hydroxyisobutyrate was detected, confirming the necessity of IBD for β-hydroxyisobutyrate formation. No labeled intermediate accumulation was observed in the pccA and pccB mutants, implying that PCC must perform an early step that is necessary for further metabolism of butyrate.
TABLE 1.
Distribution of 14C-labeled carboxy acids in M. extorquens AM1 and mutants incubated with 14C-labeled substrates
| Strain or mutant | Labeled intermediate(s) detected with the following substrate:
|
|||
|---|---|---|---|---|
| [1-14C]butyrate + methanol | [1-14C]acetate + methanol | [14C]bicarbonate + propionate | [1-14C]bicarbonate + butyrate | |
| AM1 | β-Hydroxybutyrate | β-Hydroxybutyrate, butyrate, methylsuccinate, isobutyrate, β-hydroxyisobutyrate, methylmalonate | Methylmalonate, succinate | Ethylmalonate, methylsuccinate |
| ccr | β-Hydroxybutyrate, methylsuccinate, methacrylate, β-hydroxyisobutyrate | NTa | NT | NT |
| meaA | β-Hydroxybutyrate, methylsuccinate, β-hydroxyisobutyrate, unknown compound | β-Hydroxybutyrate, butyrate, methylsuccinate, isobutyrate, β-hydroxyisobutyrate | NT | Ethylmalonate, methylsuccinate |
| ibd2 | β-Hydroxybutyrate | NT | NT | NT |
| pccA pccB | None detected | β-Hydroxybutyrate | None detected | None detected |
| meaB | NT | β-Hydroxybutyrate, butyrate, methylsuccinate, isobutyrate, β-hydroxyisobutyrate, methylmalonate | Methylmalonate | Ethylmalonate, methylsuccinate |
| mcmA | NT | NT | Methylmalonate | Ethylmalonate, methylsuccinate |
| epm | NT | NT | Methylmalonate, succinate | NT |
NT, not tested.
FIG. 2.
Autoradiograph of labeled intermediates separated by TLC, which have accumulated after exposure of whole cells of an meaA mutant to [14C]butyrate and methanol during 10, 30, 60, 90, 240, and 420 s. Intermediates: 1, methylsuccinate; 2, unknown compound; 3, β-hydroxyisobutyrate; 4, β-hydroxybutyrate.
More labeled intermediates were detected in similar short-term labeling experiments using [1-14C]acetate in place of [1-14C]butyrate. After incubation in the presence of labeled acetate and unlabeled methanol, cells of M. extorquens AM1 and meaA and meaB mutants accumulated β-hydroxybutyrate, butyrate, isobutyrate, methylsuccinate, and β-hydroxyisobutyrate (Table 1). In addition to these compounds, methylmalonate was detected in wild-type M. extorquens AM1 and the meaB mutant but not in the meaA mutant. Again, β-hydroxybutyrate was the sole 14C-labeled product identified in the pccA and pccB mutants.
Carboxylase activity detection.
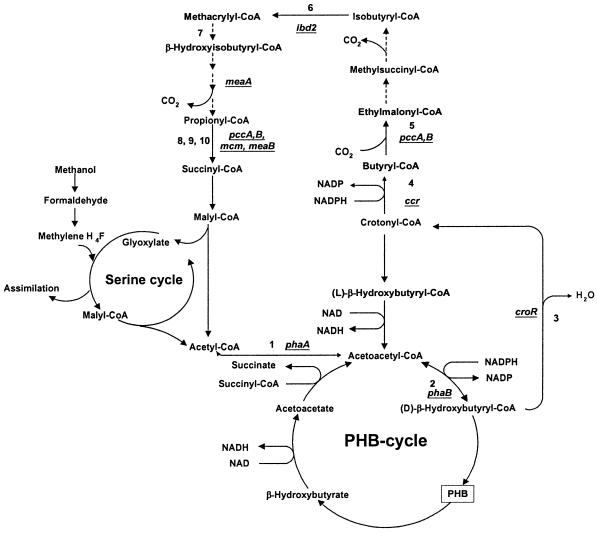
The lack of intermediates building up in the PCC mutants suggested that PCC might carry out a carboxylation step necessary for further metabolism of butyrate (Fig. 3). Therefore, we analyzed the PCC mutants in more detail. The level of PCC activity in wild-type M. extorquens AM1 was 26 mU/mg. Loss of PCC activity in the pccB mutant was confirmed in our previous study (12). We also tested PCC activity in the newly generated pccA mutant, confirming that both pccA and pccB are necessary for PCC activity. We were also interested in testing M. extorquens AM1 for the presence of butyryl-CoA carboxylase activity, predicted by the detection of methylsuccinate as an intermediate of butyrate metabolism in M. extorquens AM1 (Fig. 3). Crude cell extracts of wild-type M. extorquens AM1, and also of pccA and pccB mutants, were tested for the presence of butyryl-CoA carboxylase activity (see Materials and Methods). No butyryl-CoA carboxylase activity was found in either pccA or pccB mutants, but it was present in wild-type M. extorquens AM1, at a lower level than the activity of PCC (4 mU/mg). These results suggest that PCC catalyzes the butyryl-CoA carboxylase reaction in M. extorquens AM1.
