Abstract
Previous studies established the critical roles of AlcR and alcaligin inducer in positive regulation of alcaligin siderophore biosynthesis and transport genes in Bordetella pertussis and Bordetella bronchiseptica. Transcriptional analyses using plasmid-borne alcR genes of B. pertussis UT25 and B. bronchiseptica B013N to complement the alcR defect of B. bronchiseptica strain BRM13 (ΔalcR1 alcA::mini-Tn5 lacZ1) revealed interspecies differences in AlcR inducer requirements for activation of alcABCDER operon transcription. Whereas the B. pertussis UT25 AlcR protein retained strong inducer dependence when produced from multicopy plasmids, B. bronchiseptica B013N alcR partially suppressed the alcaligin requirement for transcriptional activation. Functional analysis of AlcR chimeras produced by interspecies domain swapping and interspecies reciprocal site-specific mutagenesis determined that the phenotypic difference in AlcR inducer dependence was due to a single amino acid difference within the proposed inducer-binding and multimerization domain of AlcR. Structural predictions guided the design of a mutant AlcR protein with a single amino acid substitution at this critical position, AlcR(S103T), that was fully constitutive not only when produced from multicopy plasmids but also at a single-copy gene dosage. These results indicate that AlcR residue 103 affects a critical determinant of alcaligin inducer dependence of AlcR-mediated transcriptional activation. The alcR(S103T) mutant allele is the first alcR(Con) mutant allele identified.
Bordetella pertussis and Bordetella bronchiseptica are gram-negative mucosal pathogens of humans and nonhuman mammals. In response to iron starvation, these bacteria produce and utilize the macrocyclic dihydroxamate siderophore alcaligin (13, 28). B. pertussis and B. bronchiseptica can also utilize some nonnative siderophores produced by other bacterial species as nutritional iron sources (3, 4). Apart from these siderophore iron sources, certain host iron sources are also readily utilized by these bacterial pathogens. Vanderpool and Armstrong provided the first genetic evidence of a functional host iron uptake system in B. pertussis and B. bronchiseptica (38). B. pertussis and B. bronchiseptica possess a specific transport system for heme iron utilization that is encoded by the bhu gene cluster; production of the required heme iron receptor protein BhuR is both iron repressible and heme inducible. Evidence suggesting that B. pertussis and B. bronchiseptica can utilize the host iron glycoproteins lactoferrin and transferrin (26, 33) has also been reported.
In B. pertussis and B. bronchiseptica, the iron starvation stress response is regulated transcriptionally by the Fur repressor protein with ferrous iron as the corepressor (7). Fur controls the expression of the alcABCDER alcaligin biosynthesis and regulation operon and the fauA ferric alcaligin outer membrane receptor gene at three promoter-operator regions within the alcaligin gene cluster (5, 7, 10, 22, 23; T. J. Brickman and S. K. Armstrong, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. B-241, p. 70, 1997). AlcR is an AraC family transcriptional regulator (5, 32) that is necessary for maximal expression of alcaligin siderophore biosynthesis and transport genes under iron starvation stress conditions (5, 10, 12). Transcriptional activation by AlcR requires the presence of alcaligin siderophore acting as the inducer (12). Since these Bordetella species are capable of utilizing multiple alternative iron sources for growth, priority regulation involving the chelator itself as the inducer ensures that alcaligin biosynthesis and uptake functions are expressed maximally only under conditions in which alcaligin is perceived in the environment (12). We hypothesize that the ability of Bordetella spp. to prioritize the expression of their different iron systems may be important for effective adaptation to changes in the host environment during the course of an infection.
AraC family regulators are also known to be involved in the regulation of other bacterial iron transport systems. The AraC-like protein PchR positively regulates expression of the pyochelin siderophore system of Pseudomonas aeruginosa; transcriptional activation is responsive to pyochelin, and induction requires a functional ferric pyochelin receptor (18, 19). Similarly, the Yersinia pestis AraC family regulator YbtA is required for maximal expression of yersiniabactin siderophore biosynthesis and transport genes (15).
In studies characterizing the role of alcaligin as the inducer of AlcR-mediated transcriptional activation, it was observed that B. bronchiseptica AlcR was partially constitutive for activation of a chromosomal alcA::mini-Tn5 lacZ1 fusion gene when AlcR was produced from multicopy plasmids. In contrast, B. pertussis AlcR exhibited normal alcaligin inducer dependence even when overproduced. Elucidation of the molecular basis of this difference in multicopy suppression phenotype, a single amino acid substitution in the predicted inducer-binding and multimerization domain of AlcR, led to the identification of a key determinant of AlcR function. Structural predictions directed the construction of a mutant AlcR protein that was fully constitutive for activation of transcription even at a single-copy gene dosage.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. bronchiseptica strains, plasmid cloning vectors, and alcR+ plasmids used in this study are listed in Table 1. The construction and phenotypic characterization of the B. bronchiseptica ΔalcR1 alcA::mini-Tn5 lacZ1 mutant strain BRM13 have been described previously (12). Escherichia coli DH5α [F− φ80d lacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 λ− gyrA96 relA1] (Gibco BRL, Gaithersburg, Md.) was used as the host strain for routine plasmid construction and propagation and as the donor strain in conjugal transfer of plasmids to Bordetella recipients. The broad-host-range plasmid cloning vectors pRK415 (24) and pBBR1MCS (25) were used in the construction of alcR+ multicopy plasmids used in complementation analyses; the suicide plasmid cloning vector pSS1129 (37) was used to deliver cloned alcR alleles to the B. bronchiseptica chromosome by homologous recombination. DH5α(pRK2013) provided plasmid-encoded mobilization functions (16) in triparental matings to transfer pRK415 and pSS1129 plasmid derivatives to B. bronchiseptica. Plasmids pRK21 and pRK15 have been described previously (5, 12). Plasmids pRK/1.6KP (Bb) and pRK/2.3EP (Bp) were constructed by subcloning the 1.6-kb KpnI-PstI alcR+ DNA region of B. bronchiseptica B013N from plasmid pRK15 (12) and the 2.3-kb EcoRI-PstI alcR+ DNA region of B. pertussis UT25 from cosmid pCP1.11 (23), respectively, to plasmid vector pRK415. The same alcR+ 1.6-kb KpnI-PstI and 2.3-kb EcoRI-PstI DNA fragments of B. bronchiseptica B013N and B. pertussis UT25 were also subcloned to plasmid vector pBBR1MCS to produce plasmids pBB/1.6KP (Bb), pBB/1.6KP (Bp), pBB/2.3EP (Bb), and pBB/2.3EP (Bp).
TABLE 1.
