Abstract
Rubredoxins (Rds) are essential electron transfer components of bacterial membrane-bound alkane hydroxylase systems. Several Rd genes associated with alkane hydroxylase or Rd reductase genes were cloned from gram-positive and gram-negative organisms able to grow on n-alkanes (Alk-Rds). Complementation tests in an Escherichia coli recombinant containing all Pseudomonas putida GPo1 genes necessary for growth on alkanes except Rd 2 (AlkG) and sequence comparisons showed that the Alk-Rds can be divided in AlkG1- and AlkG2-type Rds. All alkane-degrading strains contain AlkG2-type Rds, which are able to replace the GPo1 Rd 2 in n-octane hydroxylation. Most strains also contain AlkG1-type Rds, which do not complement the deletion mutant but are highly conserved among gram-positive and gram-negative bacteria. Common to most Rds are the two iron-binding CXXCG motifs. All Alk-Rds possess four negatively charged residues that are not conserved in other Rds. The AlkG1-type Rds can be distinguished from the AlkG2-type Rds by the insertion of an arginine downstream of the second CXXCG motif. In addition, the glycines in the two CXXCG motifs are usually replaced by other amino acids. Mutagenesis of residues conserved in either the AlkG1- or the AlkG2-type Rds, but not between both types, shows that AlkG1 is unable to transfer electrons to the alkane hydroxylase mainly due to the insertion of the arginine, whereas the exchange of the glycines in the two CXXCG motifs only has a limited effect.
Rubredoxins (Rds) are the simplest of the iron-sulfur redox active proteins and usually contain a single Fe(S-Cys)4 site. In the aerobic bacterium Pseudomonas putida GPo1, Rd is an essential component of the alkane hydroxylase system. It shuttles electrons from Rd reductase, a protein which reduces its flavin adenine dinucleotide at the expense of NADH, to alkane hydroxylase (19, 21, 26, 28, 39). The alkane hydroxylase belongs to a large class of integral membrane proteins (9, 42) that contain an oxo-bridged diiron cluster (32) and is of interest as a biocatalyst for the production of alcohols, fatty acids, and epoxides (24, 44).
Sequencing of the GPo1 alk genes, reviewed in reference 43, showed that the OCT plasmid of GPo1 encodes two Rds, AlkF and AlkG (14), the second of which has been studied in detail in earlier biochemical studies (see below). AlkG is unusual in that it is more than three times the size of other bacterial Rds. It consists of two Rd domains, AlkG1 and AlkG2, connected by a 70-amino-acid linker which shows no homology to other sequences in the Swiss-Prot database (3, 14, 27). AlkF has not been isolated or detected. The primary sequence of AlkF consists of an Rd-like N-terminal domain (AlkF1) and an 80-amino-acid C-terminal extension. AlkF1 and the N-terminal domain of AlkG (AlkG1) are more closely related to each other than to the C-terminal domain of AlkG (AlkG2) (see Fig. 3). Only plasmids which encode AlkG2 were able to restore growth on n-alkanes in P. putida carrying a CAM-OCT alkA mutant plasmid (14). Two Rd genes cloned from the P. putida strain P1 are very similar to the GPo1 alkF and alkG genes (34, 41). In contrast, the Acinetobacter sp. ADP1 Rd (RubA) (10) is similar in size to Rds from anaerobic bacteria. It is more closely related to AlkG2 than to AlkG1 or AlkF1.
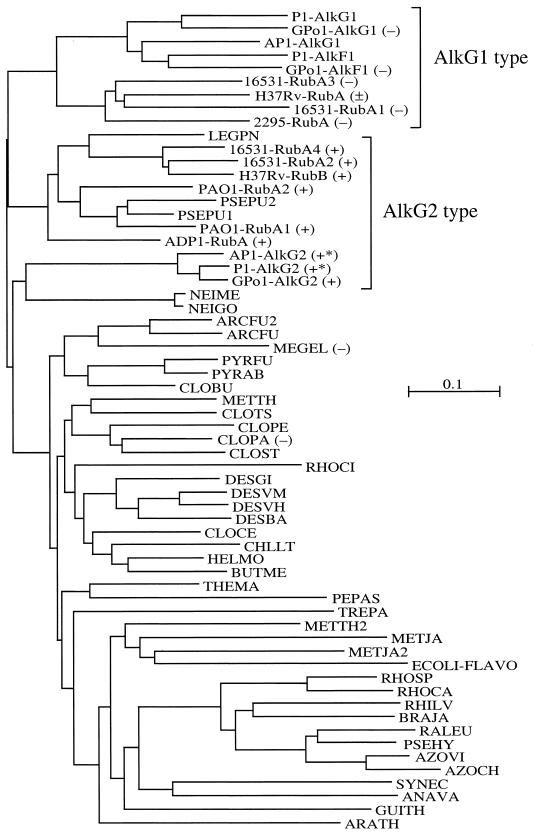
FIG. 3.