FIG. 3.
Pathways of glyoxylate regeneration in M. extorquens AM1. Enzymes: 1, β-ketothiolase; 2, NADPH-linked acetoacetyl-CoA reductase; 3, (R)-specific enoyl-CoA hydratase; 4, crotonyl-CoA reductase; 5, butyryl-CoA carboxylase; 6, IBD; 7, (S)-specific enoyl-CoA hydratase; 8, PCC; 9, methylmalonyl-CoA epimerase; 10, MCM. Dotted lines reflect enzymes not yet known or detected.
Detection of CO2 incorporation.
To test for potential products of butyryl-CoA carboxylation, we performed short-term labeling experiments with the wild type and the new mutants using 14CO2 and either butyrate or propionate. When whole cells of wild-type M. extorquens AM1 were incubated in the presence of 14CO2 and unlabeled butyrate, ethylmalonate was the early intermediate and methylsuccinate was the later intermediate. These data suggest that butyryl-CoA is carboxylated to ethylmalonyl-CoA by a carboxylase activity, presumably butyryl-CoA carboxylase catalyzed by PCC, and is later converted to methylsuccinyl-CoA, possibly by a mutase activity.
Two genes encoding potential mutases are known to be required for the glyoxylate regeneration pathway, mcmA, encoding MCM activity (this study), and meaA, predicted to encode a mutase of unknown function (12, 33, 39). Both of these were candidates for the putative mutase converting ethylmalonyl-CoA to methylsuccinyl-CoA. However, cells of mcmA and meaA mutants also were able to accumulate ethylmalonate and methylsuccinate, suggesting that neither MCM nor MeaA catalyzes isomerization of ethylmalonyl-CoA into methylsuccinyl-CoA.
Cells of pccA and pccB mutants accumulated neither ethylmalonate nor methylsuccinate under these conditions. These data provide further evidence that PCC must be responsible for butyryl-CoA carboxylation.
When cells of wild-type M. extorquens AM1 were incubated in the presence of 14C-labeled bicarbonate and unlabeled propionate, the main 14C-labeled intermediate was succinate. In addition, small amounts of methylmalonate were detected during the first 10 s of incubation.
Under the same conditions, the epm mutant accumulated both succinate and methylmalonate, suggesting that M. extorquens AM1 has multiple enzymes capable of methylmalonyl-CoA epimerization (Table 1).
Methylmalonate and succinate were not detected as labeled intermediates in pccA and pccB mutants incubated under the same conditions. This result is consistent with pccA and pccB encoding the subunits of PCC and carrying out both PCC and butyryl-CoA carboxylase activities.
The mcmA and meaB mutants also did not accumulate succinate. However, they accumulated methylmalonate. These data suggest that mcmA encodes a polypeptide required for MCM activity. These data also imply that meaB may have a function in methylmalonyl-CoA metabolism. It may function as a novel methylmalonyl-CoA epimerase, or it may be involved in the regulation of MCM and/or methylmalonyl-CoA epimerase activities.
Test for protein modification by phosphorylation.
Since meaB encodes a protein belonging to the GTPase/ATPase family, we tested the hypothesis that it might phosphorylate another protein of importance in the glyoxylate regeneration pathway. Succinate-grown cells of M. extorquens AM1 and the meaB mutant were incubated with [32P]orthophosphate in the presence of either succinate or methanol (for induction). After 3 h of incubation, cell extracts were subjected to SDS-PAGE, and labeled protein bands were detected by autoradiography (data not shown). No difference in labeling patterns was found between succinate- and methanol-grown cells of M. extorquens AM1. The patterns for succinate-grown cells of M. extorquens AM1 and the meaB mutant were also identical. Label was not incorporated into any proteins when the meaB mutant was incubated with methanol, as apparently no phosphorylation could be achieved under these conditions.
