Abstract
During colonization of the alfalfa rhizosphere, Pseudomonas fluorescens F113 undergoes phenotypic variation, resulting in the appearance of colonies with different morphology. Among phenotypic variants, three isolates, C, F, and S were selected, with the C variant showing colony morphology identical to that of the inoculated wild-type strain and F and S having a translucent and diffuse morphology. Phenotypic variants F and S were shown to preferentially colonize distal parts of the roots and showed alterations in motility, swimming faster than the C variant and swarming under conditions that did not allow swarming of the C variant. The motility behavior correlated with overproduction of the fliC-encoded protein flagellin but not with hyperflagellation. Flagella of the F and S variants were several times longer than those of the C variant, and overproduction of flagellin was regulated at the transcriptional level. Variant F showed alterations in traits that have been shown to be important for rhizosphere colonization, such as siderophore, cyanide, and exoprotease production, and these phenotypes were complemented by a cloned gacA. Sequence analysis of the gacA alelle in variant F suggested selection of the phenotype in the rhizosphere. Variant F was also affected in other phenotypes, such as lipopolysaccharide structure and flocculation in unshaken liquid medium, which were not complemented by the gacA or gacS gene. Mutation of the F113 sss gene, encoding a site-specific recombinase, showed that most of the phenotypic variation was due to the activity of this recombinase, indicating that phase variation occurs during rhizosphere colonization.
Bacterial phase variation consists of the diversification of a population into subpopulations which present genotypic and phenotypic differences. It has been described for a variety of species, mostly within gram-negative bacteria (reviewed in reference 26). Phase variation is typical of bacteria that occupy heterogeneous ecological niches and has been related to adaptation to different environmental situations and to sudden changes in the ecosystem (49). Under these conditions, phase variation would generate a mixed population able to colonize different parts of the ecosystem, and in the case of rapid environmental changes, part of the initial population would survive (17).
Phase variation predominantly affects surface components of cells, such as membrane antigens, flagella, and fimbriae, causing morphological alterations in colonies that allow easy detection (26). Typical examples are fimbrial variation in Escherichia coli, flagellar variation in Salmonella spp., and variation in surface antigens in several species of the genus Neisseria. From a genetic point of view, phase variation often results from genomic rearrangements that can be caused by several molecular mechanisms. The best-studied mechanisms are the action of a site-specific recombinase that can produce inversions or deletions of specific sites of the genome (1), mutations in homopolymeric tracts that produce frameshift mutations affecting translation (48) or transcription (44), and differential methylation affecting transcription of specific genes (57).
Phase variation in fluorescent pseudomonads has been described. When Pseudomonas fluorescens SBW25 was grown in static liquid medium, three subpopulations arose and occupied different parts of the culture: surface, middle part, and bottom (45). These subpopulations did not appear in a homogeneous environment such as a shaken liquid culture. Phase variants also appear in Pseudomonas aeruginosa during biofilm formation (15). These variants are affected in multiple traits such as motility, pigment production, and elastase secretion. Fluorescent phenotypic variants of Pseudomonas tolaasii also have altered motility and protease production (22). Phase variants of the rhizosphere-colonizing bacterium Pseudomonas brassicacearum affected in morphology and exoenzyme production have also been reported (12). Furthermore, a mutation in the P. aeruginosa sss gene, encoding a site-specific recombinase which in other species is implicated in phase variation, affects the production of the fluorescent siderophore pyoverdin (28). The mutation of the homologous gene in P. fluorescens WCS365 results in the loss of competitive root colonization (13), implying that phase variation might play an important role in the colonization of this heterogeneous and changing ecosystem.
Fluorescent pseudomonads are of great importance in biotechnology because of the ability of several strains to degrade xenobiotics, control plant pathogens, and act as human pathogens. The ability of some strains to colonize the plant rhizosphere allows its use in rhizoremediation (10, 30). Here we show that phenotypic variation occurs during the colonization of the alfalfa rhizosphere by the biocontrol strain P. fluorescens F113 and that phenotypic variants show alterations in phenotypes that have been described as important for rhizosphere colonization.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strain used in this study is P. fluorescens F113, originally isolated from the sugar beet rhizosphere (53), and its phenotypic variants recovered after rhizosphere colonization (this study). The F113 gene bank was constructed with partially EcoRI-digested genomic DNA cloned into plasmid pLAFR3 in E. coli LE392. Phase variants F and S were tagged with random insertions of mini Tn5 gus (Smr Spcr) (59) and Tn5 lux (Kmr) (60) transposons, respectively. Growth rate and relevant characteristics of the tagged derivatives including colonization capacity were tested and showed no differences from the original strains.
Plasmids expressing the gacA and gacS genes, pME3066 (34) and pEMH97 (29), respectively, were introduced by triparental mating using pRK2013 as the helper plasmid (19). Transcriptional fusions of the fliC promoter region from the C, F, and S variants with the promoterless lacZ gene, named pBG1208, pBG1209, and pBG1210, respectively, were constructed in pMP220 (55). All cultures of P. fluorescens F113 and phase variants were grown on SA medium (51) overnight at 28°C. Luria-Bertani (LB) medium (6) was used for high-iron cultures. Agar concentrations of 0.3, 0.5, 0.8, 1, and 1.5% were added to SA to test swimming and swarming abilities, and the cells were inoculated in the middle of the plate from exponentially growing cultures. Haloes were measured at 18 and 26 h after inoculation. King's B medium (31) was used for static cultures in order to observe biofilm formation. Exoprotease production was observed on skim milk plates (47) and HCN production was tested as previously described (11, 58). Pyoverdin production was observed on SA plates by fluorescence under UV light. β-Galactosidase activity was measured as described by Miller (42).
Rhizosphere colonization experiments.
Alfalfa seeds were sterilized in 70% ethanol for 2 min and in bleach for 15 min and rinsed properly with sterile distilled water. Seed vernalization was performed at 4°C overnight, and germination was for 2 days at 28°C. Germinated alfalfa seeds were sown in Leonard jar gnotobiotic systems using Perlite as the solid substrate and 8 mM KNO3-supplemented FP (18) as the mineral medium. After 2 days, alfalfa seedlings were inoculated with ca. 108 cells of the appropriate strains. In competition experiments, strains were inoculated at a 1:1 ratio. Plants were maintained in controlled conditions (16 h of light at 25°C and 8 h of dark at 18°C). Bacteria were recovered from the rhizosphere by vortexing the root for 2 min in a tube containing 10 ml of 0.9% NaCl, and plating appropriate dilutions on SA plates with antibiotics. Statistic analysis was done with Sigma Plot 4 software (Windows).