Tree of identity generated with ClustalX. The abbreviations are listed in the legend to Fig. 2. Rds involved in or associated with alkane degradation are indicated by square brackets labeled with AlkG1 and AlkG2 type. The results of the in vivo complementation assays (Table 2) are indicated by − (no growth) and + (growth). In two cases only the complete alkG gene was tested (+*). The M. tuberculosis H37Rv RubA complementation experiment resulted in weak growth (±).
Biochemical experiments have shown that the iron in the N-terminal domain of AlkG (AlkG1) is loosely bound and is lost easily from the isolated protein without much effect on the activity in vitro. AlkG could be cleaved with cyanogen bromide, resulting in a 12-kDa C-terminal peptide, which was about three times less active than the full-length AlkG. The 5-kDa N-terminal fragment still bound iron but was unstable and not active in alkane hydroxylation (22). AlkG1 reconstituted with iron was recently found to be redox active as well: it transfers electrons from Rd reductase to horse cytochrome c as efficiently as AlkG2 (25). It was, however, not tested with the alkane hydroxylase.
Microbial genome sequencing and our studies on bacterial alkane hydroxylases (33) (L. G. Whyte, T. H. M. Smits, D. Labbé, B. Witholt, C. W. Greer, and J. B. van Beilen, submitted for publication) have yielded several additional Rd genes that are likely to be involved in alkane oxidation. Sequence analysis and complementation studies show that these Rds or Rd domains can be divided into two groups: AlkG1-type Rds related to AlkG1 and AlkG2-type Rds related to AlkG2. Only the AlkG2-type Rds complement an Escherichia coli recombinant lacking a functional Rd 2 for growth on n-octane. Mutagenesis of the AlkG1- and AlkG2-type Rds shows which sequence elements determine whether the Rds can transfer electrons to the alkane hydroxylase.
MATERIALS AND METHODS
Strains, growth conditions, and materials.
Plasmids used or constructed in this study are listed in Table 1. E. coli JM101 [endA hsdR supR thi-1 Δ(lac-proAB) traD36 proAB laclq lacZM15](F′) (46) and DH10B (Gibco BRL) were used for cloning and the production of plasmid DNA for sequencing. All other strains are n-alkane degraders and are listed in Table 2. Luria-Bertani broth (LB) (29) and E2 medium (17), supplemented with carbon sources and/or antibiotics, were used throughout. For growth on n-octane, petri dishes with E2 medium were incubated at 30°C in a sealed box containing an open Erlenmeyer flask with n-octane. All cultures were grown aerobically at 30 or 37°C.
TABLE 1.
Plasmids used or constructed in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pAF1 | 3.5-kb EcoRI-BamHI alkGH fragment of A. borkumensis AP1 in pZEr0-2.1 | This study |
| pBG219 | pKK233-2, alkG from pBG214 | 40 |
| pCR14 | pKK233-2, alkG1 from pBG219 by PCR | This study |
| pCR15 | pCR14, AlkG1 N10G A43G | This study |
| pGEc47 | Tc alkBFGHJKL alkST (P. putida GPo1) in pLAFR1 | 7 |
| pGEc47ΔG | Tc alkFGHJKL alkST (P. putida GPo1) in pLAFR1, deletion in alkG | 40 |
| pGEc283 | alkBFGH in pMMB24, 125-bp deletion in alkG | 14 |
| pGEM7-Zf(+) | Cloning vector, Ap | Promega |
| pKK233-2 | Expression vector, Ptrc Ap | 2 |
| pKKPalk | Expression vector, Palk Ap | 35 |
| pKKrubA(ADP1) | pKKPalk, rubA (Acinetobacter sp. ADP1) | This study |
| pKKrubA1(PAO1) | pKKPalk, rubA1 (P. aeruginosa PAO1) | This study |
| pKKrubA2(PAO1) | pKKPalk, rubA2 (P. aeruginosa PAO1) | This study |
| pKKalkG(AP1) | pKKPalk, alkG (A. borkumensis AP1) | This study |
| pKKrubA(H37Rv) | pKKPalk, rubA (M. tuberculosis H37Rv) | This study |
| pKKrubB(H37Rv) | pKKPalk, rubB (M. tuberculosis H37Rv) | This study |
| pKKrubA1(Rer) | pKKPalk, rubA1 (R. erythropolis NRRL B-16531) | This study |