B. bronchiseptica strains and alcR+ plasmids used in this study
| Strain or plasmid | Relevant genotype, phenotype, or description | Reference or source |
|---|---|---|
| BRM13 | B. bronchiseptica B013N ΔalcR1 alcA::mini-Tn5 lacZ1; kanamycinr AlcR− alcaligin deficient | 12 |
| BRM13Ω(ΔalcR1::pSS/Bp) | pSS/Bp plasmid integrant derivative of BRM13; AlcR+ kanamycinr ampicillinr gentamicinr | This study |
| BRM13Ω(ΔalcR1::pSS/Bb) | pSS/Bb plasmid integrant derivative of BRM13; AlcR+ kanamycinr ampicillinr gentamicinr | This study |
| BRM13Ω(ΔalcR1::pSS/Bb-S103T) | pSS/Bp-S103T plasmid integrant derivative of BRM13; AlcR(Con) kanamycinr ampicillinr gentamicinr | This study |
| pRK415 | Mobilizable broad-host-range plasmid cloning vector; tetracycliner; RK2 origin | 24 |
| pRK21 | pRK415 with 1.6-kb KpnI-PstI B. pertussis UT25 alcR+ DNA insert fragment; AlcR+ tetracycliner; formerly designated pP9KP | 5, 12 |
| pRK15 | pRK415 with 2.3-kb EcoRI-PstI B. bronchiseptica B013N alcR+ DNA insert fragment; AlcR+ tetracycliner | 12 |
| pRK/1.6KP (Bb) | pRK415 with 1.6-kb KpnI-PstI B. bronchiseptica B013N alcR+ DNA insert fragment; AlcR+ tetracycliner | This study |
| pRK/2.3EP (Bp) | pRK415 with 2.3-kb EcoRI-PstI B. pertussis UT25 alcR+ DNA insert fragment; AlcR+ tetracycliner | This study |
| pBBR1MCS | Mobilizable broad-host-range plasmid cloning vector; chloramphenicolr | 25 |
| pBB/1.6KP (Bp) | pBBR1MCS with 1.6-kb KpnI-PstI B. pertussis UT25 alcR+ DNA insert fragment; AlcR+ chloramphenicolr | This study |
| pBB/1.6KP (Bb) | pBBR1MCS with 1.6-kb KpnI-PstI B. bronchiseptica B013N alcR+ DNA insert fragment; AlcR+ chloramphenicolr | This study |
| pBB/2.3EP (Bp) | pBBR1MCS with 2.3-kb EcoRI-PstI B. pertussis UT25 alcR+ DNA insert fragment; AlcR+ chloramphenicolr | This study |
| pBB/2.3EP (Bb) | pBBR1MCS with 2.3-kb EcoRI-PstI B. bronchiseptica B013N alcR+ DNA insert fragment; AlcR+ chloramphenicolr | This study |
| pRK/NpCb | pRK415 with 1.2-kb KpnI-PstI DNA insert fragment, N-terminal coding sequences of B. pertussis UT25 alcR spliced with C-terminal coding sequences of B. bronchiseptica B013N alcR by overlap extension PCR; AlcR+ tetracycliner | This study |
| pRK/NbCp | pRK415 with 1.2-kb KpnI-PstI DNA insert fragment, N-terminal coding sequences of B. bronchiseptica B013N alcR spliced with C-terminal coding sequences of B. pertussis UT25 lcR by overlap extension PCR; AlcR+ tetracyclineR | This study |
| pRK/NpCp | pRK415 with 1.2-kb KpnI-PstI DNA insert fragment, N-terminal coding sequences of B. pertussis UT25 alcR spliced with C-terminal coding sequences of B. pertussis UT25 alcR by overlap extension PCR; AlcR+ tetracycliner | This study |
| pRK/NbCb | pRK415 with 1.2-kb KpnI-PstI DNA insert fragment, N-terminal coding sequences of B. bronchiseptica B013N alcR spliced with C-terminal coding sequences of B. bronchiseptica B013N alcR by overlap extension PCR; AlcR+ tetracycliner | This study |
| pRK/Bp-G103S | pRK415 with 1.2-kb KpnI-PstI DNA insert fragment encoding B. pertussis UT25 alcR G103S mutant allele, generated by PCR mutagenesis; AlcR+ tetracycliner | This study |
| pRK/Bb-S103G | pRK415 with 1.2-kb KpnI-PstI DNA insert fragment encoding B. bronchiseptica B013N alcR S103G mutant allele, generated by PCR mutagenesis; AlcR+ tetracycliner | This study |
| pRK/Bb-S103T | pRK415 with 1.2-kb KpnI-PstI DNA insert fragment encoding B. bronchiseptica B013N alcR S103T mutant allele, generated by PCR mutagenesis; AlcR(Con) tetracycliner | This study |
| pSS1129 | Mobilizable suicide plasmid; ampicillinr gentamicinr; ColE1 origin | 37 |
| pSS/Bp | pSS1129 with 1.2-kb EcoRI-HindIII DNA insert fragment of pRK/NpCp; AlcR+ ampicillinr gentamicinr | This study |
| pSS/Bb | pSS1129 with 1.2-kb EcoRI-HindIII DNA insert fragment of pRK/NbCb; AlcR+ ampicillinr gentamicinr | This study |
| pSS/Bb-S103T | pSS1129 with 1.2-kb EcoRI-HindIII DNA insert fragment of pRK/Bb-S103T; AlcR(Con) ampicillinr gentamicinr | This study |
AlcR domain swapping by overlap extension PCR.
The B. pertussis UT25 and B. bronchiseptica B013N alcR N-terminal (amino acids 1 to 223) and C-terminal (amino acids 224 to 303) domain coding sequences (5) were exchanged by overlap extension PCR essentially as described by Horton et al. (21). The coding sequences for the AlcR N-terminal and C-terminal domains of B. pertussis UT25 and B. bronchiseptica B013N were each PCR amplified using primer pairs N1(KpnI) and N2, and C1 and C2(PstI) (Table 2), yielding domain-specific products with overlapping termini spanning the N- and C-terminal coding sequence splicing junction. PCR primers N1(KpnI) and C2(PstI) were designed to include GC clamps and restriction enzyme site adapters (Table 2) to facilitate directional cloning of the fusion products as KpnI-PstI DNA fragments to the plasmid cloning vector pRK415. PCR primers N2 and C1, which were internal to the alcR coding sequence, were complementary and spanned the alcR domain splicing junction. PCR mixtures contained 100 ng of B. pertussis UT25 or B. bronchiseptica B013N genomic DNA, 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.75), 2 mM MgSO4, 0.1% Triton X-100, 100 μg of bovine serum albumin/ml, 200 μM each deoxynucleoside triphosphate, primer pairs at 0.5 μM each, 5% dimethyl sulfoxide, and 2.5 U of Pfu Turbo DNA polymerase (Stratagene, La Jolla, Calif.) in a 100-μl reaction volume. Reactions were performed by using a thermal cycler programmed for 30 cycles of denaturation at 96°C for 1 min, primer annealing at a temperature 5°C below the primer melting temperature (Tm) for 1 min, and primer elongation at 72°C for 1 min.
TABLE 2.
Oligonucleotide primers used in alcR PCR splicing and mutagenesis
| Primer | Nucleotide sequence (5′ to 3′)a | Nucleotide positionsb |
|---|---|---|
| N1 (KpnI) | ggccGGTACCTGAGCAACATGGGAGAGCAG | 1-26 |
| C2(PstI) | ggccctgcagGAATGGGATGGTCAATGCCGCAATG | 1189-1162 |
| N2 | CAGTCGCGCCAGGTCATCCAGAGTATGG | 909-882 |
| C1 | CCATACTCTGGATGACCTGGCGCGACTG | 882-909 |
| N2P(G103) | CCGGATGAACGCCCGTCATGGCGAC | 538-514 |
| C1P(G103) | GTCGCCATGACGGGCGTTCATCCGG | 514-538 |
| N2B(S103) | CCGGATGAACGCTCGTCATGGCGAC | 538-514 |
| C1B(S103) | GTCGCCATGACGAGCGTTCATCCGG | 514-538 |
| N2B(T103) | CCGGATGAACGGTCGTCATGGCGAC | 538-514 |
| C1B(T103) | GTCGCCATGACGACCGTTCATCCGG | 514-538 |
Uppercase letters represent alcR sequences; lowercase letters represent synthetic primer GC clamp and restriction site adapter sequences.