DNA extraction, Southern blots, and amplification.
Total genomic DNA was extracted (4) from C, F, and S liquid cultures and digested with EcoRI. Electrophoresis on 1% agarose gel was performed. DNA was transferred overnight to a Hybond-N nitrocellulose membrane (Amersham). The probe labeling and detection were performed with the digoxigenin (DIG) luminescence detection kit for nucleic acids (Boehringer Mannheim) according to the manufacturer's instructions, and X-Omat LS film (Kodak) was used for detection.
Amplification of fliC was performed using primers fliF and fliR, designed after conserved flagellin sequences of other Pseudomonas spp. PCR was performed using genomic DNA of F113 as the template under the following conditions: one hold of 5 min at 94°C; 25 cycles of 30 s at 94°C, 30 s at 51°C, and 30 s at 72°C; and one hold of 7 min at 72°C. Amplification of the promoter region of fliC was performed using primers pfliF and cosR in the conditions previously indicated with an annealing temperature of 51°C. Amplification of sss was performed using primers sss F and sss R, designed after conserved site-specific recombinase sequences of other Pseudomonas spp. PCR was performed using genomic DNA of F113 as the template under the previously indicated conditions with an annealing temperature of 52°C.
Construction of an sss insertion mutant.
Directed mutagenesis was used to generate single recombination mutants of the sss gene in P. fluorescens F113. An amplified fragment of the sss gene was interrupted with a kanamycin resistance cassette, cloned into pK18mobsac (50), and introduced into wild-type F113. Single recombinants were obtained and tested by Southern blot with the sss probe.
Transmission electron and optical microscopy.
Cells from liquid and solid cultures of the phenotypic variants were deposited on a drop of water. Formvar-coated grids were placed on top of the drop for 30 s to allow adhesion of bacterial cells. Grids were stained for 1 min with a 1% solution of potassium phosphotungstate and washed for 1 min in a drop of water. Grids were air dried and examined with a transmission electron microscope. Phase contrast was used for observation of liquid and solid cultures of the phenotypic variants with an optical microscope. Measuring of flagellum length was done with Q-Win software (Leica).
Protein extraction and Western blots.
Total extracellular proteins were extracted by acetone precipitation of the spent medium (8) after vortexing cultures for 2 min to detach flagella. Proteins were electrophoresed in 12% acrylamide gels that were stained with Coomassie blue. The same electrophoretic conditions were used for Western blotting. Gels were transferred to Immobilon-P transfer membranes (Millipore) for 1 h under standard conditions. The membranes were incubated with a 1:10,000 dilution of an antiflagellin antiserum (13) for 1 h and with a peroxidase-tagged secondary antibody (anti-rabbit immunoglobulin) for 30 min. The enhanced chemiluminescence (ECL) method and Hyperfilm ECL (Amersham) were used for development.
LPS extraction and electrophoresis.
Extraction of total lipopolysaccharide (LPS) from phenotypic variants by a standard procedure and a 13% acrylamide gel was performed and silver stained after treatment with metaperiodate (38).
Nucleotide sequence accession numbers.
The fliC sequence, including the promoter region, has been deposited in the GenBank database with accession number AF399739. The sss gene sequence has been deposited in the GenBank database with accession number AF416734.
RESULTS
Phenotypic variation occurs during alfalfa rhizosphere colonization.
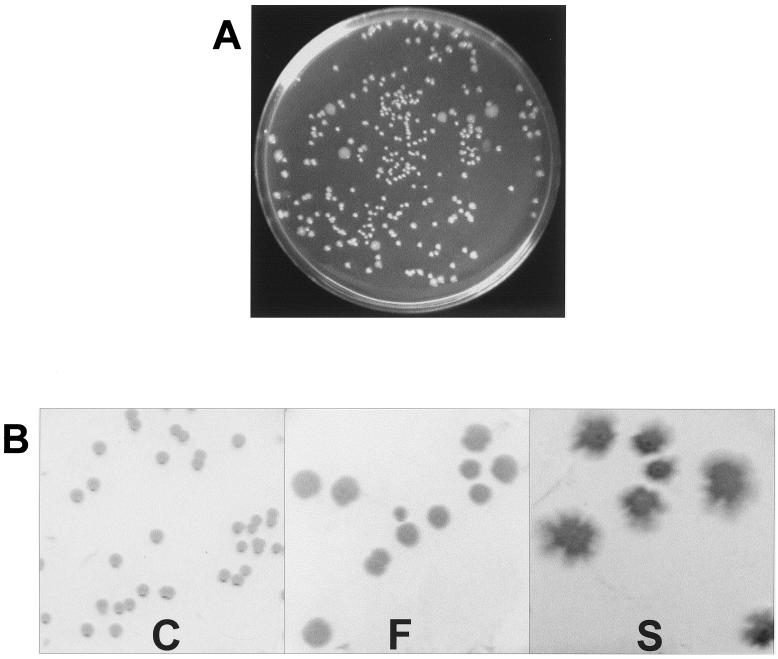
P. fluorescens F113 is a good colonizer of the alfalfa rhizosphere (unpublished results). When bacteria isolated from the rhizosphere of F113-inoculated alfalfa plants were plated (Fig. 1A), a morphologically diverse population was recovered. Arbitrarily primed PCR analysis of the different morphological variants (not shown) demonstrated that all the colonies tested corresponded to strain F113, indicating that phenotypic changes have occurred in the original homogeneous inoculated strain during its passage through the alfalfa rhizosphere, yielding a heterogeneous population.
FIG. 1.
Colony morphology of P. fluorescens F113. (A) F113 colonies recovered on SA plates after 4 weeks of alfalfa rhizosphere colonization. Colonies of different morphology are clearly observed. (B) Colonies of phenotypic variants C, F, and S after isolation and subculture on SA plates. The morphology of the colonies was maintained on subsequent cultures.
