Abstract
We describe here the functional characterization of the putative flgM gene of Pseudomonas aeruginosa. FlgM of P. aeruginosa is most similar to FlgM of Vibrio parahaemolyticus. A conserved region is present in the C-terminal half of the FlgM of P. aeruginosa and in FlgM homologues of other organisms that includes the σ28 binding domain. A role for the flgM gene of P. aeruginosa in motility was demonstrated by its inactivation. The β-galactosidase activity of a transcriptional fusion of the fliC promoter to lacZ was upregulated in the flgM mutant, suggesting that the activity of FliA, the sigma factor that regulates fliC, was increased. Consistent with these results, an increased amount of flagellin was demonstrated in the flgM mutant of P. aeruginosa strain PAK by Western blot, suggesting that FlgM negatively regulates transcription of fliC by inhibiting the activity of FliA. Direct interaction of the P. aeruginosa FlgM with the alternative sigma factor σ28 was demonstrated by utilizing the yeast two-hybrid system. Three putative consensus σ54 recognition sites and one σ28 site were found in the flgM upstream region. However, analysis of the transcriptional fusion of the flgM promoter to lacZ in different mutant backgrounds showed that the flgM promoter was not entirely dependent on either σ28 or σ54. A transcript was detected by primer extension that was 8 bp downstream of the consensus σ28-binding site. Thus, a system for the control of flagellin synthesis by FlgM exists in P. aeruginosa that is different from that in the enteric bacteria and seems to be most similar to that of V. cholerae where both σ28-dependent and -independent mechanisms of transcription exist.
Chemotactic motility in Pseudomonas aeruginosa is facilitated by a single polar flagellum whose biogenesis is dependent on a highly regulated pathway of timed gene expression and protein synthesis. More than 40 genes in P. aeruginosa are involved in the flagellum biogenesis, and they are regulated in a complex hierarchy that has been shown to be different from that described in the Salmonella system (3, 10, 30). The highest level of transcriptional regulation that has been discovered so far in P. aeruginosa is mediated by the alternative sigma factor σ54 (RpoN) and by the positive transcriptional regulator FleQ that belongs to the NtrC subfamily of response regulators (3, 35). None of the flagellar genes in Salmonella enterica serovar Typhimurium are regulated by σ54, but FleQ homologues and RpoN have been shown to be involved in the regulation of flagellar biogenesis in other organisms such as Vibrio parahaemolyticus (20), Vibrio cholerae (9), Helicobacter pylori (32), Caulobacter crescentus (2), and Campylobacter jejuni (19). FleQ and σ54 regulate the expression of other regulatory genes in P. aeruginosa, including the fleSR operon that encodes a sensor kinase (FleS) and a transcriptional regulator (FleR), members of a two-component regulatory system (3) that are involved in the regulation of flagellin synthesis. FleN is an additional flagellar regulator that controls flagellar number by exerting an antagonistic effect on FleQ activity (10). Another flagellar gene regulator of P. aeruginosa is the alternative sigma factor σ28 or FliA that controls flagellin synthesis (33). In contrast to gene regulation described in the Salmonella system, where expression of class 3 genes, including flagellin, is positively regulated only by σ28, fliC encoding flagellin in P. aeruginosa is regulated by both σ28 and σ54 (35). However, a major regulator of flagellum biogenesis in a variety of organisms (serovar Typhimurium, Escherichia coli, and Bacillus subtilis [13, 21, 27]), the anti-sigma factor FlgM, has not been demonstrated to date in P. aeruginosa. Furthermore, it has been demonstrated in the Salmonella flagellar system that negative regulation of σ28 by FlgM couples transcription of class 3 promoters such as fliC with assembly of the hook basal body complex by secretion of FlgM upon hook basal body completion (14, 17). The existence of similar mechanisms of control in P. aeruginosa, while suspected, is unproven at this point.
In this report we functionally characterized the putative flgM gene of P. aeruginosa that is flanked by the flgA gene on the 5′ end and the flgN gene at the 3′ end. The flgM gene encodes a protein with very limited homology to FlgM of E. coli and serovar Typhimurium but demonstrates significant homology to the putative FlgM of V. parahaemolyticus. Two lines of evidence strongly suggest that the putative flgM gene is indeed the flagellar anti-sigma factor in P. aeruginosa. First, the expression of flagellin was upregulated in the flgM mutant PAK-M both at the transcriptional level and at the translational level. Second, the putative FlgM of P. aeruginosa was shown to interact directly with the alternative sigma factor σ28 in the yeast two-hybrid system. Analysis of the promoter region of the P. aeruginosa flgM gene demonstrated that the transcription of the flgM gene was not entirely dependent on σ28, which is similar to that described for V. cholerae (29).
MATERIALS AND METHODS
Bacteria, yeast strains, plasmids, and media.
All bacterial strains, yeast strains, and plasmid vectors used in this study are described in Table 1. The bacterial cultures were grown in liquid Luria broth (L broth) (25) at 37°C with shaking at 250 rpm or on L agar plates (1.7% agar) with or without antibiotics. The antibiotic concentrations used were as follows: for E. coli, ampicillin at 200 μg/ml and gentamicin at 10 μg/ml, and for P. aeruginosa, carbenicillin at 300 μg/ml, gentamicin at 100 μg/ml, tetracycline at 100 μg/ml, and streptomycin at 300 μg/ml. The yeast strain AH109 was cultured in yeast extract-peptone-dextrose medium at 30°C with shaking at 250 rpm or on synthetic dropout minimal medium agar plates omitting one or both of the amino acids l-leucine and l-tryptophan (Sigma, St. Louis, Mo.).
TABLE 1.