Nucleotide positions correspond to positions in the reported alcR region nucleotide sequence (GenBank accession number AF018255) of B. pertussis strain UT25 (5).
To splice the alcR domain coding sequences by overlap extension PCR, the N- and C-terminal domain-specific PCR products were combined in equimolar proportions, denatured, and reannealed to allow strand reassortment via base pairing within the 28-nucleotide (nt) overlap region to form partial duplex molecules. The recessed termini of the partial duplex molecules were extended with Pfu Turbo DNA polymerase and deoxynucleoside triphosphates to yield full-length 1.2-kb alcR DNA duplexes that were PCR amplified by using primers N1(KpnI) and C2(PstI). PCR conditions were essentially the same as those for production of the individual alcR domain coding sequences. Combining B. pertussis UT25 and B. bronchiseptica B013N N- and C-terminal domain coding sequences yielded interspecies hybrid alcR genes with an in-frame fusion at the splicing junction. As controls for the splicing procedure, the wild-type alcR genes of both species were reconstituted by joining N-terminal and C-terminal domain coding sequences derived from the same-species alcR gene.
alcR DNA fragments produced by the splicing reactions were purified, digested with KpnI and PstI to produce cohesive termini at the synthetic adapter sequences, and ligated with KpnI-PstI-digested pRK415 plasmid vector DNA. The cloning procedures resulted in two interspecies hybrid alcR+ plasmids, pRK/NpCb and pRK/NbCp, and two reconstituted wild-type alcR+ plasmids, pRK/NpCp and pRK/NbCb (Table 1).
Site-specific PCR mutagenesis.
The alcR genes of B. pertussis UT25 and B. bronchiseptica B013N were reciprocally altered to the nucleotide sequence of the other species' gene by an overlap extension PCR mutagenesis procedure (20). Site-specific PCR mutagenesis was performed essentially as described for AlcR domain swapping by overlap extension PCR using B. pertussis UT25 and B. bronchiseptica B013N genomic DNA templates, but with complementary pairs of mutagenic primers [N2P(G103) and C1P(G103) or N2B(S103) and C1B(S103) (Table 2)] which overlapped the alcR DNA region spanning the codon specifying amino acid position 103 in the B. pertussis UT25 or B. bronchiseptica B013N alcR open reading frame. The spliced products were cloned to plasmid vector pRK415 using the N1(KpnI) and C2(PstI) primer-encoded KpnI and PstI adapter sites to produce alcR mutant plasmids pRK/Bp-G103S and pRK/Bb-S103G (Table 1) for phenotypic analysis in BRM13. Similarly, PCR mutagenesis using the mutagenic primers N2B(T103) and C1B(T103) (Table 2) was performed to mutate the B. bronchiseptica alcR gene to produce the alcR(S103T) mutant allele, which was cloned to produce plasmid pRK/Bb-S103T. Predictions of the functional consequences of amino acid substitutions at AlcR position 103, leading to the choice of a threonine substitution, were made by using Dayhoff's mutation odds matrix (14), the amino acid substitution matrices of Bordo and Argos (6) for the identification of structurally tolerated substitutions, and the SIFT (30) sequence homology-based protein engineering tool (Fred Hutchinson Cancer Research Center [http://blocks.fhcrc.org/∼pauline/SIFT_seq_submit.html]) for the prediction of tolerated and deleterious amino acid substitutions. All mutations were confirmed by nucleotide sequencing.
Bacterial culture conditions.
E. coli and B. bronchiseptica strains were cultured on Luria-Bertani agar plates supplemented as required with antibiotics. Modified Stainer-Scholte (SS) medium (36) was used for broth cultures of B. bronchiseptica; iron-replete and iron-depleted SS culture conditions were achieved by the methods of Armstrong and Clements (2). Tetracycline was used at 15 μg/ml to select for pRK415 plasmid derivatives, and kanamycin was used at 50 μg/ml for maintenance of pRK2013 and for selection of the kanamycin resistance marker of mini-Tn5 lacZ1 mutant strain BRM13. Ampicillin was used at 100 μg/ml, and gentamicin was used at 10 μg/ml, for maintenance of plasmid vector pSS1129 derivatives in E. coli and for selection for insertions of pSS1129-borne alcR alleles at the chromosomal ΔalcR1 locus of B. bronchiseptica strain BRM13. In analyses of induction of alcABCDER operon transcription by alcaligin, SS culture medium was supplemented with purified alcaligin siderophore at a 20-μg/ml final concentration, which approximates the inducer requirement for maximal activation of transcription of the alcABCDER operon by AlcR under iron-depleted SS culture conditions (12). All glassware was acid cleaned and rinsed repeatedly in distilled deionized water prior to use.
Bordetella alcaligin siderophore purification.
Alcaligin was purified from B. bronchiseptica culture supernatants by a modification of the benzyl alcohol/ether extraction method of Neilands (29) as previously described by Brickman and coworkers (13) and was recrystallized from ethanolic solution.
Routine DNA procedures.
General genetic techniques were performed essentially as described previously (34). E. coli was transformed by electroporation using standard methods (Bio-Rad, Hercules, Calif.). Conjugal transfer of pRK415 and pSS1129 plasmid derivatives to B. bronchiseptica involved triparental matings using E. coli DH5α as the plasmid donor host strain and DH5α(pRK2013) as the source of mobilization functions as described previously (9). Transconjugants or plasmid integrants were selected on agar plates containing the appropriate selective antibiotics and crude colicin B (8).
β-Galactosidase assays.
B. bronchiseptica alcA::mini-Tn5 lacZ1 fusion strains were assayed for β-galactosidase activity by the method of Miller (27) as modified by Brickman and coworkers (11) after culture in iron-replete or iron-depleted SS medium, or in iron-depleted SS medium supplemented with purified alcaligin at a 20-μg/ml final concentration. β-Galactosidase activities represent means from triplicate assays (n = 3) ± 1 standard deviation.
Nucleotide sequencing.
Nucleotide sequencing of B. bronchiseptica alcR used the dideoxy chain termination method (35) and double-stranded plasmid DNA templates, with nucleotide sequencing services provided by the University of Minnesota Microchemical Facility and the University of Minnesota Advanced Genetic Analysis Center. DNA oligonucleotide primers were purchased from Invitrogen Life Technologies (Grand Island, N.Y.). Management and analysis of nucleotide sequence data used the Lasergene sequence analysis software system for the Macintosh PowerPC computer (DNASTAR, Inc., Madison, Wis.).