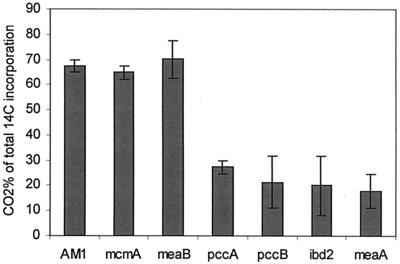
Distribution of 14C from [1-14C]butyrate between CO2 and biomass.
Our failure to demonstrate the appearance of 14C from [1-14C]butyrate in methylmalonate in the ccr mutant implies a scenario in which the labeled carbon is lost during the unknown steps in the pathway leading from β-hydroxyisobutyryl-CoA to propionyl-CoA. To test this hypothesis, the percentages of radioactivity incorporated in cell biomass and CO2 were determined for pccA, pccB, ibd2, meaA, mcmA, and meaB mutants and for wild-type M. extorquens AM1. Incorporation of [14C]butyrate into CO2 and cells increased linearly with time in M. extorquens AM1 and all the mutants (data not shown). The majority of 14C (65 to 70%) was detected in CO2 in the mcmA and meaB mutants and wild-type M. extorquens AM1 (Fig. 4). The pccA, pccB, ibd2, and meaA mutants incorporated only 18 to 27% of radioactivity in CO2. These results suggest that the carbon atom in position 1 of butyrate is lost as CO2 during oxidation into propionyl-CoA. The labeling patterns in the pccA and pccB mutants support the hypothesis that PCC catalyzes an early step in the metabolism of butyrate. A similar labeling pattern in the meaA mutant confirms that the unknown reaction catalyzed by MeaA precedes the decarboxylation step.
FIG. 4.
Production of 14CO2 from 1-14C-labeled butyrate in wild-type and mutant M. extorquens AM1, as a percentage of total incorporation.
Microbial genome screening for genes involved in this pathway.
In order to assess the occurrence of this novel pathway for acetyl-CoA oxidation beyond representatives of methylotrophic groups, we searched genomic databases of microbes belonging to various groups of bacteria and archaea. Amino acid sequences translated from the genes for glyoxylate regeneration identified here and in our previous studies were used as queries for BLASTP searches against the NCBI nonredundant database or against genomic databases of fully sequenced microbes. We have found that orthologs of all the genes demonstrated to participate in the acetyl-CoA oxidation pathway in M. extorquens AM1 are present in the genomes of C. crescentus, Streptomyces coelicolor, Rhodobacter sphaeroides, and R. capsulatus. meaA, ccr, and ibd2 are closely linked on the chromosome of C. crescentus, and ibd2, croR, meaA, and ccr apparently form an operon in S. coelicolor. meaA and ccr are linked on the chromosomes of R. sphaeroides and R. capsulatus.
DISCUSSION
Serine cycle methylotrophs exemplified by M. extorquens AM1 do not possess the glyoxylate shunt, as they lack isocitrate lyase (ICL) (1, 2). For glyoxylate regeneration in the serine cycle, such bacteria utilize an alternative pathway, having butyryl-CoA and propionyl-CoA as intermediates (12, 23). The same pathway operates during growth of ICL-minus methylotrophs on C2 compounds (15, 16, 30). The same or a similar pathway operates in Streptomyces spp. during growth on C2 compounds, and ICL is not expressed under these conditions (19). Although a number of genes and enzymes participating in this novel pathway have been identified (12, 23), a major part of this pathway for conversion of acetyl-CoA to glyoxylate remained unknown. In this study we have used a genome analysis approach to select candidate genes for this pathway, and we have tested these genes by mutation. We also monitored the fates of labeled acetate, butyrate, and bicarbonate in these mutants and in the wild type to detect potential intermediates in this pathway and to assess the roles of the participating genes more precisely.