Yeast and bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotypea | Source of reference |
|---|---|---|
| S. cerevisiae AH109 | MATatrp1-901 leu2-3, 112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MELIUAS-MEL1TATA-lacZ | Clontech |
| E. coli DH5α | hsdR recA lacZYA φ80 lacZΔM15 | Gibco-BRL |
| P. aeruginosa | ||
| PAK | Wild-type clinical isolate | D. Bradley |
| PAK-M | PAK flgM::Gmr | This study |
| PAK-N1G | PAK rpoN::Gmr | 18 |
| PAK-Q | PAK fleQ::Gmr | 3 |
| MS540 | PAK fliA::Gmr | 33 |
| Plasmids | ||
| pCR2.1 | Cloning vector, Ampr Kanr, LacZα | Invitrogen |
| pCR2.1-M | pCR2.1 containing a 2.0-kb SstI/HindIII fragment including the flgM gene from PAK | This study |
| pCR2.1-MG | pCR2.1-M with a Gmr gene inserted into a unique BamHI site of the flgM gene | This study |
| pET15bVP | Expression vector, T7 promoter, His tag coding sequence, Ampr, pBR322 origin, contains a broad-host-range origin of replication oriV | 3 |
| pET15bVP-M | pET15bVP with a 324-bp NdeI/BamHI PCR fragment of the PAK flgM gene | This study |
| pET15bVP-A | pET15bVP with a 744-bp NdeI/BamHI PCR fragment of the PAK fliA gene | This study |
| pMMB67HE | Broad-host-range cloning vector, tac promoter, Carbr | 30 |
| pMMB67HE-M | pMMB67HE with a 1.2-kb EcoRI/SstI fragment from pCR2.1-FM containing the flgM gene | This study |
| pDN19lacΩ | Promoterless lacZ oriV oriT Tcr Strr Ω fragment | 34 |
| pDN19lacΩ-M | pDN19lacΩ with a 614-bp EcoRI/BamHI fragment of the flgM gene promoter region | This study |
| pMS565 | pDN19lacΩ containing the fliA promoter region | 33 |
| pPT269 | pDN19lacΩ containing the fliC promoter region | 34 |
| pGADT7 | Cloning vector, GAL4768-881 AD, LEU2, Ampr, HA epitope tag | Clontech |
| pGBKT7 | Cloning vector, GAL41-147 DNA-BD, LEU2, Kanr, c-Myc epitope tag | Clontech |
| pGADT7-M | PAK flgM gene of 324-bp inserted as a PCR product into the NdeI/BamHI sites of pGADT7 | This study |
| pGADT7-A | PAK fliA gene of 744-bp inserted into the NdeI/BamHI sites of pGBKT7 | This study |
| pGBKT7-M | PAK flgM gene of 324-bp inserted as a PCR product into the NdeI/BamHI sites of pGBKT7 | This study |
| pGBKT7-A | PAK fliA gene of 744-bp inserted into the NdeI/BamHI sites of pGBKT7 | This study |
Gmr, gentamicin resistance; Kanr, kanamycin resistance; Carbr, carbenicillin resistance; Strr, streptomycin resistance; Tcr, tetracycline resistance; Ampr, ampicillin resistance; HA, hemagglutinin.
Computer analyses.
The amino acid sequence of the putative FlgM of V. parahaemolyticus was used to search the Pseudomonas genome database (www.pseudomonas.com) by using BLASTP (1). A protein annotated as FlgM was found in the Pseudomonas genome that was used in a BLASTP search for FlgM homologues in other organisms at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov). The deduced amino acid sequences of the P. aeruginosa FlgM and its homologues in other organisms were subjected to a PILEUP- and PRETTYBOX-generated alignment by using the GCG package (SeqWeb version 1.2, Wisconsin Package version 10.1). An evolutionary analysis that used the Kimura protein distance as correction method, which is based on the number of substitutions per 100 amino acids, was also performed on these proteins by using the same GCG package.
Electroporations and transformations.
E. coli transformations were performed by a standard procedure (31). The DNA used for electroporation in P. aeruginosa was prepared by the alkaline lysis procedure (6), and electroporations were performed as described previously (10).
PCR amplification and site-directed mutagenesis.
PCR was performed with Taq DNA polymerase (Gibco-BRL, Inc., Gaithersburg, Md.) as described previously (10). Briefly, DNA was initially denatured for 5 min at 95°C, followed by 35 cycles consisting of denaturation for 1 min at 95°C, annealing for 1 min at 55°C, and extension at 72°C for 3 min with primers RER51 and RER52 to generate a 744-bp fragment carrying the fliA gene. Annealing and extension at 70°C for 3 min with primers P6Sst and P9Hind were used to generate a 2.0-kb DNA fragment carrying the flgM gene, for 2 min 30 s with primers P10Eco and P11Bam to generate the 614-bp flgM promoter region, or for 1 min with primers P14Nde and P15Bam to generate the 324-bp flgM gene. Site-directed mutagenesis was performed by using the QuickChange Mutagenesis Kit (Stratagene, La Jolla, Calif.) according to the instructions in the user's manual. Specifically, the site-directed mutagenesis PCR was performed with Pfu DNA polymerase and primers P12Bam and P13Bam to generate a unique BamHI site at the start of flgM, under the following conditions: an initial denaturation for 0.5 min at 95°C, followed by 18 cycles of denaturation for 0.5 min at 95°C, annealing for 1 min at 55°C, and extension for 12 min at 68°C. The nucleotide sequences of the primers will be available upon request.
Plasmid constructions.