In order to test the distribution pattern of the phenotypic variants, the roots of inoculated plants were divided into three parts: the upper region until the beginning of secondary roots, the central part containing secondary roots, and the last centimeter. It was observed that the wild-type compact colonies were recovered from throughout the root system, while the phenotypic variants were recovered mostly from the central part and last centimeter of the root, suggesting that the different subpopulations reflect specialization in colonizing different parts of the root (Table 1).
TABLE 1.
Phenotypic variants isolated from different parts of alfalfa roots after rhizosphere colonization by P. fluorescens F113a
| Root partb | CFU (104) of phenotypic variants | Total CFU (104) per root | % of phenotypic variants per root |
|---|---|---|---|
| Basal part | <1 | 69.6 | <1 |
| Medium part | 87.7 | 896.4 | 9.8 |
| Root tip | 13.1 | 55.7 | 23.5 |
Roots were collected 3 weeks after inoculation. Experiments were done in triplicate, and results showed less than 15% standard variation.
Basal part, from the stem base to the beginning of secondary roots; medium part, the rest of the root system, including secondary roots; root tip, the last centimeter of the root.
From the phenotypic variants with different morphologies that arose in the rhizosphere colonization experiments, we isolated three colonies for further studies (Fig. 1B) and termed them variants C, S, and F. Type C colonies showed a morphology indistinguishable from that of the inoculated F113 strain, while variants S and F presented a clearly different morphology and produced diffuse colonies, S colonies being more irregular than F colonies. The phenotypes of the three variants were stable and maintained in subsequent cultures. The biochemical profiles of variants C, F, and S were analyzed by using API 20 NE strips. The phenotypes of the three variants were identical except that variant F did not possess a protease activity and variant S was unable to reduce nitrate. When grown on asparagine as the sole nitrogen source, variant S excreted ammonia, alkalinizing the spent medium to pH 9.
The appearance of phenotypic variants during alfalfa rhizosphere colonization might indicate that the F and S variants were better adapted to colonize this ecosystem. To test this hypothesis, colonization by the F and S variants was compared to colonization by the wild-type strain F113, variant C, in separate gnotobiotic systems. Experiments were done in triplicate, and the results showed less than 15% standard deviation. Average root weight was 137 ± 17 mg. Variant C was recovered from the alfalfa rhizosphere at the same level as F and S variants, 106 CFU per root, showing no significant difference (P < 0.15). These results indicated that none of the F or S variants colonized the rhizosphere better than variant C when tested individually. It is important to note that while colonies from F113-inoculated plants again showed phenotypic variation, reversion to the wild-type morphology in plants inoculated with variants F and S was not observed.
To test if variants F and S were more competitive than the wild type in rhizosphere colonization, plants were coinoculated in a 1:1 ratio with the C variant and one antibiotic resistance-tagged variant, F or S. Neither of the variants was recovered at a significantly higher level than the wild-type strain, indicating that variants F and S are not more competitive for rhizosphere colonization than wild-type F113.
Phenotypic variants S and F are affected in motility.
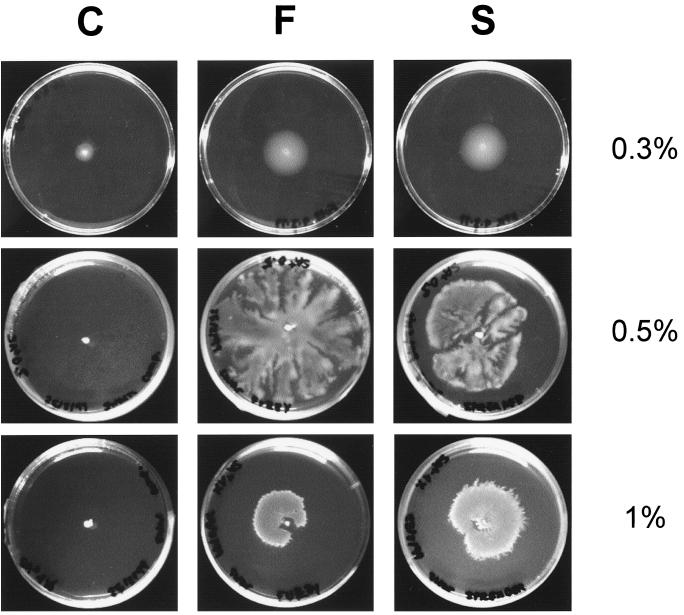
The diffuse morphology shown by variants S and F and their distribution pattern in rhizosphere colonization suggested that they might be affected in motility on solid substrates. To test this hypothesis, the motility behavior of the three variants was investigated on several agar concentrations. As shown in Fig. 2, swimming (0.3% agar concentration) was faster for variants F and S, and these variants produced swimming haloes with a diameter twice the size of that of the C form after 26 h of incubation. When higher agar concentrations were used, the F and S variants showed swarming motility, even at agar concentrations of 1.5%. Swarming under these conditions was never observed for the C variant. Furthermore, variants S and F showed different motility behavior, with variant S swimming faster than variant F and variant F swarming faster than S (not shown).
FIG. 2.
Swimming (0.3% agar) and swarming (0.5 and 1% agar) of C, F, and S variants on SA plates. Swimming motility displayed on 0.3% agar plates presented concentric haloes, while swarming had a dendritic pattern. The C variant showed no swarming ability at any agar concentration.
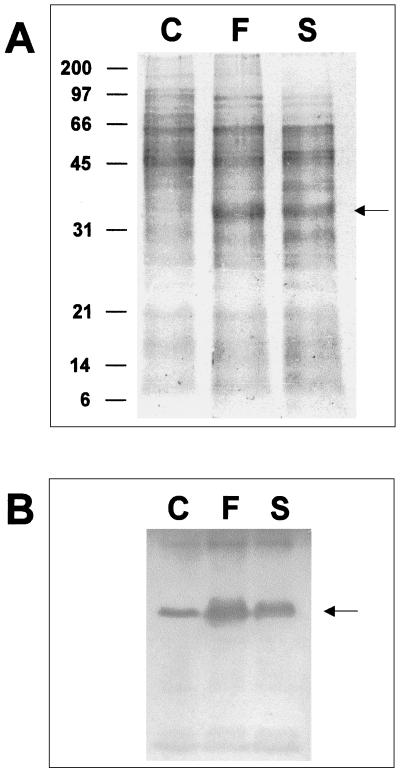
Swimming and swarming motility depends on flagella (25). To test whether the motility behavior of variants S and F was due to flagellin overproduction, proteins from liquid culture supernatants of the three variants were precipitated and electrophoresed. As shown in Fig. 3A, a protein of about 35 kDa was overproduced in variants S and F. No differences were observed for other proteins in the three variants. Western blot analysis with an antiflagellin antiserum for P. fluorescens (14) demonstrated that the overproduced protein corresponded to flagellin (Fig. 3B).