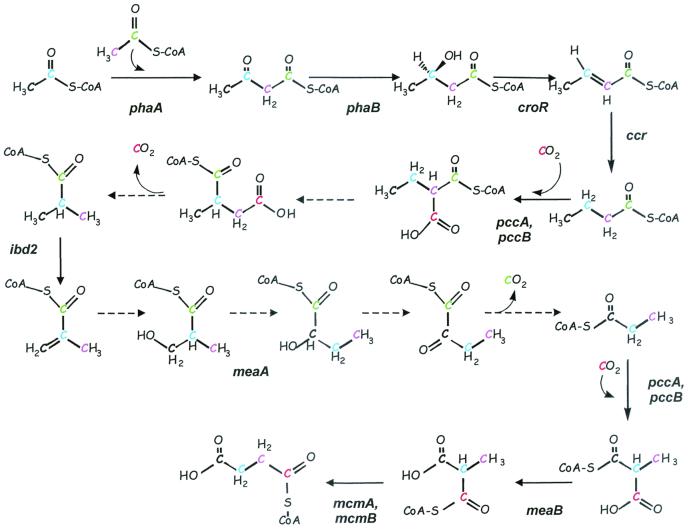
Five new genes participating in the pathway have been identified, mutations in which produced the characteristic mutant phenotype: pccA, mcmA, croR, ibd2, and meaB. In addition, the following intermediates were identified (after removal of CoA moieties): ethylmalonate, methylsuccinate, isobutyrate, methacrylate, and β-hydroxyisobutyrate. Our data suggest the scheme shown in Fig. 3, which involves some of the intermediates and enzymes of valine degradation shown in Fig. 1 but also involves other steps.
Identification of the (R)-specific crotonase (encoded by croR) completes the first phase of acetyl-CoA oxidation, via acetoacetyl-CoA (catalyzed by PhaA), d-β-hydroxybutyryl-CoA (catalyzed by PhaB), crotonyl-CoA (catalyzed by CroR), and butyryl-CoA (catalyzed by Ccr [Fig. 3]). Analysis of labeled intermediates originating from 1-14C-labeled butyrate, acetate, or bicarbonate suggests that carboxylation of butyryl-CoA is the next step in the pathway. Labeling patterns in the pccA and pccB mutants suggest that PCC must be responsible for the carboxylation of butyryl-CoA to produce ethylmalonyl-CoA, since these mutants do not accumulate ethymalonyl-CoA or any other further intermediates of the pathway.
The enzymes that catalyze the steps between ethylmalonyl-CoA and isobutyryl-CoA remain unclear. The labeling data suggest that ethylmalonyl-CoA is transformed into methylsuccinyl-CoA, possibly by a mutase reaction. However, neither MCM nor MeaA (an unknown mutase) is required for this transformation, as methylsuccinyl-CoA accumulates in both mutants. It is possible that an as yet undetected mutase carries out this reaction. Methylsuccinyl-CoA is probably decarboxylated by an unknown decarboxylase to produce isobutyryl-CoA. M. extorquens AM1 does not possess recognizable isobutyryl-CoA mutase genes, and isobutyryl-CoA mutase, although present, has been shown not to take part in this pathway in Streptomyces (38, 39); therefore, this enzyme is unlikely to be a participant in the pathway in M. extorquens AM1.
ibd2 is responsible for the conversion of isobutyryl-CoA into methacrylyl-CoA. Mutants of Streptomyces avermitilis defective in this enzyme were also affected in growth on acetate and butyrate, further supporting the hypothesis that Streptomyces employs the same or a similar pathway for acetyl-CoA oxidation (41).
The next step appears to be the conversion of methacrylyl-CoA into β-hydroxyisobutyryl-CoA, probably by an enoyl-CoA hydratase. Several candidates for this enzyme were found in the genome sequence, but none of the mutants with mutations in these genes exhibited a phenotype. It is possible that this step is carried out by redundant enzymes or that this step is performed by an (R)-specific crotonase.
The conversion of β-hydroxyisobutyryl-CoA into propionyl-CoA is unclear. The conversions taking place in the known valine degradation pathway (Fig. 1) do not appear to be involved at this stage, as mutants with mutations in the candidates for β-hydroxyisobutyrate dehydrogenase and β-hydroxyisobutyryl-CoA hydrolase did not show a phenotype, and no candidate for methylmalonic semialdehyde dehydrogenase could be identified in the genome sequence. On the other hand, based on the analysis of labeled metabolite accumulation patterns, MeaA, a putative mutase, must be involved in this transformation.