A 2.0-kb amplification product was obtained by PCR with the primers P6Sst and P9Hind, with PAK genomic DNA as a template, and inserted in cloning vector pCR2.1 (Invitrogen, Carlsbad, Calif.). The resulting plasmid pCR2.1-M was sequenced and demonstrated to contain a complete open reading frame tentatively identified as the P. aeruginosa flgM and flanking DNA, including the complete putative flgN gene downstream and part of the putative flgA gene upstream. PCR2.1-M was digested with HindIII, followed by religation which removed a BamHI site, and was then used as a template in site-directed mutagenesis PCR. A unique BamHI site was generated in the 5′ end of the flgM gene by site-directed mutagenesis that facilitated the insertion of a gentamicin resistance (Gmr) gene, yielding pCR2.1-MG. Plasmids pET15bVP-M and pET15bVP-A were obtained, respectively, by cloning the flgM gene as a 324-bp PCR fragment and the fliA gene as a 744-bp PCR fragment into the NdeI and BamHI sites of a low-copy-number expression vector pET15bVP (3). PET15bVP-M was used for complementation of the flgM mutation in PAK-M. An additional construct containing flgM on a broad-host-range plasmid pMMB67HE was made to assess the dose effect of flgM on the expression of FliC in the flgM mutant PAK-M. A 1.2-kb EcoRI/SstI fragment was obtained from the vector pCR2.1-M and cloned into the EcoRI and SstI sites of pMMB67HE. The resulting construct pMMB67HE-M included an intact flgM gene and part of the flgA gene. A 614-bp EcoRI/BamHI PCR fragment containing the putative flgM promoter region was obtained with the primers P10Eco and P11Bam and with PAK genomic DNA as a template. This PCR fragment was cloned into the EcoRI and BamHI sites of the vector pDN19lacΩ (34) to engineer the promoter fusion construct pDN19lacΩ-M.
Motility assay and EM.
Chemotactic motility was assessed by qualitative analysis of the zone formed by the motile bacteria on a 0.3% L agar plate. The plates were inoculated by stabbing the plates with different P. aeruginosa strains with a sterile toothpick. The plates were incubated at 37°C for 8 h. For electron microscopy (EM), static cultures were grown overnight at 37°C with the appropriate antibiotics. A drop of the culture was allowed to adhere to a carbon-coated grid for 10 s, and excess culture was drained off; the grid was then rinsed in a drop of saline, and adherent cells were negatively stained with a 2% aqueous solution of phosphotungstic acid for 10 s. Samples were examined with a Hitachi H-7000 transmission electron microscope.
β-Galactosidase assay.
Plasmid pPT269 (34) containing the fliC promoter region fused to a promoterless lacZ was electroporated into P. aeruginosa wild-type PAK and the flgM mutant PAK-M. The flgM-lacZ fusion plasmid pDN19lacΩ-M was electroporated into P. aeruginosa strains PAK, PAK-Q, PAK-RG, PAK-N1G, and MS540 (Table 1). β-Galactosidase assays were performed as described previously (25). Cells were grown in L broth with streptomycin to an A600 of 0.7 to 1.0, harvested, and assayed for β-galactosidase activity. Two independent experiments were performed in triplicate when the β-galactosidase activity was assessed.
Yeast two-hybrid system.
To examine whether there was a direct in vivo interaction between the anti-sigma factor FlgM of P. aeruginosa and the alternative sigma factor FliA, a yeast transcriptional assay, the Matchmaker GAL4 Two-Hybrid System 3 (Clontech Laboratories, Inc., Palo Alto, Calif.) was used. Two fusion vectors were used, pGADT7 that expresses proteins fused to the GAL4 activation domain and pGBKT7 that expresses proteins fused to the GAL4 DNA-binding domain. The yeast strain AH109 utilizes three reporter genes that are under the control of GAL4 upstream activating sequences. Construction of gene fusions was made possible by obtaining NdeI/BamHI fragments of the entire flgM gene (324 bp) and the entire fliA gene (744 bp) from the vectors pET15bVP-M and pET15bVP-A, respectively. These fragments were cloned in-frame into the NdeI and BamHI sites of both the fusion vectors pGADT7 and pGBKT7, giving the following constructs: pGADT7-M, pGADT7-A, pGBKT7-M, and pGBKT7-A. These constructs, along with proper control vectors, were transformed into the recipient yeast strain AH109 according to the manufacturer's instructions. Transcription of the reporter genes lacZ, ADE2, and HIS3 facilitated the assessment of protein interaction by detecting the color reaction of β-galactosidase activity (lacZ) and by nutritional selection omitting the amino acids l-adenine (ADE2) or l-histidine (HIS3) (Sigma). β-Galactosidase activity was detected as described below. Briefly, yeast colonies were patched on filter paper (Whatman no. 1; Whatman International, Ltd., Maidstone, England), allowed to dry, and subsequently immersed in liquid nitrogen for 1 min. The filter paper was placed onto an additional filter paper presoaked with appropriate substrate and buffer and then incubated at 30°C for color reaction.
Western blots.
To demonstrate a translated product from the fliC gene in the flgM mutant PAK-M, Western blotting was performed. After overnight growth of the cultures, identical optical densities of cell suspensions were adjusted, and cells were centrifuged. Proteins from whole-cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 10% gel by using Bio-Rad's Mini-Protean II system (23) and then electrophoretically transferred as described previously (10). Alkaline phosphatase-conjugated immunoglobulin G whole molecule from Sigma was used as secondary antibody, and the alkaline phosphatase activity was detected by color reaction.
Primer extension analysis.