Phenotypic characterization of AlcR expressed from wild-type and mutant alcR alleles in single copy.
alcR+ integrant strains were constructed by integration of pSS1129 suicide plasmid-borne alcR alleles at the ΔalcR1 locus of BRM13 by homologous recombination. The wild-type alcR genes of B. pertussis UT25 and B. bronchiseptica B013N, as well as the B. bronchiseptica alcR(S103T) mutant allele, were subcloned from their respective pRK415 derivatives as 1.2-kb EcoRI-HindIII fragments to suicide plasmid pSS1129 to produce plasmids pSS/Bp, pSS/Bb, and pSS/Bb-S103T (Table 1). Each pSS1129 suicide plasmid derivative was conjugally transferred to B. bronchiseptica BRM13, and plasmid integrants were selected and maintained on the basis of ampicillin and gentamicin resistance associated with the plasmid vector. Transcriptional activation by AlcR was monitored as β-galactosidase activity produced from the BRM13 alcA::mini-Tn 5 lacZ1 fusion gene in response to iron starvation and the presence of alcaligin inducer.
Nucleotide sequence accession number.
The GenBank accession number assigned to the B. bronchiseptica B013N alcR gene is AF426103.
RESULTS
Interspecies variation in the alcaligin inducer requirement of AlcR for activation of alcABCDER operon transcription.
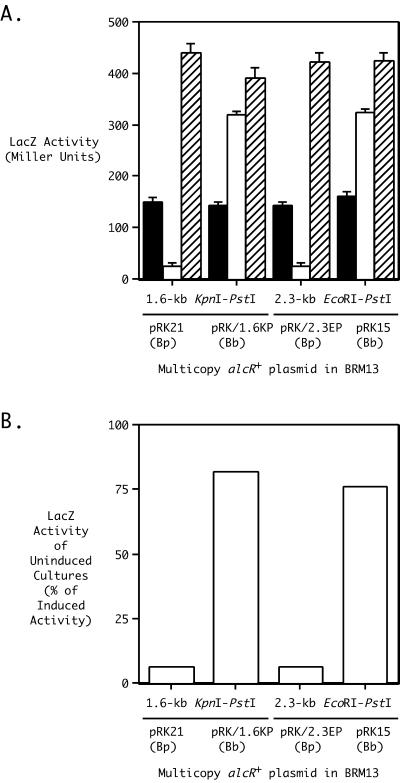
AlcR is a Fur-regulated AraC-like transcriptional regulator encoded within the alcaligin gene cluster (Fig. 1) of B. pertussis and B. bronchiseptica (5, 32). The alcR gene of B. pertussis was first identified by phenotypic complementation of the alcaligin utilization and production defects of a B. bronchiseptica mutant selected on the basis of streptonigrin resistance (5). Genetic and biochemical studies established the requirement of AlcR for maximal expression of the alcABCDER operon (5, 12, 22) and the fauA gene encoding the outer membrane receptor for ferric alcaligin under iron starvation stress conditions (10). Transcriptional activation by AlcR was shown to be dependent on the presence of the cognate siderophore alcaligin acting as the inducer (12). Subsequent studies aimed at further analysis of the inducer requirement of AlcR involved construction and comparative phenotypic characterization of AlcR produced from B. pertussis and B. bronchiseptica alcR+ plasmids. Gene fusion analyses using alcR+ plasmids to activate alcABCDER operon transcription in the AlcR− and alcaligin-deficient B. bronchiseptica strain BRM13 revealed that the AlcR proteins of B. pertussis UT25 and B. bronchiseptica B013N exhibited different apparent requirements for inducer when produced from multicopy plasmids (Fig. 2). B. bronchiseptica AlcR displayed a partial constitutive [AlcR(Con)] phenotype; expression of B. bronchiseptica B013N alcR from the pRK415-based multicopy plasmid pRK15 partially suppressed the alcaligin inducer requirement of AlcR normally needed for maximal alcABCDER operon transcription under iron starvation stress conditions. In iron-depleted cultures, comparison of alcABCDER operon transcriptional activities of uninduced cultures with those of cultures supplemented with alcaligin provides a useful measure of the degree of inducer dependence of transcriptional activation by AlcR (Fig. 2B). In iron-depleted cultures of BRM13(pRK15) without alcaligin inducer supplementation, alcA::mini-Tn5 lacZ1 transcriptional activity was 76% of the maximal transcriptional activity that was associated with alcaligin-induced cultures of the same strain. In contrast, B. pertussis AlcR produced from plasmid pRK21 exhibited strong dependence on alcaligin inducer when produced in BRM13; transcription of the alcA::mini-Tn5 lacZ1 fusion gene in uninduced BRM13(pRK21) cultures was only 6% of the transcriptional activity of alcaligin-induced cultures.
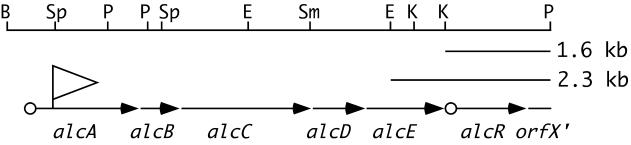
FIG. 1.
Genetic organization of the Bordetella alcABCDER alcaligin siderophore operon. The diagram represents an approximately 8-kb BamHI-PstI chromosomal DNA subregion of the alcaligin gene cluster of B. bronchiseptica B013N. Filled arrows indicate the genetic limits and transcriptional orientations of cistrons comprising the alcABCDER operon, and open circles represent the locations of known Fur-regulated promoter-operator regions upstream of alcA and alcR. The large open arrowhead indicates the position and transcriptional orientation of the promoterless lacZ reporter gene associated with the alcA::mini-Tn5 lacZ1 transposon insertion of alcaligin-deficient B. bronchiseptica mutant strain BRM1 (2) and its ΔalcR1 derivative strain BRM13 (12). The spatial limits of the 1.6-kb KpnI-PstI and 2.3-kb EcoRI-PstI subregions used to construct alcR+ multicopy plasmids are indicated. Abbreviations: B, BamHI; Sp, SphI; P, PstI; E, EcoRI; Sm, SmaI; K, KpnI.
FIG. 2.
Interspecies allelic variation in AlcR requirements for alcaligin inducer: partial constitutivity of B. bronchiseptica AlcR associated with production from multicopy plasmids. (A) β-Galactosidase reporter activities associated with expression of the alcA::mini-Tn5 lacZ1 fusion element of B. bronchiseptica ΔalcR1 mutant strain BRM13 complemented either with the multicopy B. bronchiseptica alcR+ plasmids pRK/1.6KP(Bb) and pRK15 or with the analogous B. pertussis alcR+ plasmids pRK21 and pRK/2.3EP(Bp). The genetic limits of the cloned DNAs (1.6-kb KpnI-PstI and 2.3-kb EcoRI-PstI) carried by B. pertussis and B. bronchiseptica alcR+ plasmids are indicated. β-Galactosidase activities were measured for cells cultured in iron-replete SS medium (solid bars), iron-depleted SS medium (open bars), or iron-depleted SS medium supplemented with 20 μg of purified alcaligin/ml (hatched bars). β-Galactosidase activities are expressed in Miller units (mean ± 1 standard deviation; n = 3). (B) The relative degree of constitutivity of AlcR produced from multicopy B. bronchiseptica or B. pertussis alcR+ plasmids in BRM13 under iron-depleted culture conditions was quantified by expressing the β-galactosidase activities produced without alcaligin inducer as percentages of the induced activities.