FIG. 3.
Flagellin production by P. fluorescens F113 phenotypic variants. (A) Polyacrylamide gel electrophoresis of total extracellular proteins. The overproduced FliC, of approximately 35 kDa, is marked with an arrow. Molecular size markers are indicated on the left. (B) Western blot analysis using an antiflagellin antiserum. Only the band corresponding to flagellin (marked with an arrow) was detected. Staining shows overproduction of FliC in variants F and S.
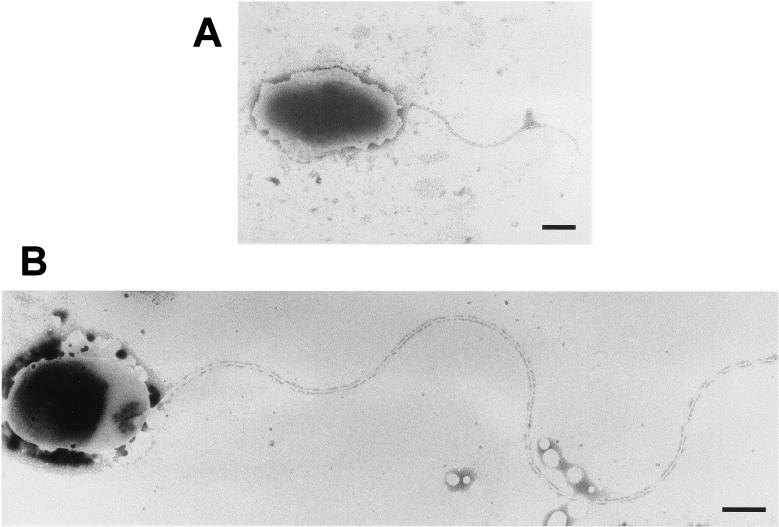
Cells of the three variants were observed by both optical and transmission electron microscopy. Cells from the swarming edges of the F and S variants were elongated in comparison to cells from the center of the swarm plates (not shown), as has been observed previously in P. aeruginosa (33). Besides, when cells from liquid cultures of the three variants were observed by phase-contrast microscopy, the F and S variants displayed significant greater movement than C variant cells (not shown). However, we have not observed hyperflagellation. Cells with one and two polar flagella were observed by transmission electron microscopy from cultures of the three variants, not only from swarming plates but also from exponentially growing liquid cultures. Transmission electron microscopy also showed that cells from the F and S variants grown in all conditions possessed flagella that were much longer than the flagella from the C variant (Fig. 4). Flagella from C cells measured 2.4 ±0.9 μm, while flagella from F were 7.0 ± 0.8 μm and those from S were 6.6 ± 1.3 μm, indicating that the result of flagellin overproduction was longer flagella rather than more flagella, as is the case for other swarming bacteria (25).
FIG. 4.
Transmission electron microscopy of phenotypic variants. (A) C variant possesses one or two normal polar flagella. Bar, 500 nm. (B) F variant, and also S variant (not shown), possesses flagella more than twofold longer than those of variant C. Bar, 300 nm.
Two primers derived from conserved regions of the fliC gene, which encodes flagellin in other fluorescent pseudomonads, were used to amplify genomic DNA from strain F113. An amplification product of 563 bp was obtained and sequenced, showing 85% identity with the Pseudomonas putida type b flagellin (GenBank accession number AF034765). This fragment was used to probe genomic DNA from the three variants, and a single band from the three variants hybridized with the fliC probe. This probe was also used to screen an F113 cosmid library, and overlapping cosmids were isolated from all the positive colonies. These results strongly indicated that P. fluorescens F113 produces a single flagellin and that flagellin overproduction and therefore swarming motility is not due to the production of an alternative flagellin. New primers were designed to obtain the complete sequence of fliC from the cosmid isolated from the library. The fliC sequence, including the promoter region, has been deposited in the GenBank database with accession number AF399739.
To test whether flagellin overproduction in the F and S variants was transcriptionally regulated, new primers were designed to amplify the promoter region of fliC from each variant. The amplified promoter regions from the three variants were cloned into an expression vector containing the promoterless lacZ gene, and these constructs were introduced into different backgrounds and β-galactosidase activity was measured (Table 2). When constructs containing promoters were introduced into the respective variants, overexpression was observed for variants F and S compared to variant C. Overexpression of fliC at the transcriptional level was therefore correlated with overproduction of flagellin. When constructs containing each promoter fused to lacZ were introduced into variant C, no differences in expression were observed, indicating that the differences of expression observed before were not due to changes in the promoter region. Finally, the promoter region amplified from variant C fused to lacZ was introduced into each variant, and the expression measured was higher for variants F and S, indicating that the differences are due to the particular regulation of fliC in each variant. Subsequently, the promoter regions of fliC from the C, F, and S variants were sequenced and showed no differences (not shown), indicating that differences in fliC expression among the variants were not produced by sequence differences in the promoters.
TABLE 2.
Expression of fliC promoters isolated from variants C, F, and S, measured as β-galactosidase activitya of lacZ fusions
| Constructb | β-Galactosidase activity (Miller units)
|
||
|---|---|---|---|
| C | F | S | |
| pBG1208 | 840 | 3,139 | 1,754 |
| pBG1209 | 963 | 2,408 | —c |
| pBG1210 | 604 | — | 1,432 |
The experiments were done in triplicate in each case and showed less than 10% variation.