RNA was prepared from P. aeruginosa strains PAK, PAK-N1G, PAK-Q, and MS540 by using Trizol reagent (Gibco-BRL/Life Technologies, Grand Island, N.Y.) according to the manufacturer's protocol. The primer (7.5 μM) was 5′ end labeled by using [γ-32P]ATP (25 μCi) and T4 polynucleotide kinase at 37°C for 30 min. The enzyme was heat inactivated after the addition of 40 mM EDTA (pH 7.5) and purified by elution through a G-25 spin column (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). Labeled primer was annealed to 50 μg of RNA in first-strand buffer and RNaseOUT RNase inhibitor at 65°C for 1 h. After a slow cooling to room temperature, deoxynucleoside triphosphate (0.5 mM), dithiothreitol (10 mM), RNaseOUT, and Superscript II (RNase H− RT) were added for reverse transcription at 42°C for 1 h. After cDNA synthesis, RNase H treatment was given at 37°C for 20 min, followed by phenol-chloroform extractions and ethanol precipitations. Sequencing was done with the same primer (unlabeled) by using [α-32P]dATP and the Sequenase version 2.0 DNA Sequencing kit (U.S. Biochemicals, Cleveland, Ohio). DNA was resolved on an 8 M urea-8% polyacrylamide gel. Two primers were used that were 91 and 64 bp downstream of the consensus σ28 binding site, respectively. The start site shown in Fig. 5 was detected by using the primer that was 91 bp downstream of the σ28 binding site. In addition to that, six more primers were used in the primer extension analysis to detect any additional start sites. The nucleotide sequences of these primers are available upon request.
FIG. 5.
(A) Partial nucleotide sequence of the 614 bp upstream flgM promoter region used for β-galactosidase assays. In this promoter sequence three putative σ54 binding sites (GGCACG-N4-TTGC) and one σ28 binding site (TAAAGTTT-N11-GCCGATAA) are indicated in boldface and underlined. The transcriptional start site 8 bp downstream of the σ28 consensus sequence is shown as +1, with an arrow showing the direction of transcription. (B) Primer extension analysis of the flgM gene. Sequencing ladders GATC are on the left, and primer extension products from P. aeruginosa strains PAK (lane 1), PAK-N1G (σ54 mutant) (lane 2), MS540 (σ28 mutant) (lane 3), and PAK-Q (fleQ mutant) (lane 4) RNAs are shown to the right. The arrow denotes the transcriptional start site.
Sequencing and nucleotide sequence accession number.
DNA sequencing was performed as described previously (10). The nucleotide sequence consisting of 2,048 nucleotides containing the partial sequence of flgA and the complete sequences of flgM and flgN genes have been submitted to GenBank (accession no. AY029221).
RESULTS
Identification and computer analysis of P. aeruginosa flgM.
Attempts to identify a FlgM homologue in P. aeruginosa based on its homology to FlgM in the enteric bacteria E. coli and S. enterica serovar Typhimurium were unsuccessful. Later, the existence of a putative FlgM protein in V. parahaemolyticus was reported (20), and the amino acid sequence of this protein was used to perform a BLASTP search in the Pseudomonas genome database. A P. aeruginosa homologue was discovered that was initially considered to be a hypothetical protein and then, later during these studies, was deemed to be a putative FlgM protein (confidence level 4) with an unknown function. The putative P. aeruginosa FlgM protein consisting of 107 amino acids was further used in a BLASTP search for FlgM homologues in other organisms. The P. aeruginosa protein showed 61% similarity and 31% identity to FlgM of V. parahaemolyticus, 61% similarity and 28% identity to FlgM of V. cholerae, and 57% similarity and 38% identity to FlgM of Proteus mirabilis. The flgM gene mapped downstream of a chemotaxis gene cheV and a flagellar gene flgA and upstream of flgN. Thus, the location of the flgM gene between flgA and flgN in the P. aeruginosa genome is consistent with the gene organization and transcriptional direction in the flagellar system of V. parahaemolyticus (20) and in V. cholerae (29). The deduced amino acid sequence of flgM of P. aeruginosa was aligned with FlgM homologues from several other organisms by using the GCG multiple sequence analysis programs PILEUP and PRETTYBOX. As shown in Fig. 1, FlgM of P. aeruginosa had weak homology to FlgM proteins of some bacteria, particularly with E. coli, S. enterica serovar Typhimurium, Yersinia enterocolitica, and Proteus mirabilis throughout the open reading frame except for a short stretch of amino acids in the C-terminal part of the sequence (amino acids 75 to 101), which was conserved in all bacteria. These results correlated with a computer-generated evolutionary analysis that demonstrated FlgM of P. aeruginosa to have the largest genetic distance from that of E. coli, serovar Typhimurium, Y. enterocolitica, and Proteus mirabilis and the closest genetic distance to FlgM of Vibrio species (data not shown).
FIG. 1.
Computer-generated alignment (PILEUP and PRETTYBOX) based on the deduced amino acid sequence of FlgM from P. aeruginosa (7_pae) and those of its homologues from E. coli (1_eco), serovar Typhimurium (2_sty), Y. enterocolitica (3_yen), Proteus mirabilis (4_pmi), V. cholerae (5_vch), V. parahaemolyticus (6_vpa), and B. subtilis (8_bsu) by using GCG multiple alignment sequence programs (SeqWeb version 1.2, Wisconsin Package version 10.1). Black shading indicates identity, and gray shading (light and dark) indicate degrees of similarities among amino acid residues.
Insertional inactivation and complementation of flgM.