The B. bronchiseptica alcR+ plasmid pRK15, with a 2.3-kb EcoRI-PstI DNA insert fragment (Fig. 1), includes an additional 0.7 kb of alcR upstream DNA sequence compared with the cloned 1.6-kb KpnI-PstI B. pertussis alcR+ DNA region of pRK21. When multicopy alcR+ plasmids pRK/2.3EP (Bp) and pRK/1.6KP (Bb), which carry the corresponding 2.3-kb EcoRI-PstI and 1.6-kb KpnI-PstI genetic regions of these species as plasmids pRK15 and pRK21, were constructed and analyzed in BRM13, the same interspecies difference in the AlcR inducer requirement was observed (Fig. 2). Similarly, the partial constitutivity of AlcR produced from plasmid-borne copies of B. bronchiseptica B013N alcR was associated with the same 1.6-kb KpnI-PstI and 2.3-kb EcoRI-PstI cloned alcR+ genetic regions carried in a different plasmid vector, pBBR1MCS, that are present in plasmids pBB/1.6KP (Bb) and pBB/2.3EP (Bb) (data not shown). The corresponding pBBR1MCS-derived B. pertussis UT25 plasmids, pBB/1.6KP (Bp) and pBB/2.3EP (Bp), did not suppress the alcaligin inducer requirement of AlcR. Thus, the interspecies difference in AlcR function was apparently not related to differences in the spatial limits of the cloned alcR sequences, nor was it specific to expression of alcR from pRK415-based plasmids.
Despite this consistent interspecies variation in AlcR inducer dependence associated with production of AlcR from multicopy plasmids, AlcR of B. pertussis UT25 and AlcR of B. bronchiseptica B013N were found to activate alcA::mini-Tn5 lacZ1 transcription in BRM13 to approximately the same levels in alcaligin-induced cultures (Fig. 2A). Thus, although the two species' AlcR proteins exhibit different degrees of inducer dependence when produced in this genetic context, both proteins are equally capable of activating alcABCDER operon transcription to the maximal levels normally associated with growth of Bordetella under iron-depleted conditions in the presence of inducer.
AlcR domain swapping: role of the AlcR N-terminal domain in interspecies variation in the alcaligin inducer requirement associated with alcR expression from multicopy plasmids.
Given the strong conservation of structural features among members of the AraC family of transcriptional regulators (17), AlcR determinants responsible for the observed interspecies variation in the inducer requirement were hypothesized to be associated with the N-terminal domain of the activator protein. This hypothesis was tested by examining the inducer requirements of interspecies AlcR chimeras for activation of alcABCDER operon transcription in strain BRM13. Chimeras were generated from the alcR genes of B. pertussis UT25 and B. bronchiseptica B013N by reciprocal exchange of the N- and C-terminal coding sequences by overlap extension PCR. Four spliced alcR products were generated and cloned to the multicopy plasmid vector pRK415 to yield the chimeric alcR+ plasmids pRK/NpCb and pRK/NbCp, as well as wild-type alcR+ plasmids of both species, pRK/NpCp and pRK/NbCb, as splicing reaction controls (Table 1).
In BRM13, the chimeric plasmid pRK/NbCp encoding AlcR with the B. bronchiseptica N-terminal domain conferred the partial constitutive regulatory phenotype associated with production of the wild-type B. bronchiseptica AlcR protein from multicopy plasmids (Fig. 3). The transcriptional activity of the alcA::mini-Tn5 lacZ1 fusion gene in uninduced iron-depleted cultures of BRM13(pRK/NbCp) was 83% of the alcaligin-induced transcriptional activity. Concordantly, control plasmid pRK/NbCb, bearing a spliced version of the wild-type B. bronchiseptica alcR gene, also conferred the partial AlcR(Con) phenotype in BRM13. In contrast, AlcR proteins with the B. pertussis N-terminal domain, produced from the chimeric plasmid pRK/NpCb or the wild-type B. pertussis plasmid pRK/NpCp, exhibited the normal inducer dependence observed previously for the wild-type B. pertussis AlcR protein, with uninduced iron-depleted cultures displaying only 5 and 6% of the alcaligin-induced transcriptional activities, respectively. This phenotypic characterization of AlcR interspecies chimeras strongly suggested that the interspecies variation in the AlcR inducer requirement for transcriptional activation was attributable to undefined determinants associated with the N-terminal portions of the AlcR proteins.
FIG. 3.
Alcaligin inducer requirements of wild-type and interspecies chimeric AlcR proteins produced from alcR alleles borne on multicopy plasmids. (A) alcA::mini-Tn5 lacZ1 transcriptional activities were monitored in the alcR mutant strain BRM13 complemented with wild-type and interspecies chimeric alcR+ plasmids constructed by overlap extension PCR. β-Galactosidase activities were measured for cells cultured in iron-replete SS medium (solid bars), iron-depleted SS medium (open bars), or iron-depleted SS medium supplemented with 20 μg of purified alcaligin/ml (hatched bars). β-Galactosidase activities are expressed in Miller units (mean ± 1 standard deviation; n = 3). (B) The relative degree of constitutivity of AlcR is depicted as in Fig. 2B. Multicopy alcR+ plasmids: NpCb, pRK/NpCb; NbCp, pRK/NbCp; NpCp, pRK/NpCp; NbCb, pRK/NbCb.
The alcR coding sequences of B. pertussis UT25 and B. bronchiseptica B013N differ by a single nucleotide.
The nucleotide sequence of the alcR gene of B. pertussis strain UT25 (GenBank accession number AF018255) was reported previously by Beaumont and coworkers (5). The UT25 alcR coding sequence spans residues 220 to 1131, numbered as in the GenBank database record, and specifies a 304-amino-acid protein with a molecular mass of approximately 34 kDa. In an effort to define the molecular basis of the interspecies variation in AlcR function, the nucleotide sequence of the alcR gene of B. bronchiseptica B013N was determined and compared with the known alcR sequence of B. pertussis strain UT25. The alcR coding sequence of B. bronchiseptica B013N was found to differ from alcR of B. pertussis UT25 by a single nucleotide, corresponding to position 526 in the B. pertussis UT25 alcR GenBank record. As a result, B. bronchiseptica B013N AlcR carries a serine residue (encoded by an AGC triplet) at amino acid position 103, whereas a glycine residue (encoded by a GGC triplet) is found at that position in the N-terminal domain of the UT25 protein, as deduced from the nucleotide sequence. The alcR upstream sequences of B. bronchiseptica B013N, including the Fur- and iron-regulated promoter-operator control region defined in previous studies (5; T. J. Brickman and S. K. Armstrong, Abstr. 97th Gen. Meet. Am. Soc. Microbiol.) as well as putative AlcR translation initiation control sequences, were found to be identical to the known B. pertussis UT25 alcR region sequence extending to the upstream KpnI recognition site (Fig. 1). By analogy with other members of the AraC family of transcriptional regulators, this amino acid sequence difference at position 103 resides in the structural domain putatively involved in inducer-binding and multimerization and thus might be predicted to influence these functions of the AlcR protein. Since no other nucleotide sequence differences were identified, these data suggested that the identity of amino acid residue 103 had a profound influence on AlcR function when the activator was produced in this genetic context.
Phenotypic characterization of the inducer requirements of mutant AlcR proteins produced from reciprocal interspecies alcR mutant alleles borne on multicopy plasmids.