The MeaA reaction then appears to be followed by a decarboxylation step. The hypothesis consistent with these data would be the rearrangement of β-hydroxyisobutyryl-CoA to α-hydroxybutyryl-CoA catalyzed by MeaA, with subsequent oxidative decarboxylation via α-ketobutyryl-CoA to propionyl-CoA (Fig. 5). In the pathway we are proposing here, PCC encoded by pccAB carries out two steps, the early butyryl-CoA carboxylase reaction and the later PCC reaction. A similar dual role for PCC has been postulated in Streptomyces, in which butyryl-CoA carboxylase is necessary for the production of ethylmalonyl-CoA, an intermediate in polyketide biosynthesis, while PCC is involved in valine degradation (28).
FIG. 5.
Schematic diagram showing intermediates of the proposed route for conversion of acetyl-CoA to glyoxylate in M. extorquens AM1. Colors trace carbons from the acetyl moiety of acetyl-CoA. The CO2 fixed early in the pathway (red carbon) is released, while the second CO2 released is derived from the C-1 carbon of butyryl-CoA (green carbon). Dotted lines reflect enzymes not yet known or detected.
MCM would be predicted to convert l-methylmalonyl-CoA into succinate, and labeling patterns in mcmA mutants support that role. The fate of succinyl-CoA is not exactly clear at this point, but it likely that it is converted into malate via reactions of the tricarboxylic acid cycle, and then to malyl-CoA, which is cleaved by malyl-CoA lyase into glyoxylate and acetyl-CoA.
One step in the putative conversion of propionyl-CoA into succinyl-CoA remains unclear. Only one recognizable gene potentially encoding a methylmalonyl-CoA epimerase (epm) was found in the genome of M. extorquens AM1, but mutants with mutations in this gene had a wild-type phenotype, suggesting the presence of a nonorthologous methylmalonyl-CoA epimerase in addition to Epm. Since the meaB mutant has lost its ability to convert methylmalonyl-CoA to succinyl-CoA, meaB might be responsible for a novel methylmalonyl-CoA epimerase. Some coenzyme B12-dependent enzymes (propanediol and 1,2-propanediol dehydratase) are reactivated by proteins in the presence of ATP (31). Thus, MeaB might also have a role in reactivation of MCM. Another possible function for MeaB would be regulation of MCM or/and Epm activity via protein phosphorylation. One example of this type of regulation is the reversible phosphorylation of isocitrate dehydrogenase in E. coli controlling the flux of carbon through the glyoxylate cycle (25). However, comparisons of protein phosphorylation patterns in wild-type M. extorquens AM1 and meaB mutants did not confirm this hypothesis. Lastly, MeaB could be a novel transcriptional regulator for MCM and/or methylmalonyl-CoA epimerase. However, no difference in transcription from the mcmA promoter was observed between the meaB mutant and the wild type (data not shown).
In summary, the combined results of the genome searches, mutant phenotypes, and intermediate accumulation patterns strongly suggest that the glyoxylate regeneration pathway in M. extorquens AM1 follows the route shown in Fig. 3, involving intermediates and enzymes of PHB synthesis, valine degradation, and polyketide biosynthesis. Although it is not clear at this time why a pathway that requires only a few steps in most bacteria, via the glyoxylate shunt, is so complicated in this bacterium, it may reflect branch points to other parts of metabolism.
Analysis of available bacterial genomes for the presence of genes showing significant identity to the genes specifically involved in glyoxylate regeneration in M. extorquens AM1 and acetate metabolism in Streptomyces suggests the presence of this pathway in C. crescentus, R. capsulatus, and R. sphaeroides. Thus, the pathway seems to be more widespread in the bacterial world than previously thought, and not limited to ICL-minus methylotrophs.
Acknowledgments
This work was supported by grants from the NIH (GM39646 and GM98933).
We are grateful to Cliff Unkefer for valuable discussions and for help with drawings.
Footnotes
Dedicated to J. R. Quayle.