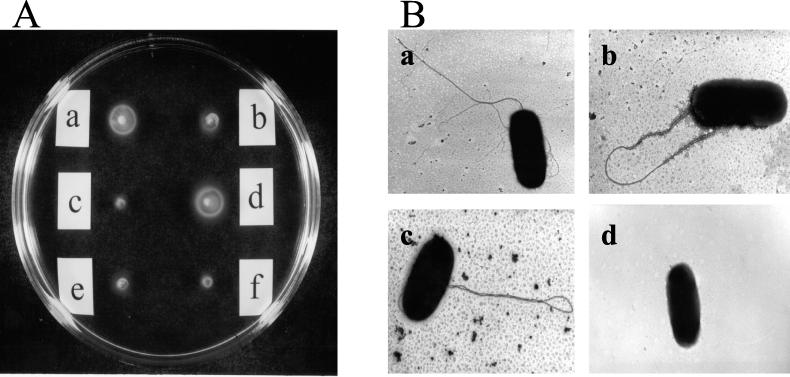
To examine the function of the putative flgM gene, a chromosomal flgM mutation was generated in P. aeruginosa PAK by allelic replacement. The plasmid pCR2.1-MG that included the inactivated flgM gene was electroporated into PAK and replaced the corresponding wild-type gene by double reciprocal recombination, giving rise to the mutant strain, PAK-M. Inactivation of the flgM gene was confirmed by PCR with primers P6Sst and P9Hind which gave rise to an ∼3.8-kb product in the mutant (data not shown). The flgM mutant strain PAK-M and the corresponding wild-type strain were examined on motility plates (0.3% agar). The flgM mutant strain PAK-M was weakly motile, as assessed by the significantly lower size of the swarming zone compared with the wild-type PAK after 8 h of incubation (Fig. 2Aa and b). To rule out that this defective phenotype was a result of inactivating downstream genes, an intact copy of the flgM gene was cloned as a 324-bp NdeI/BamHI fragment into the plasmid pET15bVP, and this construct (pET15bVP-M) was introduced into PAK-M by electroporation. The motility function was restored to that of the wild-type PAK by providing the flgM gene in trans, while the vector control (pET15bVP) did not restore this phenotype (Fig. 2Ac and d). When the flgM gene, carried on the plasmid pMMB67HE-M under the stronger tac promoter, was introduced into the flgM mutant PAK-M, a nonmotile phenotype was evident, a finding consistent with an explanation that FlgM in excess inhibited transcription of the σ28-dependent fliC promoter (Fig. 2Ae and f). Electron microscopy studies revealed that the flgM mutant strain PAK-M possessed a single polar flagellum as the wild-type PAK, while the PAK-M carrying the flgM gene on plasmid pMMB67HE-M was not flagellated (Fig. 2B). In contrast to this observation, when the flgM gene was expressed from the plasmid pET15bVP-M, the bacteria showed a single polar flagellum. These data suggested a dose effect of flgM in motility and flagellin accumulation.
FIG. 2.
Dose effect of flgM in motility. (A) Chemotactic motility of P. aeruginosa strains PAK (a); PAK-M (flgM mutant) (b); PAK-M (pET15bVP), which is the flgM mutant strain containing empty vector (c); PAK-M (pET15bVP-M) expressing the flgM gene at very low levels (d); PAK-M (pMMB67HE), which is another vector control (e); and PAK-M (pMMB67HE-M), which has flgM under the tac promoter (f). (B) Electron micrographs of wild-type PAK (a), PAK-M (flgM mutant) (b), PAK-M (pET15bVP-M) (c), and PAK-M (pMMB67HE-M) (d). Magnifications: a, ×42,500; b, ×50,000; c and d, ×37,500.
Negative regulation of flagellin synthesis by FlgM.
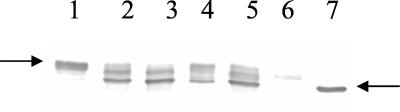
Transcription of the class 3 fliC promoter has been demonstrated to be repressed by FlgM in serovar Typhimurium (17, 28). By analogy, it is reasonable to expect that the fliC promoter would be upregulated in P. aeruginosa flgM mutant PAK-M, if FlgM acts as a negative regulator of FliA. Indeed, a ca. 11.5-fold upregulation of the β-galactosidase activity of the flagellin gene (fliC) promoter fusion was demonstrated in the flgM mutant PAK-M (Table 2). In addition, when an intact copy of the flgM gene was introduced in the PAK-M containing the fliC-lacZ fusion, the β-galactosidase activity of the fliC promoter fusion was completely abrogated (Table 2). These results suggested that an excess of FlgM suppresses transcription of fliC. To verify that the demonstrated upregulation of the fliC promoter in the flgM mutant background also correlated with accumulation of translated fliC, whole-cell lysates of wild-type PAK and PAK-M were analyzed by Western blot for their flagellin content. As shown in Fig. 3 (lane 2), an excess of flagellin was demonstrated in the flgM mutant PAK-M compared with the wild-type PAK (lane 1). However, flagellin was not expressed when the flgM gene was introduced on the plasmid pMMB67HE-M under the control of the tac promoter, as shown in Fig. 3, lane 6. This result is consistent with that of the β-galactosidase assays showing no activity of the fliC promoter fusion in PAK-M when flgM was introduced on pMMB67HE-M, as well as with the results of the motility assay and EM demonstrating a nonmotile, nonflagellated phenotype of PAK-M carrying the same plasmid. The PAK-M strain carrying the empty vector pMMB67HE appeared the same as PAK-M (Fig. 3, lane 5). However, when flgM was expressed from pET15bVP-M, flagellin was detected in amounts similar to the wild-type PAK (Fig. 3, lane 4). These results confirmed the findings from the motility and EM analyses that the amount of FlgM affects the amount of intracellular flagellin and thus the presence or absence of a flagellum and chemotactic motility on the plates.
TABLE 2.
Assessment of transcriptional activities of the fliC promoter in strains PAK and PAK-M
| Host strain | Plasmid construct | β-Galactosidase activity (Miller units; mean ± SD) |
|---|---|---|
| PAK, wild type | placΩ | 278 ± 36 |
| PAK, wild type | placΩfliC | 2,080 ± 87 |
| PAK-M, flgM mutant | placΩ | 164 ± 107 |
| PAK-M, flgM mutant | placΩfliC | 23,948 ± 1065 |
| PAK-M, flgM mutant | placΩfliC, pMMB67HE | 13,655 ± 677 |
| PAK-M, flgM mutant | placΩfliC, pMMB67HE-M | 247 ± 64 |
FIG. 3.