To confirm the hypothesized role of amino acid 103 in the interspecies variation in AlcR inducer dependence, site-directed mutagenesis was used to generate mutant B. bronchiseptica B013N and B. pertussis UT25 alcR alleles carrying single reciprocal nucleotide changes associated with the wild-type genes of the alternate species. That is, the mutated alcR genes were designed to produce a mutant B. bronchiseptica AlcR protein with a serine-to-glycine change at position 103, Bb-S103G, and a mutant B. pertussis AlcR protein with a glycine-to-serine change at position 103, Bp-G103S; thus, each mutant corresponded to the opposing species' AlcR protein sequence. The alcR mutant alleles generated by site-directed mutagenesis were cloned to the pRK415 multicopy plasmid for phenotypic characterization of the mutant AlcR proteins in strain BRM13. Phenotypic analysis supported the predicted role of amino acid residue 103 in interspecies differences in alcaligin inducer requirements for AlcR-mediated transcriptional activation (Fig. 4); the mutant AlcR proteins each exhibited the degree of constitutivity that was normally associated with the alternate species' wild-type AlcR protein. In BRM13, the B. pertussis AlcR(G103S) mutant protein produced from plasmid pRK/Bp-G103S activated alcA::mini-Tn5 lacZ1 transcription under uninduced iron-depleted conditions to a level equivalent to 71% of the induced level of transcription, similar to the partial AlcR(Con) phenotype associated with overproduction of the B. bronchiseptica wild-type AlcR protein. Conversely, alcABCDER operon transcriptional activities in uninduced cultures of BRM13(pRK/Bb-S103G) producing the B. bronchiseptica AlcR(S103G) mutant protein averaged only 6% of induced levels, similar to the phenotype associated with the wild-type B. pertussis AlcR regulator. Since each of the mutated alcR alleles differed from the corresponding wild-type alcR sequence by only a single nucleotide in the codon for amino acid 103, the interspecies phenotypic variation in AlcR inducer requirements was attributed to the amino acid difference at position 103 in the N-terminal protein domain.
FIG. 4.
Alcaligin inducer requirements of B. pertussis UT25 and B. bronchiseptica B013N mutant AlcR proteins produced from reciprocally mutated alcR alleles borne on multicopy plasmids. (A) alcA::mini-Tn5 lacZ1 transcriptional activities were monitored in strain BRM13 complemented with wild-type and mutated alcR+ plasmids constructed by overlap extension PCR. Each species' alcR genes were altered by a single nucleotide to the nucleotide sequence of the wild-type alcR gene of the other species. β-Galactosidase activities were measured for cells cultured in iron-replete SS medium (solid bars), iron-depleted SS medium (open bars), or iron-depleted SS medium supplemented with 20 μg of purified alcaligin/ml (hatched bars). β-Galactosidase activities are expressed in Miller units (mean ± 1 standard deviation; n = 3). (B) The relative degree of constitutivity of AlcR is depicted as in Fig. 2B. Multicopy alcR+ plasmids: Bp wt, pRK/NpCp; Bb wt, pRK/NbCb; Bp G103S, pRK/Bp-G103S; Bb S103G, pRK/Bb-S103G.
A threonine substitution at amino acid position 103 in the B. bronchiseptica AlcR protein renders it fully constitutive for transcriptional activation.
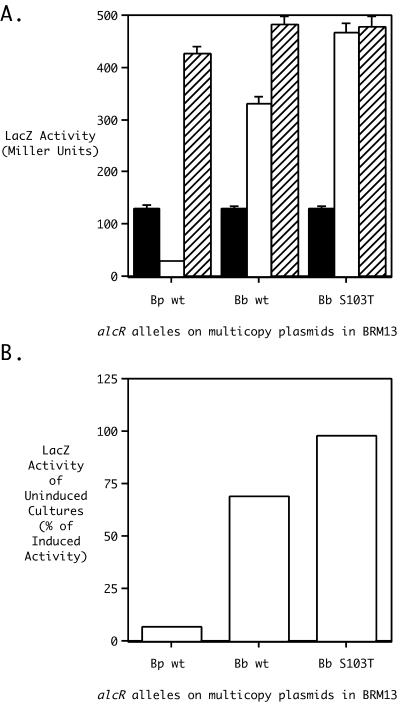
The striking interspecies difference in the AlcR inducer requirement suggested that amino acid position 103 was associated with an important functional determinant of inducer dependence. The functional importance of AlcR amino acid residue 103 was further probed by site-directed mutagenesis. Prediction of the functional consequences of amino acid substitutions at AlcR position 103 using Dayhoff's mutation odds matrix (14) suggested that a threonine substitution for either species' naturally occurring serine or glycine would be well tolerated structurally, since these three amino acids have been observed to share similar properties in proteins. The amino acid substitution matrices of Bordo and Argos (6) were applied for the prediction of amino acid substitutions that would be least likely to disrupt local protein structure or protein folding, so as to allow informative probing of the functional significance of the substituted residue. These matrices predicted that threonine could be substituted for serine at B. bronchiseptica AlcR amino acid position 103 with 95% confidence that local protein structure would not be radically perturbed, yet the substitution might influence specific aspects of protein function potentially related to inducer requirements for transcriptional activation. A relatively new tool for protein engineering and functional analysis, SIFT (30), predicts structurally tolerated amino acid substitutions in proteins based on sequence homologies and also predicts whether a particular amino acid substitution will be likely to specifically affect protein function. SIFT analysis of AlcR predicted that a serine-to-threonine change at position 103 would be well tolerated and not deleterious to protein function yet was likely to alter the phenotype associated with the protein. Based on the general concurrence of the three prediction tools and the partial AlcR(Con) phenotype associated with the wild-type B. bronchiseptica AlcR protein carrying a serine residue at position 103, it was hypothesized that a physicochemically similar threonine substitution at position 103 would alter B. bronchiseptica AlcR inducer dependence. PCR-based site-specific mutagenesis was performed using mutagenic primers N2B(T103) and C1B(T103) with B. bronchiseptica genomic DNA as the template to generate the alcR(S103T) mutant allele, which was cloned to plasmid vector pRK415 to produce plasmid pRK/Bb-S103T. Phenotypic analysis of the AlcR(S103T) mutant protein produced from pRK/Bb-S103T in BRM13 confirmed that the threonine substitution, as predicted, had a significant effect on the inducer requirement of the protein for transcriptional activation (Fig. 5). The AlcR(S103T) mutant protein exhibited a greater degree of constitutivity than the partially constitutive (69%) wild-type B. bronchiseptica AlcR protein assayed in parallel (Fig. 5B); the threonine substitution rendered the mutant AlcR activator fully constitutive (98%). Notably, the AlcR(S103T) mutant protein activated alcABCDER operon transcription to approximately the same maximal level as the wild-type activators of both species (Fig. 5A); thus, the phenotypic effect of the mutation appeared to be confined to the inducer dependence function of the protein.
FIG. 5.
Alcaligin inducer requirement of the AlcR(S103T) mutant protein produced from the alcR(S103T) mutant allele borne on a multicopy plasmid. (A) alcA::mini-Tn5 lacZ1 transcriptional activities were monitored in strain BRM13 complemented with wild-type and mutated alcR+ plasmids constructed by overlap extension PCR. β-Galactosidase activities were measured for cells cultured in iron-replete SS medium (solid bars), iron-depleted SS medium (open bars), or iron-depleted SS medium supplemented with 20 μg of purified alcaligin/ml (hatched bars). β-Galactosidase activities are expressed in Miller units (mean ± 1 standard deviation; n = 3). (B) The relative degree of constitutivity of AlcR is depicted as in Fig. 2B. Multicopy alcR+ plasmids: Bp wt, pRK/NpCp; Bb wt, pRK/NbCb; Bb S103T, pRK/Bb-S103T.