Constructs pBG1208, pBG1209, and pBG1210 carry the fliC promoter region from variants C, F, and S, respectively, fused to promoterless lacZ.
—, not determined.
Variant F is affected in multiple traits relevant to rhizosphere colonization.
To further characterize the three phenotypic variants, characteristics that have been shown to be important in rhizosphere colonization (39) were tested. Variant F produced a lipopolysaccharide (LPS) with a different electrophoretic profile, showing an extra band for the O antigen (not shown) compared with variants C and S. The ability to form biofilms was tested by inoculating each variant in unshaken liquid medium. Variants C and S formed a biofilm in the liquid-air interface, while variant F grew in the whole volume and eventually flocculated. The flocculation behavior observed for variant F grown in static liquid culture is probably related to the different LPS structures.
P. fluorescens F113 produces the siderophore pyoverdin under iron limitation (28). The F variant overproduced pyoverdin in iron-limited medium SA, as judged from the fluorescence of the culture after excitation with UV light. On iron-rich medium, pyoverdin production is inhibited by the Fur repression system (36). However, variant F produced pyoverdin on the iron-rich medium LB, although at a much lower level than in SA medium.
To test whether the overproduction of pyoverdin was related to increased transcription of the genes responsible for pyoverdine synthesis, a fusion of the pvdA promoter with a promoterless lacZ (pPV51) (35) was introduced into the three variants. Higher expression in the F variant background was observed in both SA and LB media (3,575 and 252 Miller units, respectively), corresponding to low and high iron availability. The results for variants C and S were 1,344 and 828 Miller units, respectively, in SA medium and 25 and 0 Miller units, respectively, in LB medium. The results show that variant F overexpresses genes coding for the pathway of pyoverdin biosynthesis in high- and low-iron conditions.
Variant F is also affected in the production of secondary metabolites and secreted proteins. Unlike C and S, the F variant was unable to produce cyanide and exoprotease (Table 3).
TABLE 3.
Summary of the phenotypic features of C, F and S variants
| Characteristic | C | F | S |
|---|---|---|---|
| Colony morphology | Compact, opaque | Diffuse, translucent | Diffuse, translucent |
| Motility | Normal | Enhanced | Enhanced |
| fliC expression | Normal | Overexpressed | Overexpressed |
| Avg flagellum length (μm) | 2.3 | 7.0 | 6.6 |
| LPS profilea | 3 | 4 | 3 |
| Flocculation in liquid medium | − | + | − |
| Pyoverdin production on LB | − | + | − |
| Pyoverdin production on SA | + | ++ | + |
| Cyanide production | + | − | + |
| Exoprotease production | + | − | + |
Number of bands in the LPS O antigen.
A point mutation in gacA is responsible for changes in secondary metabolism of variant F.
GacA and GacS are the activator and sensor, respectively, of a two-component regulatory system that regulates secondary metabolism. The production of extracellular proteases and cyanide in P. fluorescens is coordinately regulated by the gacS/gacA system (7). It has recently been reported that mutations in the gacA or gacS gene cause overproduction of pyoverdin on iron-limited medium (16), suggesting that the gacA/gacS system may also regulate the production of this siderophore.
To test whether the multiple alterations in the F variant were due to a mutation in either of these genes, plasmids containing the cloned gacA and gacS genes were introduced into this variant. The cloned gacA gene complemented pyoverdin, cyanide, and exoprotease production but not the morphology, motility, LPS pattern, or flocculation on static liquid culture of the F variant. The plasmid containing gacS did not complement any of the phenotypes. These results indicated that the F variant is affected in the gacA gene. The sequencing of the gacA allele in the F variant showed a point mutation resulting in changing a glycine to a serine (position 26).
Summarizing the characteristics of the three variants (Table 3), F and S differ from C in colony shape and enhanced motility, which is correlated with longer flagella, overproduction of flagellin, and overexpression of fliC. For other traits tested, variants C and S were identical except for nitrate reduction, in which variant S was deficient. Apart from all the traits related to motility, variant F displayed two more classes of different phenotypic characteristics, one related to production of secondary metabolites (pyoverdin, exoprotease, and HCN), which was complemented by the gacA gene, and another not related to the gacA/gacS system, consisting of two traits that might be linked, a different LPS pattern and flocculation in static liquid cultures.
Site-specific recombinase is the major factor influencing phenotypic variation of F113 during rhizosphere colonization.
A site-specific recombinase, encoded by the sss gene in Pseudomonas spp., has been shown to be important for rhizosphere colonization (13, 40). Primers were designed after the published sequences of the sss gene in other Pseudomonas strains to amplify this gene from F113. An amplification product of 696 bp was obtained and sequenced, corresponding to a fragment of the sss gene. This fragment was used to probe genomic DNA from the three variants and to screen the F113 cosmid library. The same cosmid was isolated from all positive colonies, and new primers were designed to obtain the complete sequence of the sss gene, which has been deposited in the GenBank database with accession number AF416734.
The amplified 696-bp fragment was interrupted with a kanamycin resistance marker and cloned into pK18mobsac. This construct was introduced into wild-type F113 to obtain an sss insertion mutant. Putative mutants were obtained and tested by Southern blotting (not shown). An sss mutant was selected (F113-sss2) and used to perform rhizosphere colonization experiments. Alfalfa seedlings were inoculated with F113-sss2 and with wild-type F113 separately. CFU were recovered from the rhizosphere, and phenotypic variants were counted. From the rhizosphere of plants inoculated with F113, 10% of the total CFU recovered were phenotypic variants, as was established in previous experiments. From the rhizosphere of plants inoculated with F113-sss2, very few phenotypic variants were recovered, representing less than 1% of total CFU. These reductions in phenotypic variants recovered from the rhizosphere indicated that sss plays a major role in the phenotypic variation occurring during rhizosphere colonization and that phase variation due to the action of this site-specific recombinase is the major mechanism of phenotypic variation.
It has been described that spontaneous gacA/gacS mutants show a diffuse morphology (20), which is similar to that of the F113 phenotypic variants that arose after rhizosphere experiments. The high frequency of spontaneous gacA mutants has been observed particularly on rich media (16). To test if the gacA mutation was related to phenotypic variation during rhizosphere colonization, we used the exoprotease production phenotype because it is an easily detectable trait reflecting gacA/gacS function. We checked the frequency of colonies that did not show protease activity among morphological variants and wild-type colonies isolated from the rhizosphere.