Flagellin content in whole-cell lysates of wild-type PAK and flgM mutant PAK-M, analyzed by Western blot with polyclonal anti-FliC rabbit serum. Lanes: 1, wild-type PAK; 2, PAK-M (flgM mutant); 3, pET15bVP vector control in PAK-M; 4, pET15bVP-M plasmid in PAK-M; 5, pMMB67HE vector control in PAK-M; 6, pMMB67HE-M plasmid in PAK-M; and 7, orfA mutant that makes nonglycosylated flagellin. The arrow on the left indicates modified flagellin (45 kDa). The arrow on the right indicates nonglycosylated flagellin (∼40 kDa).
FlgM-FliA in vivo protein interaction.
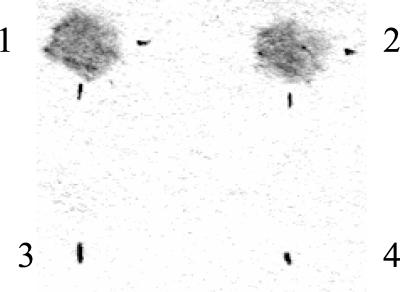
One of the criteria for anti-σ28 activity of FlgM is the physical association of FlgM with the alternative sigma factor σ28 (FliA) described in serovar Typhimurium (8, 28). In order to confirm this interaction of FlgM with FliA in P. aeruginosa, a two-hybrid yeast transcriptional assay was used that allowed in vivo protein interaction to be detected by activating transcription of the reporter genes lacZ, ADE2, and HIS3. A 324-bp NdeI/BamHI fragment containing the entire flgM gene and a 744-bp NdeI/BamHI fragment containing the entire fliA gene were cloned in both of the vectors pGADT7 and pGBKT7, giving pGADT7-M, pGADT7-A, pGBKT7-M, and pGBKT7-A. β-Galactosidase activity was detected when both plasmids pGADT7-M and pGBKT7-A, as well as pGADT7-A and pGBKT7-M, were transformed in the yeast strain AH109 (Fig. 4, panels 1 and 2), indicating induced transcription of the lacZ gene due to interaction between the fusion proteins FlgM and FliA. No activity was detected in the yeast AH109 transformants with the flgM or fliA gene fusion constructs with the corresponding vector controls, pGADT7-M with pGBKT7 or pGADT7-A with pGBKT7 (Fig. 4, panels 3 and 4). In addition, growth of yeast strain AH109 on medium that was devoid of l-histidine or l-adenine occurred only when AH109 was transformed with the gene fusion constructs pGADT7-M and pGBKT7-A or the gene constructs pGADT7-A and pGBKT7-M. These results indicate that the putative FlgM protein physically interacts with the alternative sigma factor FliA in P. aeruginosa, in agreement with the mechanism of action of FlgM in other organisms.
FIG. 4.
FlgM-FliA in vivo fusion protein interaction in yeast AH109 transformants. Positive interaction is based on expression of β-galactosidase activity after transcription of the reporter gene lacZ. Panels 1 and 2 show positive β-galactosidase activity in yeast strain AH109 carrying plasmids pGADT7-M and pGBKT7-A and plasmids pGADT7-A and pGBKT7-M, indicating positive interaction between the FlgM and FliA proteins. Panels 3 and 4 show pGADT7-M or pGADT7-A carrying the fliA or flgM, respectively, with the vector control pGBKT7 where no β-galactosidase activity was detected.
Transcriptional regulation of the flgM gene.
The flgM upstream region was inspected for the presence of consensus σ54 (YTGGCACG-N4-TTGCW) (5) and σ28 (TAAAGTTT-N11-GCCGATAA) (21) recognition sites. Downstream of the flgA gene, three putative σ54 binding sequences were identified which were located 119, 128, and 143 bp upstream of the translational start site of the flgM gene (Fig. 5A). The putative σ54 binding sequence located 119 bp upstream of the flgM gene matched 12 of 17 residues with the consensus σ54 binding site, and the additional two upstream putative σ54 binding sites matched 11 of 17 and 10 of 17 residues. In addition, a putative σ28 consensus site having only a single mismatch with the consensus σ28 binding site was located 8 bp upstream of the transcriptional start site (Fig. 5A). To gain further insight into the transcriptional regulation of the flgM gene, the putative flgM promoter region comprising 614 bp was fused to the promoterless lacZ gene. The β-galactosidase activity of the flgM promoter was measured in several P. aeruginosa strains. As shown in Table 3, the existence of a flgM promoter in the flgM upstream region was demonstrated in wild-type PAK since β-galactosidase activity was ca. 81 times higher than that of the pDN19lacΩ vector control. P. aeruginosa strains MS540 (σ28 mutant), PAK-NIG (σ54 mutant), and PAK-Q (fleQ mutant) had high activities of β-galactosidase, although they were reduced compared to that of the wild-type PAK. These data suggested the existence of a promoter element that was not entirely dependent on σ28, σ54, or FleQ. Further analysis of the flgM promoter by primer extension revealed a transcriptional start site (Fig. 5) 8 bp downstream of the consensus σ28 binding site. However, no σ54-dependent transcripts could be detected when several primers downstream of the three putative σ54 consensus binding sites were used. A faint band was, however, found at the same location in all four lanes upstream of the σ28 binding site, but there was no recognizable consensus binding site for any known sigma factor just upstream of this location. These results suggest that, in P. aeruginosa, the anti-sigma 28 factor FlgM is regulated by a mechanism that is somewhat different from that existing in enteric bacteria, in which flgM transcription is solely dependent on σ28. Another unknown sigma factor may also be involved in P. aeruginosa. This mode of flgM regulation is the closest to that observed in V. cholerae (29).