Since the alcR(S103T) mutation had a strong influence on AlcR inducer requirements when produced from multicopy plasmids, it was predicted that the alcR(S103T) mutant allele might confer an altered inducer dependence phenotype even at a single-copy gene dosage. The B. pertussis UT25 and B. bronchiseptica B013N wild-type alcR genes and the alcR(S103T) mutant allele were delivered to the chromosome of BRM13 by homologous recombination as described in Materials and Methods and were assayed for inducer dependence. All integrated alcR alleles were capable of activating alcA::mini-Tn5 lacZ1 transcription to similar maximal levels under inducing conditions (Fig. 6). Analysis of the AlcR inducer requirements for transcriptional activation confirmed that the wild-type AlcR proteins of both species displayed strong inducer dependence for transcriptional activation when expressed from single-copy genes, in contrast to the pronounced interspecies variation in AlcR function associated with production from multicopy plasmids. When produced from integrated alcR+ alleles in strain BRM13 during iron starvation in the absence of inducer, B. pertussis UT25 AlcR activated alcA::mini-Tn5 lacZ1 transcription to a level that averaged only 6% of induced transcriptional activity, and B. bronchiseptica B013N AlcR activated transcription to a level that averaged only 17% of induced levels. Remarkably, the AlcR(S103T) mutant protein was fully constitutive at a single-copy gene dosage, with uninduced transcriptional activity averaging 110% of induced transcriptional activity. Thus, the alcR(S103T) mutant allele is a true alcR(Con) allele that encodes a mutant AlcR(Con) protein capable of activating alcABCDER operon transcription in an inducer-independent manner at a single-copy gene dosage.
FIG. 6.
Alcaligin inducer requirement of the AlcR(S103T) mutant protein produced from the alcR(S103T) mutant allele at a single-copy gene dosage. (A) alcR+ integrant strains were constructed by integration of pSS1129 suicide plasmid-borne alcR alleles at the ΔalcR1 locus of BRM13 by homologous recombination, and alcA::mini-Tn5 lacZ1 transcriptional activities were monitored by β-galactosidase assays of cells cultured in iron-replete SS medium (solid bars), iron-depleted SS medium (open bars), or iron-depleted SS medium supplemented with 20 μg of purified alcaligin/ml (hatched bars). β-Galactosidase activities are expressed in Miller units (mean ± 1 standard deviation; n = 3). (B) The relative degree of constitutivity of AlcR is depicted as in Fig. 2B. Plasmid integrant strains: Bp wt, BRM13Ω(ΔalcR1::pSS/Bp); Bb wt, BRM13Ω(ΔalcR1::pSS/Bb); Bb S103T, BRM13Ω(ΔalcR1::pSS/Bb-S103T).
DISCUSSION
Expression of alcaligin siderophore biosynthesis and transport genes of B. pertussis and B. bronchiseptica is subject to both negative and positive transcriptional control. Transcription of the alcABCDER operon and the ferric alcaligin outer membrane receptor gene fauA is Fur and iron repressible (7, 10, 22, 23; Brickman and Armstrong, Abstr. 97th Gen. Meet. Am. Soc. Microbiol.) but is activated by the alcaligin-inducible AraC-like regulator AlcR under iron starvation stress conditions (5, 12). In studies detailed in this report, gene fusion analyses revealed that the AlcR regulator proteins of B. pertussis and B. bronchiseptica, when produced from multicopy plasmids, exhibited different apparent requirements for the inducer to activate transcription of the alcABCDER operon. Further examination of the basis of this phenomenon led to the identification of a key protein determinant of AlcR inducer responsiveness. This discovery guided the design and construction of a mutated gene encoding a fully constitutive AlcR mutant protein.
When produced from either pRK415- or pBBR1MCS-based multicopy plasmids, B. bronchiseptica AlcR activity was found to be partially constitutive, whereas B. pertussis AlcR retained its normal alcaligin inducer dependence. Phenotypic characterization of interspecies AlcR chimeras generated by domain swapping confirmed the predicted involvement of the AlcR N-terminal domain in the interspecies variation in inducer dependence. Nucleotide sequencing of the B. bronchiseptica B013N alcR gene revealed that it differed from the known B. pertussis UT25 alcR nucleotide sequence by a single nucleotide, resulting in a substitution at amino acid position 103 in the N-terminal domain. Site-directed mutagenesis was used to generate mutant B. bronchiseptica and B. pertussis alcR genes carrying single reciprocal nucleotide changes associated with the wild-type genes of the other species. Phenotypic analysis of the mutant AlcR proteins confirmed the hypothesized role of AlcR amino acid residue 103 in the interspecies difference in alcaligin inducer requirements for activation.
In this study, the alcR coding sequence of B. bronchiseptica strain B013N was found to be identical to the reported alcR coding sequence of B. bronchiseptica strain BB1015 (32). In the published report of the BB1015 alcR nucleotide sequence (GenBank accession number AJ000061), the authors noted the same single-nucleotide sequence difference between the alcR coding sequences of B. bronchiseptica BB1015 and B. pertussis strain BPSM, and they deduced the serine-to-glycine substitution in the B. pertussis BPSM protein sequence. The conservative nature of this serine-glycine amino acid substitution led to the prediction that the AlcR proteins would have the same function (32). Based on the findings obtained in the present study, we propose that there are significant phenotypic effects of the serine-glycine substitutions in the B. pertussis UT25 and B. bronchiseptica B013N AlcR proteins that influence their inducer requirements.
The functional importance of AlcR amino acid residue 103 was further analyzed by site-directed mutagenesis. The mutagenesis strategy was assisted primarily by the SIFT protein engineering and functional analysis tool of Ng and Henikoff (30) for prediction of structurally tolerated amino acid substitutions likely to have phenotypic effects on protein function. SIFT analysis predicted that a serine-to-threonine substitution at B. bronchiseptica AlcR position 103 would be well tolerated structurally yet likely to alter the phenotype associated with the protein; thus, it was hypothesized that the protein in which threonine was substituted would display altered inducer dependence for activation of transcription. In phenotypic analyses, the AlcR(S103T) mutant protein was found to be fully constitutive for transcriptional activation not only at a multicopy but also at a single-copy gene dosage in BRM13 integrant strains. In addition, differences in the inducer dependence of the wild-type B. pertussis and B. bronchiseptica AlcR proteins were measurable even when the proteins were produced from single-copy genes in plasmid integrant strains.