REFERENCES
- 1.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
- 2.Bellion, E., and J. C. Spain. 1975. The distribution of the isocitrate lyase serine pathway amongst one-carbon utilizing organisms. Can. J. Microbiol. 22:404-408. [DOI] [PubMed] [Google Scholar]
- 3.Bobik, T. A., and M. E. Rasche. 2001. Identification of the human methylmalonyl-CoA racemase gene based on the analysis of prokaryotic gene arrangements: implications for decoding the human genome. J. Biol. Chem. 276:37194-37198. [DOI] [PubMed] [Google Scholar]
- 4.Celis, R. T. F., P. F. Leadlay, I. Roy, and A. Hansen. 1998. Phosphorylation of the periplasmic binding protein in two transport systems for arginine incorporation in Escherichia coli K-12 is unrelated to the function of the transport system. J. Bacteriol. 180:4828-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chistoserdov, A. Y., L. V. Chistoserdova, W. S. McIntire, and M. E. Lidstrom. 1994. Genetic organization of mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J. Bacteriol. 176:4052-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chistoserdova, L., and M. E. Lidstrom. 1997. Identification and mutation of a gene for glycerate kinase activity from a facultative methylotroph, Methylobacterium extorquens AM1. J. Bacteriol. 179:4946-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chistoserdova, L. V., and M. E. Lidstrom. 1991. Purification and characterization of hydroxypyruvate reductase from the facultative methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 173:7228-7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chistoserdova, L. V., and M. E. Lidstrom. 1992. Cloning, mutagenesis, and physiological effect of a hydroxypyruvate reductase gene from Methylobacterium extorquens AM1. J. Bacteriol. 174:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chistoserdova, L. V., and M. E. Lidstrom. 1994. Genetics of the serine cycle in Methylobacterium extorquens AM1: identification of sgaA and mtdA and sequences of sgaA, hprA, and mtdA. J. Bacteriol. 176:1957-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chistoserdova, L. V., and M. E. Lidstrom. 1994. Genetics of the serine cycle in Methylobacterium extorquens AM1: cloning, sequence, mutation, and physiological effect of glyA, the gene for serine hydroxymethyltransferase. J. Bacteriol. 176:6759-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chistoserdova, L. V., and M. E. Lidstrom. 1994. Genetics of the serine cycle in Methylobacterium extorquens AM1: identification, sequence, and mutation of three new genes involved in C1 assimilation, orf4, mtkA, and mtkB. J. Bacteriol. 176:7398-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chistoserdova, L. V., and M. E. Lidstrom. 1996. Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extorquens AM1. Microbiology 142:1459-1468. [DOI] [PubMed] [Google Scholar]
- 13.Dawes, I. W., and I. W. Sutherland. 1994. Microbial physiology, 2nd ed. Blackwell Scientific, Oxford, United Kingdom.
- 14.Ditta, G., T. Schmidhauser, F. Yakobson, P. Lu, X. Liang, D. Finlay, D. Guiney, and D. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 15.Dunstan, P. M., C. Anthony, and W. T. Drabble. 1972. Microbial metabolism of C1 and C2 compounds. The role of glyoxylate, glycollate and acetate in the growth of Pseudomonas AM1 on ethanol and on C1 compounds. Biochem. J. 128:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunstan, P. M., and C. Anthony. 1973. Microbial metabolism of C1 and C2 compounds. The role of acetate during growth of Pseudomonas AM1 on C1 compounds, ethanol and β-hydroxybutyrate. Biochem. J. 132:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulton, G. F., D. N. Nunn, and M. E. Lidstrom. 1984. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J. Bacteriol. 160:718-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haller, T., T. Buckel, J. Retey, and J. A. Gerlt. 2000. Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli. Biochemistry 39:4622-4629. [DOI] [PubMed] [Google Scholar]
- 19.Han, L., and K. A. Reynolds. 1997. A novel alternative anaplerotic pathway to the glyoxylate cycle in streptomycetes. J. Bacteriol. 179:5157-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harder, W., M. Attwood, and J. R. Quayle. 1973. Methanol assimilation by Hyphomicrobium spp. J. Gen. Microbiol. 78:155-163. [Google Scholar]