TABLE 3.
Assessment of transcriptional regulation of the flgM promoter in P. aeruginosa
| Host strain | Genetic background | β-Galactosidase activity (Miller units; mean ± SD)
|
|
|---|---|---|---|
| Vector alone | flgM promoter | ||
| PAK | Wild type | 172 ± 4 | 13,961 ± 379 |
| PAK-N1G | rpoN mutant | 88 ± 18 | 9,809 ± 857 |
| PAK-Q | fleQ mutant | 34 ± 3 | 6,831 ± 721 |
| MS540 | fliA mutant | 47 ± 26 | 8,698 ± 1,030 |
DISCUSSION
One major control mechanism in flagellar biosynthesis that has not yet been accounted for in P. aeruginosa is the control of σ28-dependent transcription of fliC by the anti-sigma factor FlgM. Here we describe the identification and functional analysis of the anti-sigma factor FlgM in P. aeruginosa.
Transcription of fliC has been shown to be dependent on the fliA-encoded sigma factor σ28 in P. aeruginosa and in other organisms such as serovar Typhimurium, E. coli, and B. subtilis (24, 26, 28, 33). Inhibition of σ28 by the anti-sigma factor FlgM has also been demonstrated (7, 13, 21). In this context, it was reasonable for us to pursue the identification of and to ascertain whether FlgM was present and acted as the negative regulator of σ28 in P. aeruginosa. While our previous efforts based on homology to the FlgM of E. coli and serovar Typhimurium were unsuccessful in identifying FlgM, we were now able to identify a putative FlgM candidate by searching the Pseudomonas genome database by using the annotated FlgM of V. parahaemolyticus (20). The PILEUP- and PRETTYBOX-generated alignment of the FlgM of different organisms showed a low degree of homology throughout the open reading frame except for a short stretch of ca. 26 amino acids positioned from amino acids 75 to 101 in the C-terminal half of the protein. Interestingly, it has been demonstrated that this part of FlgM of serovar Typhimurium contains the σ28 binding domain that is positioned from amino acids 41 to 97. Mutations in the C-terminal part of FlgM disrupt the activity of the anti-sigma factor (8, 11), indicating that the most conserved region of the FlgM protein is involved in its interaction with σ28 in several organisms. Furthermore, in an evolutionary analysis of FlgM of different organisms, P. aeruginosa FlgM was most distant from the FlgM of E. coli and serovar Typhimurium but was closest to FlgM in V. parahaemolyticus and V. cholerae, findings correlating with those from the PILEUP- and PRETTYBOX-generated alignment.
Insertional inactivation and complementation of the putative flgM gene in P. aeruginosa made it possible to analyze the function of flgM. Even though EM analysis revealed that the flgM mutant PAK-M possessed a single polar flagellum, this mutant was only weakly motile, as observed by the smaller swarming zone compared to wild-type PAK. This may be due to intracellular accumulation of FliC or to the fact that the transcription of genes controlled by σ28-dependent promoters other than fliC, e.g., chemotaxis genes, may be affected in this mutant. Thus far, the transcription of chemotaxis genes in P. aeruginosa has not been shown to be σ28 dependent, although in B. subtilis and in the enteric bacteria chemotaxis genes are σ28 dependent (12, 16). Motility was restored to normal when the flgM gene was provided to the flgM mutant on the plasmid pET15bVP-M. However, a nonmotile phenotype was observed when flgM was introduced into this strain under the control of the tac promoter on the plasmid pMMB67HE-M. Complementation of the motility defect in studies with the low-copy expression vector pET15bVP under noninduced conditions in P. aeruginosa has previously been demonstrated (10). The fact that the phenotype was not restored when the flgM gene was introduced with the plasmid pMMB67HE-M may be explained by the amount of FlgM exceeding the threshold level leaving no free σ28 available for flagellin expression. A Western blot of this complemented mutant supported this conclusion in that the amount of flagellin seen was markedly diminished from the wild-type strain. This repressive effect of FlgM has been suggested in wild-type serovar Typhimurium when FlgM was overexpressed (17). In addition, observations were made that the flgM mutant in serovar Typhimurium was weakly motile, possessed longer flagella, and expressed flagella in increased numbers (13, 22). However, the flagellum of the P. aeruginosa flgM mutant PAK-M appeared normal under EM.
Besides characterizing the morphology and phenotype of the flgM mutant PAK-M, β-galactosidase assays and Western blots were used to assess σ28-dependence of the fliC promoter in P. aeruginosa at both transcriptional and translational levels. Our results manifested upregulated β-galactosidase activity of the flagellin gene (fliC) promoter lacZ fusion in the flgM mutant PAK-M, which correlated with an increased amount of the translated fliC product. In addition, the overexpression of FlgM not only downregulated the fliC promoter in the flgM mutant PAK-M but also reduced flagellin synthesis. Although it is still unknown if extracellular transportation of FlgM to initiate σ28-dependent transcription of late gene promoters is also a prerequisite in P. aeruginosa, identification of flgM will facilitate further investigations of this phenomenon to clarify this mechanism. Another unexpected observation was made in the flgM mutant that a greater proportion of the flagellin migrated faster than the wild-type strain. This band migrated at the same location as nonglycosylated flagellin (Fig. 3). This is consistent with an interpretation that excess of flagellin accumulates in the cytoplasm and cannot be dealt with by the glycosylation pathway or that there is an actual defect in posttranslational glycosylation of the flagellin, a phenomenon recently described in P. aeruginosa (4).