The difference in inducer dependence between B. bronchiseptica B013N and B. pertussis UT25 AlcR regulators produced from multicopy plasmids is remarkable given their single amino acid difference. Both species' AlcR proteins require alcaligin for activation of transcription when produced from chromosomal genes. The partial constitutivity of AlcR results when the B. bronchiseptica alcR copy number is elevated to an estimated 5 to 8 copies/cell (31) on pRK415-based plasmids. However, despite the striking suppression of the inducer requirement associated with this relatively modest increase in the B. bronchiseptica alcR gene dosage, strict inducer dependence is retained with B. pertussis alcR, even at approximately 30 to 40 copies/cell (1) on the pBBR1MCS-based plasmids. These results suggest that natural levels of AlcR production are critical for the controlled induction of AlcR-mediated transcriptional activation by the siderophore. In an exhaustive review of the AraC/XylS family of transcriptional regulators and their mechanisms of action, Gallegos et al. (17) proposed an explanation for suppression of the regulators' inducer requirements, based largely on detailed analyses of the Pseudomonas putida XylS regulator. Consistent with the proposed mechanisms of regulation by AraC type proteins and the role of the inducer in activation (17), a tentative hypothesis explaining the suppression of the inducer requirement associated with B. bronchiseptica AlcR overproduction can be proffered. We propose that an inactive AlcR protein conformation exists in equilibrium with an active AlcR conformation that is proficient for transcriptional activation. The postulated role of alcaligin inducer is to shift that equilibrium toward the active AlcR conformation by binding to the inactive regulator protein. Overproduction of the regulator protein would also be predicted to increase the concentration of the active conformer, thus suppressing the requirement for the inducer.
Since overproduction of B. bronchiseptica AlcR suppresses its inducer requirement but overproduction of the B. pertussis protein does not, it is also reasonable to hypothesize that the B. bronchiseptica AlcR protein shifts from the inactive conformation to the active conformation more readily than B. pertussis AlcR. This might explain the observed difference in the AlcR inducer requirement between these species at a single-copy gene dosage with normal levels of AlcR production. These hypothetical differences in AlcR structure result from substitutions at amino acid position 103. In the absence of crystallographic data, it is not known whether the inducer actually contacts AlcR amino acid position 103 in the active regulator.
We speculate that the interspecies difference in AlcR signal requirements for activation may have biological relevance for signal perception and transduction in priority regulation of the alcaligin siderophore system in the two related species. The fact that a modest increase in copy number results in inducer-independent AlcR function in B. bronchiseptica suggests that alcaligin inducer may normally be required only to “prime the pump” for initial activation of transcription of the alcaligin system genes in B. bronchiseptica. Once activated, elevated levels of AlcR production resulting from increased expression of the alcABCDER operon may suppress any additional need for inducer. In contrast, B. pertussis AlcR may require the persistence of the signal from the inducer to maintain AlcR in a conformation that is proficient to activate transcription. Thus, if the inducer is no longer perceived by B. pertussis, activation of alcaligin system gene transcription ceases, and the bacterium's energy and precursors can be appropriately channeled to the utilization of alternative sources of nutritional iron. It is an intriguing possibility that the stringent genetic regulation of the alcaligin siderophore system by the AlcR protein of B. pertussis has relevance for the success of B. pertussis as an obligate pathogen. Ongoing genetic and biochemical analyses are aimed at further elucidation of structure-function relationships of AlcR.
Acknowledgments
This work was supported by Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Antoine, R., and C. Locht. 1992. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6:1785-1799. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, S. K., and M. O. Clements. 1993. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J. Bacteriol. 175:1144-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall, B., and G. N. Sanden. 1995. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology 141:3193-3205. [DOI] [PubMed] [Google Scholar]
- 4.Beall, B., and T. Hoenes. 1997. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology 143:135-145. [DOI] [PubMed] [Google Scholar]
- 5.Beaumont, F. C., H. Y. Kang, T. J. Brickman, and S. K. Armstrong. 1998. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport genes in Bordetella pertussis and Bordetella bronchiseptica. J. Bacteriol. 180:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordo, D., and P. Argos. 1991. Suggestions for “safe” residue substitutions in site-directed mutagenesis. J. Mol. Biol. 217:721-729. [DOI] [PubMed] [Google Scholar]
- 7.Brickman, T. J., and S. K. Armstrong. 1995. Bordetella pertussis fur gene restores iron-repressibility of siderophore and protein expression to deregulated Bordetella bronchiseptica mutants. J. Bacteriol. 177:268-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickman, T. J., and S. K. Armstrong. 1996. Colicins B and Ia as novel counterselective agents in interspecies conjugal DNA transfers from colicin-sensitive Escherichia coli donors to other gram-negative recipient species. Gene 178:39-42. [DOI] [PubMed] [Google Scholar]
- 9.Brickman, T. J., and S. K. Armstrong. 1996. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J. Bacteriol. 178:54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brickman, T. J., and S. K. Armstrong. 1999. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J. Bacteriol. 181:5958-5966. [DOI] [PMC free article] [PubMed]
- 11.Brickman, T. J., B. A. Ozenberger, and M. A. McIntosh. 1990. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J. Mol. Biol. 212:669-682. [DOI] [PubMed] [Google Scholar]
- 12.Brickman, T. J., H. Y. Kang, and S. K. Armstrong. 2001. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J. Bacteriol. 183:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brickman, T. J., J.-G. Hansel, M. J. Miller, and S. K. Armstrong. 1996. Purification, spectroscopic analysis, and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. BioMetals 9:191-203. [DOI] [PubMed] [Google Scholar]
- 14.Dayhoff, M. O., R. M. Schwartz, and B. C. Orcutt. 1978. A model of evolutionary change in proteins, p. 345-352. In M. O. Dayhoff (ed.), Atlas of protein sequence and structure, vol. 5, suppl. 3. National Biomedical Research Foundation, Washington, D.C. [Google Scholar]
- 15.Fetherston, J. D., S. W. Bearden, and R. D. Perry. 1996. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol. Microbiol. 22:315-325. [DOI] [PubMed] [Google Scholar]
- 16.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinrichs, D. E., and K. Poole. 1993. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J. Bacteriol. 175:5882-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrichs, D. E., and K. Poole. 1996. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol. 178:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 21.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 22.Kang, H. Y., and S. K. Armstrong. 1998. Transcriptional analysis of the Bordetella alcaligin siderophore biosynthesis operon. J. Bacteriol. 180:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang, H. Y., F. C. Beaumont, T. J. Brickman, and S. K. Armstrong. 1996. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J. Bacteriol. 178:4877-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 25.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 26.Menozzi, F. D., C. Gantiez, and C. Locht. 1991. Identification and purification of transferrin- and lactoferrin-binding proteins of Bordetella pertussis and Bordetella bronchiseptica. Infect. Immun. 59:3982-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Moore, C. H., L. A. Foster, J. G. Gerbig, D. W. Dyer, and B. W. Gibson. 1995. Identification of alcaligin as the siderophore produced by Bordetella pertussis and Bordetella bronchiseptica. J. Bacteriol. 177:1116-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neilands, J. B. 1952. A crystalline organo-iron pigment from a rust fungus (Ustilago sphaerogena). J. Am. Chem. Soc. 74:4846-4847. [Google Scholar]
- 30.Ng, P. C., and S. Henikoff. 2001. Predicting deleterious amino acid substitutions. Genome Res. 11:863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pogliano, J., T. Q. Ho, Z. Zhong, and D. R. Helinski. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pradel, E., N. Guiso, and C. Locht. 1998. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J. Bacteriol. 180:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redhead, K., T. Hill, and H. Chart. 1987. Interaction of lactoferrin and transferrins with the outer membrane of Bordetella pertussis. J. Gen. Microbiol. 133:891-898. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider, D. R., and C. D. Parker. 1982. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect. Immun. 38:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stibitz, S. 1994. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 235:458-465. [DOI] [PubMed] [Google Scholar]
- 38.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]