When colonies recovered from plants inoculated with F113 were tested, colonies lacking protease activity represented 6% of the phenotypic variants, compared to only 0.3% of the wild-type colonies, indicating that mutations in the gacA/gacS system were responsible for a minor part of the phenotypic variation. When the same test was performed with the few colonies (26 colonies) derived from the sss mutant that showed a variant phenotype, 89% did not produce exoprotease, indicating that mutations in gacA/gacS were responsible for the majority of phenotypic variation in this background. These results indicate that both selection of the gacA/gacS mutant phenotype and phase variation due to the action of a site-specific recombinase occur during alfalfa rhizosphere colonization, phase variation being the main factor responsible for the phenotypic diversity.
DISCUSSION
Phase variation is a process that allows the diversification of a single bacterial population into differentiated subpopulations. Here we have shown that this process takes place during alfalfa rhizosphere colonization by P. fluorescens F113. Three phase variants, C, F, and S, have been observed and isolated, with the C variant presenting the wild-type phenotype.
Variants F and S are affected in motility on surfaces. Three types of movement on surfaces have been described in Pseudomonas spp.: swimming, swarming, and twitching (27). Swimming and swarming are flagellum dependent (25), and twitching depends on type IV pili (9), although pili have been implicated in the swarming of a P. aeruginosa strain (33). Variants F and S have shown differences in swimming and swarming compared with the wild-type C variant, and these differences can be attributed to overexpression of the fliC gene, leading to the overproduction of flagellin. Both the F and S variants showed swimming behavior stronger than that of the wild type and were able to swarm under experimental conditions that did not allow swarming of the wild-type strain.
Swarming of the F and S variants is atypical for several reasons. Together with other pseudomonads, the F and S variants of P. fluorescens F113 are the only polarly flagellated bacteria that have been shown to swarm, but in contrast to P. aeruginosa and other swarming bacteria, we did not observe an increase in the number of flagella. Instead, the F and S variants produced flagella that were more than twofold longer than the wild-type flagella. To our knowledge, this increase in the length of flagella has not been observed in other swarming bacteria. The F and S variants can swarm in our experimental conditions in a medium that contains a single amino acid, asparagine, as the nitrogen source. Most bacteria require a mixture of amino acids to swarm (25), and only for Proteus mirabilis has it been shown that a single amino acid, glutamine in this case, supports swarming (3). F and S variants can swarm on agar concentrations that inhibit swarming of other bacteria (46). Swarming on agar concentrations higher than 1% has been shown only for strong swarmers such as P. mirabilis (2, 41) and Vibrio parahaemolyticus (5). The increased motility in variants F and S correlates with longer flagella, flagellin overproduction, and overexpression of fliC due to its regulation in both variants and not to changes in the promoter region.
We observed differences in the swarming behavior of the F and S variants, particularly at high agar concentrations, where the F variant swarms better than the S variant. As fliC expression and FliC production were similar for both strains, it is reasonable that differences are due to factors other than flagella that are implicated in swarming, such as surfactant production (23, 37) and LPS structure (see below).
Aside from morphology, nitrate reduction, and motility behavior, the C and S variants were identical for all other phenotypes tested. However, the F variant differs in all of the traits tested. We observed that while the C and S variants form a film on the surface of static liquid medium, variant F flocculates and sinks. This flocculation behavior has been observed previously in a spontaneous P. fluorescens mutant that is selected under static culture conditions (45). The observation that the flocculating F variant shows alterations in the LPS electrophoretic profile suggests that these two phenotypes are related, as it has been shown that LPS mutants of several bacterial species flocculate on liquid media (21). Different flocculation behavior has also been observed in biofilms formed by P. aeruginosa phase variants (15). Differences in swarming between the F and S variants might also be related to differences in the LPS. It has recently been shown than in Salmonella enterica serovar Typhimurium, the LPS plays a critical role in swarming, probably by acting as a wetting agent (56).
The F variant also produced larger amounts of the siderophore pyoverdin under low- and high-iron conditions, correlated with overexpression in the same conditions of pvdA, encoding an enzyme of the pyoverdin biosynthesis pathway. This wild-type phenotype was restored with the cloned gacA gene. It has recently been shown that gacA and gacS mutants overproduce pyoverdin under iron limitation (16). P. tolaasii phase variants affected in the gacS homologue pheN form green fluorescent colonies, compared to the white colonies formed by the wild type (22). All these results suggest that the gacA/gacS system downregulates pyoverdin production in a variety of fluorescent pseudomonads. It has been reported that the gacA gene is necessary for swarming of Pseudomonas syringae (32). However, the gacA-defective F variant is able to swarm, indicating either that swarming in P. fluorescens F113 does not require gacA or that swarming in these phase variants is different from the swarming of the wild-type strain.
The gacA/gacS system coordinately regulates the production of secondary metabolites, namely, HCN and secreted proteins such as exoprotease (7). The F variant is also affected in these phenotypes and does not produce HCN or exoprotease activity. These phenotypes were also complemented by the cloned gacA gene but not by the gacS gene, demonstrating that the F variant is affected in that gene. The phenotypes related to motility, LPS, and flocculation were not restored by the cloned gacA gene. Sequencing of the gacA allele in the F variant showed that a point mutation, resulting in the change of a glycine to a serine, was responsible for the lack of function.
Spontaneous point mutations in gacA have also been described in other pseudomonads (20). Spontaneous gacA/gacS mutants present a diffuse morphology, similar to what we have observed in the phenotypic variants recovered from rhizosphere. However, the introduction of gacA gene into the F variant did not complement morphology. Although the appearance of gacA/gacS-related phenotypes among phase variants in different species of fluorescent pseudomonads (12, 15, 24, 54) and the alteration in pyoverdin production in a P. aeruginosa sss mutant suggest that the gacA gene might be subjected to phase variation, phenotype selection is also possible. Our results, the sequencing of gacA allele in variant F showing point mutations, and the appearance of gacA mutants from plants inoculated with F113-sss2 demonstrated that during rhizosphere colonization, this gene is not subjected to phase variation, but a selection of the phenotype is occurring. Phenotype selection of gacA/gacS mutants has been described for Pseudomonas spp. growing on rich culture media and under certain conditions can represent 60% of the bacteria in a culture (16, 20). We have shown that gacA mutants also appear in the alfalfa rhizosphere, which is not as rich as a culture medium, and they might play a role in rhizosphere colonization.