Transcription of the flgM gene in serovar Typhimurium has been shown to be dependent on the alternative sigma factor σ28 and FlhCD, the master regulator (15). The situation in P. aeruginosa appears to be somewhat different in that flgM appears to be regulated by σ28 and an additional factor(s). The transcriptional start site studies definitively implicate σ28, and the absence of a start site in the fleQ and rpoN mutants suggests that they are also involved. Furthermore, the fliA, fleQ, and rpoN mutants containing a flgM-lacZ fusion showed similar reductions in β-galactosidase activity compared to the wild-type strain, suggesting that these regulators may be linked in some way. One explanation may be that fliA is regulated by fleQ and rpoN. However, it has been suggested that fliA is not regulated by rpoN in P. aeruginosa, a suggestion which may have to be reexamined in more extensive studies. Regardless of the role of RpoN and FliA, the residual β-galactosidase activity in these mutants suggests that other regulators are involved. P. aeruginosa does not have flhCD homologues; therefore, there are as yet no known regulatory candidates. A similar situation was reported for the control of the flgM promoter of V. cholerae (29), in which the flgM promoter exhibited high levels of transcription even in the absence of σ28 and σ54. Taken together, identification of flgM in P. aeruginosa illustrates resemblance to the flagellar system of enterics and other organisms in that FlgM represents a major regulator at a critical point in flagellin synthesis. On the other hand, the regulatory hierarchy of the flagellar system in P. aeruginosa has developed differently from that of enteric bacteria and is probably closest to that in V. cholerae.
Acknowledgments
We acknowledge the Interdisciplinary Center for Biotechnology Research at the University of Florida for assistance with DNA sequence and EM analyses. The polyclonal PAK anti-FliC antibody was kindly provided by Daniel Wozniak.
This work was supported by NIH grant AI45014 to R.R.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. K., N. Ohta, J. Wu, and A. Newton. 1995. Regulation of the Caulobacter crescentus rpoN gene and function of the purified σ54 in flagellar gene transcription. Mol. Gen. Genet. 246:697-706. [DOI] [PubMed] [Google Scholar]
- 3.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora, S. K., M. Bangera, S. Lory, and R. Ramphal. 2001. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc. Natl. Acad. Sci. USA 98:9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 22:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caramori, T., D. Barailla, C. Nessi, L. Sacchi, and A. Galizzi. 1996. Role of FlgM in σD-dependent gene expression in Bacillus subtilis. J. Bacteriol. 178:3113-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chadsey, M. S., J. E. Karlinsey, and K. T. Hughes. 1998. The flagellar anti-σ factor FlgM actively dissociates Salmonella typhimurium σ28 RNA polymerase holoenzyme. EMBO J. 17:3123-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Correa, N. E., C. M. Lauriano, R. McGee, and K. E. Klose. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35:743-755. [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta, N., S. K. Arora, and R. Ramphal. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daughdrill, G. W., M. S. Chadsey, J. E. Karlinsey, K. T. Hughes, and F. W. Dahlquist. 1997. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target σ28. Nat. Struct. Biol. 4:285-291. [DOI] [PubMed] [Google Scholar]
- 12.Fredrick, K. L., and J. D. Helmann. 1994. Dual chemotaxis signaling pathways in Bacillus subtilis: a σD-dependent gene encodes a novel protein with both CheW and CheY homologous domains. J. Bacteriol. 176:2727-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillen, K. L., and K. T. Hughes. 1991. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J. Bacteriol. 173:6453-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillen, K. L., and K. T. Hughes. 1991. Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J. Bacteriol. 173:2301-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillen, K. L., and K. T. Hughes. 1993. Transcription from two promoters and autoregulation contribute to the control of expression of the Salmonella typhimurium flagellar regulatory gene flgM. J. Bacteriol. 175:7006-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmann, J. D., and M. J. Chamberlin. 1987. DNA sequence analysis suggests that expression of flagellar and chemotaxis genes in Escherichia coli and Salmonella typhimurium is controlled by an alternative sigma factor. Proc. Natl. Acad. Sci. USA 84:6422-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsey. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277-1280. [DOI] [PubMed] [Google Scholar]
- 18.Ishimoto, K. S., and S. Lory. 1989. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc. Natl. Acad. Sci. USA 86:1954-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, Y.-K., and L. L. McCarter. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 182:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutsukake, K., and T. Iino. 1994. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J. Bacteriol. 176:3598-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Liu, X., and P. Matsumara. 1995. An alternative sigma factor controls transcription of flagellar class-III operons in Escherichia coli: gene sequence, overproduction, purification and characterization. Gene 164:81-84. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Mirel, D. B., and M. J. Chamberlin. 1989. The Bacillus subtilis flagellin gene (hag) is transcribed by the sigma 28 form of RNA polymerase. J. Bacteriol. 174:3095-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirel, D. B., P. Lauer, and M. J. Chamberlin. 1994. Identification of flagellar synthesis regulatory and structural genes in a σD-dependent operon of Bacillus subtilis. J. Bacteriol. 176:4492-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohnishi, K., K. Kutsukake, H. Suzuki, and T. Iino. 1992. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an anti-sigma factor inhibits the activity of the flagellum-specific factor, σF. Mol. Microbiol. 6:3149-3157. [DOI] [PubMed] [Google Scholar]
- 29.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 30.Ritchings, B. W., E. C. Almira, S. Lory, and R. Ramphal. 1995. Cloning and phenotypic characterization of FleS and FleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 63:4868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homologue. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starnbach, M. N., and S. Lory. 1992. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes alternative sigma factor required for flagellin synthesis. Mol. Microbiol. 6:459-469. [DOI] [PubMed] [Google Scholar]
- 34.Totten, P. A., and S. Lory. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172:188-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Totten, P. A., J. Cano-Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]