- 21.Hunaiti, A., and P. E. Kolattukudy. 1982. Isolation and characterization of acyl-coenzyme A carboxylase from erythromycin-producing Streptomyces erythraeus. Arch. Biochem. Biophys. 216:362-371. [DOI] [PubMed] [Google Scholar]
- 22.Kalb, V. F., and R. W. Bernlohr. 1977. A new spectrophotometric assay for protein in cell extracts. Anal. Biochem. 82:362-371. [DOI] [PubMed] [Google Scholar]
- 23.Korotkova, N., and M. E. Lidstrom. 2001. A connection between poly -β-hydroxybutyrate biosynthesis and growth on C1 and C2 compounds in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 183:1038-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.LaPorte, D. C., and J. E. Cronan. 1996. Tricarboxylic acid cycle and glyoxalate bypass, p. 206-216. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 26.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez, E., and H. Gramajo. 1999. Genetic and biochemical characterization of the alpha and beta components of propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2). Microbiology 145:3109-3119. [DOI] [PubMed] [Google Scholar]
- 29.Saito, H., and K.-I. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochem. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 30.Salem, A. R., and J. R. Quayle. 1971. Mutants of Pseudomonas AM1 that require glycollate or glyoxylate for growth on methanol and ethanol. Biochem. J. 124:74P. [DOI] [PMC free article] [PubMed]
- 31.Seifert, C., S. Bowien, G. Gottschalk, and R. Daniel. 2001. Identification and expression of the genes and purification and characterization of the gene products involved in reactivation of coenzyme B12-dependent glycerol dehydratase of Citrobacter freundii. Eur. J. Biochem. 268:2369-2378. [DOI] [PubMed] [Google Scholar]
- 32.Simon, R., U. Priefer, and A. Puhler. 1983. Vector plasmids for in vivo manipulations of gram-negative bacteria, p. 98-106. In A. Puhler (ed.), Molecular genetics of bacteria-plant interactions. Springer-Verlag, Berlin, Germany.
- 33.Smith, L. M., W. G. Meijer, L. Dijkhuizen, and P. Goodwin. 1996. A protein having similarity with methylmalonyl-CoA mutase is required for the assimilation of methanol and ethanol by Methylobacterium extorquens AM1. Microbiology 142:657-684. [DOI] [PubMed] [Google Scholar]
- 34.Steele, M. L., D. Lorenz, K. Hatter, A. Park, and J. R. Sokatch. 1992. Characterization of the mmsAB operon of Pseudomonas aeruginosa PAO encoding methylmalonate-semialdehyde dehydrogenase and 3-hydroxyisobutyrate dehydrogenase. J. Biol. Chem. 267:13585-13592. [PubMed] [Google Scholar]
- 35.Vrijbloed, J. W., K. Zerbe-Burkhardt, A. Ratnatilleke, A. Grubelnik-Leiser, and J. A. Robinson. 1999. Insertional inactivation of methylmalonyl coenzyme A (CoA) mutase and isobutyryl-CoA mutase genes in Streptomyces cinnamonensis: influence on polyketide antibiotic biosynthesis. J. Bacteriol. 181:5600-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitaker, J. R., and P. E. Granum. 1980. An absolute method for protein determination based on the difference in absorbance at 235 and 280 nm. Anal. Biochem. 109:156-159. [DOI] [PubMed] [Google Scholar]
- 37.Wolf, D. A., and H. A. Akers. 1986. Uncertainties remain in the catabolism of valine. Trends Biochem. Sci. 11:390-392.
- 38.Zhang, W., L. Yang, W. Jiang, G. Zhao, and J. Chiao. 1999. Molecular analysis and heterologous expression of the gene encoding methylmalonyl-coenzyme A mutase from rifamycin SV-producing strain Amicolatopsis mediterranei U32. Appl. Biochem. Biotechnol. 82:209-225. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, W., and K. A. Reynolds. 2001. MeaA, a putative coenzyme B12-dependent mutase, provides methylmalonyl coenzyme A for monensin biosynthesis in Streptomyces cinnamonensis. J. Bacteriol. 183:2071-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y. X., L. Tang, and C. R. Hutchinson. 1996. Cloning and characterization of a gene (msdA) encoding methylmalonic acid semialdehyde dehydrogenase from Streptomyces coelicolor. J. Bacteriol. 178:490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Y. X., C. D. Denoya, D. D. Skinner, R. W. Fedechko, H. A. McArthur, M. R. Morgenstern, R. A. Davies, S. Lobo, K. A. Reynolds, and C. R. Hutchinson. 1999. Genes encoding acyl-CoA dehydrogenase (AcdH) homologues from Streptomyces coelicolor and Streptomyces avermitilis provide insights into the metabolism of small branched-chain fatty acids and macrolide antibiotic production. Microbiology 145:2323-2334. [DOI] [PubMed] [Google Scholar]