Phase variation is likely to be important for rhizosphere colonization, as the appearance of new phenotypes can allow the establishment of specialized subpopulations to colonize a heterogeneous ecological niche. It has been shown that P. fluorescens mutants affected in a site-specific recombinase are defective in competitive rhizosphere colonization (13). We have shown here that the sss-encoded site-specific recombinase produces phase variation during rhizosphere colonization, being the major factor in phenotypic diversification.
The advantages of flagellar variants for rhizosphere colonization are obvious, as a subpopulation with enhanced surface motility is likely to colonize distal parts of the rhizosphere that are not easily reached by the wild-type strain. The accumulation of these variants that we have found in the distal part of the root system supports this hypothesis. Ammonia excretion by variant S could also give a selective advantage for this variant by local alkalinization that might inhibit the growth of other bacteria or fungi. The advantages of gacA mutations are less obvious. It has been shown that gacA mutants are not affected in root colonization (43), although they have limited persistence in the rhizosphere environment (52). However, if gacA mutants represent a subpopulation, they can save energy by not producing a set of secondary metabolites and proteins that will be produced by other subpopulations, and from which the gacA subpopulation will benefit. In turn, these gacA variants will secrete a larger amount of the siderophore pyoverdin that will also be useful for the rest of the population.
The results that we have presented showed that none of the phase variants is superior to the wild type in terms of rhizosphere colonization or competitive colonization, although the distribution pattern showed a specialization in rhizosphere colonization. However, it has to be considered that the wild type showed variation during colonization experiments, while the F and S forms were apparently locked in a single population. Considering that P. fluorescens sss mutants are affected in competitive rhizosphere colonization (13), it is likely that the ability to diversify in specialized subpopulations, and not a specific phenotype, is the important trait for rhizosphere colonization.
Acknowledgments
We are grateful to Carmen Gutierrez Rueda for helping with electron microscopy and to Ine Mulders and Ben Lugtenberg for the antiflagellin antiserum.
This work was supported by grants from the European Union (BIO4-CT97-2227 and QLK3-CT2000-31759) and Comunidad Autónoma de Madrid (07 M-0062-2000). M.M. is the recipient of a Ramon y Cajal contract from the Spanish Ministry of Science and Technology.
REFERENCES
- 1.Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:5724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, C., and C. Hughes. 1991. Bacterial swarming: an example of prokaryotic differentiation and multicellular behavior. Sci. Prog. 75:403-422. [PubMed] [Google Scholar]
- 3.Allison, C., H. C. Lai, D. Gygi, and C. Hughes. 1993. Cell diferentiation of Proteus mirabilis is initiated by glutamine, a specific chemoattractant for swarming cells. Mol. Microbiol. 8:53-60. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Belas, M. R., and R. R. Colwell. 1982. Scanning electron microscope observation of the swarming phenomenon of Vibrio parahaemolyticus. J. Bacteriol. 150:956-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumer, C., S. Heeb, G. Pessi, and D. Haas. 1999. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. USA 96:14073-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bollag, D. M., and S. Edelstein. 1991. Protein methods, p. 74. Wiley-Liss, New York, N.Y.
- 9.Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. [DOI] [PubMed] [Google Scholar]
- 10.Brazil, G. M., L. Kenefick, M. Callanan, A. Haro, V. de Lorenzo, D. N. Dowling, and F. O'Gara. 1995. Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bph gene expression in the rhizosphere. Appl. Environ. Microbiol. 61:1946-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castric, K. F., and P. A. Castric. 1983. Method for rapid detection of cyanogenic bacteria. Appl. Environ. Microbiol. 45:701-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chabeaud, P., A. de Groot, W. Bitter, J. Tommassen, T. Heulin, and W. Achouak. 2001. Phase-variable expression of an operon encoding extracellular alkaline protease, a serine protease homolog, and lipase in Pseudomonas brassicacearum. J. Bacteriol. 183:2117-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekkers, L. C., C. C. Phoelich, L. Van der Fits, and B. J. J. Lugtenberg. 1998. A site-specific recombinase is required for competitive root colonization by Pseudomonas fluorescens WCS365. Proc. Natl. Acad. Sci. USA 95:7051-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Weger, L. A., C. I. M. van der Vlugt, A. H. M. Wijfjes, P. A. H. M. Bakker, B. Schippers, and B. J. J. Lugtenberg. 1987. Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J. Bacteriol. 169:2769-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Déziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duffy, B. K., and G. Défago. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 66:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dybvig, K. 1993. DNA rearrangements and phenotypic switching in prokaryotes. Mol. Microbiol. 10:465-471. [DOI] [PubMed] [Google Scholar]
- 18.Fåhraeus, G. 1957. The infection of clover root hairs by nodule bacteria studied by simple glass technique. J. Gen. Microbiol. 16:374-381. [DOI] [PubMed] [Google Scholar]
- 19.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaffney, T. D., S. T. Lam, J. Ligon, K. Gates, A. Frazelle, J. Di Maio, S. Hill, S. Goodwin, N. Torkewitz, A. M. Allhouse, H. J. Kempf, and J. O. Becker. 1994. Global regulation of expression of antifungal factors by a Pseudomonas fluorescens biological control strain. Mol. Plant-Microbe Interact. 7:455-463. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg, J. B., M. J. J. Coyne, A. N. Neely, and I. A. Holder. 1995. Avirulence of a Pseudomonas aeruginosa algC mutant in a burned-mouse model of infection. Infect. Immun. 63:4166-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grewal, S. I. S., B. Han, and K. Johnstone. 1995. Identification and characterization of a locus which regulates multiple functions in Pseudomonas tolaasii, the cause of brown blotch disease of Agaricus bisporus. J. Bacteriol. 177:4658-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gygi, D., M. M. Rahman, H. C. Lai, R. Carlson, J. Guard-Peter, and C. Hughes. 1995. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol. Microbiol. 17:1167-1175. [DOI] [PubMed] [Google Scholar]
- 24.Han, B., A. Pain, and K. Johnstone. 1997. Spontaneous duplication of a 661bp element within a two component sensor regulator gene causes phenotypic switching in the colonies of Pseudomonas tolaasii, cause of brown blotch disease of mushrooms. Mol. Microbiol. 25:211-218. [DOI] [PubMed] [Google Scholar]
- 25.Harshey, R. M. 1994. Bees aren't the only ones: swarming in gram-negative bacteria. Mol. Microbiol. 13:389-394. [DOI] [PubMed] [Google Scholar]
- 26.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the on and off of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 27.Henrichsen, J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36:478-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Höfte, M., Q. Dong, S. Kourambas, V. Krishnapillai, D. Sherratt, and M. Mergeay. 1994. The sss gene product, which affects pyoverdin production in Pseudomonas aeruginosa 7NSK2, is a site-specific recombinase. Mol. Microbiol. 14:1011-1020. [DOI] [PubMed] [Google Scholar]
- 29.Hrabak, E. M., and D. K. Willis. 1992. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 174:3011-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlson, U., D. Dowling, F. O'Gara, R. Rivilla, M. Bittens, S. Francesconi, H. Pritchard, and H. C. Pedersen. 1998. Development of self-contained plant/GMM systems for soil bioremediation, p. 23-31. In G. E. de Vries (ed.), Past, present and future risk assessment when using GMOs. Overschild, The Netherlands.
- 31.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 32.Kinscherf, T. G., and D. K. Willis. 1999. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J. Bacteriol. 181:4133-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köhler, T., L. K. Curty, F. Barja, C. van Delden, and J.-C. Pechère. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laville, J., C. Voisard, C. Keel, M. Maurhofer, G. Défago, and D. Haas. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. USA 89:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leoni, L., A. Ciervo, N. Orsi, and P. Visca. 1996. Iron-regulated transcription of the pvdA gene in Pseudomonas aeruginosa: effect of fur and PvdS on promoter activity. J. Bacteriol. 178:2299-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leoni, L., N. Orsi, V. de Lorenzo, and P. Visca. 2000. Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa. J. Bacteriol. 182:1481-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindum, P. W., U. Anthoni, C. Christophersen, L. Eberl, S. Molin, and M. Givskov. 1998. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lloret, J., L. Bolaños, M. M. Lucas, J. Peart, N. J. Brewin, I. Bonilla, and R. Rivilla. 1995. Ionic stress and osmotic pressure induce different alterations in the lipopolysaccharide of a Rhizobium meliloti strain. Appl. Environ. Microbiol. 61:3701-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lugtenberg, B. J. J., and L. C. Dekkers. 1999. What makes pseudomonas bacteria rhizosphere competent? Environ. Microbiol. 1:9-13. [DOI] [PubMed] [Google Scholar]
- 40.Lugtenberg, B. J. J., L. C. Dekkers, M. Bansraj, G. V. Bloemberg, M. Camacho, T. Chin, A. Woeng, K. van den Hondel, L. Kravchenko, I. Kuiper, A. L. Lagopodi, I. Mulders, C. Phoelich, A. Ram, I. Tikhonovich, S. Tuinman, C. Wijffelman, and A. Wijfjies. 1999. Pseudomonas genes and traits involved in tomato root colonization, p. 9-13. In P. de Wit, T. Bisseling, and W. Stiekema (ed.), Biology of plant-microbe interactions. International Society for Molecular Plant-Microbe Interactions, St. Paul, Minn.
- 41.McCarter, L., and M. Silverman. 1990. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 4:1057-1062. [DOI] [PubMed] [Google Scholar]
- 42.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Natsch, A., C. Keel, H. A. Pfirter, D. Haas, and G. Défago. 1994. Contribution of the global regulation gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl. Environ. Microbiol. 60:2553-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park, S. F., D. Purdy, and S. Leach. 2000. Localized reversible frameshift mutation in the flhA gene confers phase variability to flagellin gene expression in Campylobacter coli. J. Bacteriol. 182:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in heterogeneous environment. Nature 394:69-72. [DOI] [PubMed] [Google Scholar]
- 46.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sacherer, P., G. Défago, and D. Haas. 1994. Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 116:155-160. [DOI] [PubMed] [Google Scholar]
- 48.Sarkari, J., N. Pandit, E. R. Moxon, and M. Achtman. 1994. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of promoter containing poly-cytidine. Mol. Microbiol. 13:207-217. [DOI] [PubMed] [Google Scholar]
- 49.Saunders, J. R. 1994. Population genetics of phase variable antigens, p. 247-268. In Population genetics of bacteria. Cambridge University Press, New York, N.Y.
- 50.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 51.Scher, F. M., and R. Baker. 1982. Effect of Pseudomonas putida and a synthetic iron chelator on induction of soil suppressiveness to Fusarium wilt pathogens. Phytopathology 72:1567-1573. [Google Scholar]
- 52.Schmidli-Sacherer, P., C. Keel, and G. Defago. 1997. The global regulator GacA of Pseudomonas fluorescens CHA0 is required for suppression of root diseases in dicotyledons but not in Gramineae. Plant Pathol. 46:80-90. [Google Scholar]
- 53.Shanahan, P., D. O'Sullivan, P. Simpson, J. D. Glennon, and F. O'Gara. 1992. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinha, H., A. Pain, and K. Johnstone. 2000. Analysis of the role of recA in phenotypic switching of Pseudomonas tolaasii. J. Bacteriol. 182:6532-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 56.Toguchi, A., M. Siano, M. Burkart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Woude, M., B. Braaten, and D. Low. 1996. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 4:5-9. [DOI] [PubMed] [Google Scholar]
- 58.Voisard, C., C. Keel, D. Haas, and G. Défago. 1989. Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 8:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson, K. J., A. Sessitsch, J. C. Corbo, K. E. Giller, A. D. Akkermans, and R. A. Jefferson. 1995. Beta-glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology 141:1691-1705. [DOI] [PubMed] [Google Scholar]
- 60.Wolk, C. P., Y. Cai, and J. M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. USA 88:5355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]