| pKKrubA2(Rer) | pKKPalk, rubA2 (R. erythropolis NRRL B-16531) | This study |
| pKKrubA3(Rer) | pKKPalk, rubA3 (R. erythropolis NRRL B-16531) | This study |
| pKKrubA4(Rer) | pKKPalk, rubA4 (R. erythropolis NRRL B-16531) | This study |
| pKKrubG4W(Aru) | pKKPalk, rub domain (P. rugosa NRRL B-2295) | This study |
| pKKrubG4W(Aru) | pKKPalk, rub domain (P. rugosa NRRL B-2295) | This study |
| pKK-G1 | pKKPalk, alkG1 domain (P. putida GPo1) | This study |
| pKK-G1mut1 | pKK-G1, N10 changed to G | This study |
| pKK-G1mut2 | pKK-G1, H21 changed to A | This study |
| pKK-G1mut3 | pKK-G1, PWHL29-32 changed to RWED | This study |
| pKK-G1mut4 | pKK-G1, AVRD43-46 changed to GAT | This study |
| pKK-G1mut5 | pKK-G1, R45 changed to A | This study |
| pKK-G1mut6 | pKK-G1, N10 changed to G and AVRD43-46 changed to GAT | This study |
| pKK-G1mut7 | pKK-G1, N10 changed to G, PWHL29-32 changed to RWED, and AVRD43-46 changed to GAT | This study |
| pKK-G2 | pKKPalk, alkG2 domain (P. putida GPo1) | This study |
| pKK-G2mut1 | pKK-G2, G10 changed to N | This study |
| pKK-G2mut2 | pKK-G2, A21 changed to H | This study |
| pKK-G2mut3 | pKK-G2, RFED29-32 changed to PFHL | This study |
| pKK-G2mut4 | pKK-G2, GAT43-46 changed to AVRD | This study |
| pP1EH | pBR322, alkBFGHJ (P. putida P1) | 34 |
| pZErO-2.1 | Cloning vector, Km ccdB | Invitrogen |
TABLE 2.
Rds in strains able to grow on n-alkanes, results of complementation experiments, and sources of strains
| Strain | Growth on n-alkanes | Rd name | Type | Expression plasmid | Complementationa | Reference |
|---|---|---|---|---|---|---|
| Acinetobacter sp. ADP1 | C12-C16 | RubA | AlkG2 | pKKrubA(ADP1) | + | 13 |
| A. borkumensis AP1 | C6-C16 | AlkG | AlkG1 + AlkG2 | pKKalkG(AP1) | + | 45 |
| M. tuberculosis H37Rv | Unknown | RubA | AlkG1 | pKKrubA(H37Rv) | ± | 5 |
| RubB | AlkG2 | pKKrubB(H37Rv) | + | |||
| P. rugosa NRRL B-2295 | C10-C16 | AlkB | AlkG1 | pKKrub(Aru) | − | 12 |
| pKKrubG4W(Aru) | − | |||||
| P. aeruginosa PAO1 | C12-C16 | RubA1 | AlkG2 | pKKrubA1(PAO1) | + | 37 |
| RubA2 | AlkG2 | pKKrubA2(PAO1) | + | |||
| P. putida GPo1b | C5-C12 | AlkF | AlkG1 | − | 30 | |
| AlkG | AlkG1 + AlkG2 | pBG219 | + | 14 | ||
| P. putida P1 | C5-C8 | AlkF | AlkG1 | NT | 34 | |
| AlkG | AlkG1 + AlkG2 | pP1EH | + | |||
| R. erythropolis NRRL B-16531 | C6-C30 | RubA1 | AlkG1 | pKKrubA1(Rer) | − | 12 |
| RubA2 | AlkG2 | pKKrubA2(Rer) | + | |||
| RubA3 | AlkG1 | pKKrubA3(Rer) | − | |||
| RubA4 | AlkG2 | pKKrubA4(Rer) | + |
+, −, or ±, growth, no growth, or weak growth of an E. coli GEc137(pGEc47ΔG) recombinant containing the different expression plasmids on E2 minimal medium plates with n-octane vapor as the C source; NT, not tested.
Previously known as Pseudomonas oleovorans GPo1, TF4-1L, and ATCC 29347.
Molecular genetics.
PCRs were carried out using a Perkin-Elmer GeneAmp PCR System 9600. To amplify Rd genes from alkane-degrading strains, forward primer DegRUBfw (TGYGGIGAGCTCTAYGAYGARGC), containing a SacI site, and reverse primer DegRUBrv (GGAATTCCRCARTCIGGRCA), containing an EcoRI site, were used. In these primers, I represents inosine, which base pairs with all four nucleotides, Y represents T+C, and R represents A+G. For PCRs with these primers, the following program was used: 4 min at 95°C; 25 cycles of 45 s at 95°C, 1 min at 40°C, and 1 min at 72°C; and 5 min at 72°C. Then the mixture was transferred at 4°C. After a first round of PCR, 1 μl (out of 20 μl) of product was used in a second round of PCR with the same primers. Chromosomal DNA isolated according to standard procedures (29) was used as template DNA. PCR fragments of the expected size were purified over a 3% NuSieve gel, cloned in pGEM7-Zf(+) by using SacI and EcoRI, and sequenced. PCR products obtained with other primers (see below) were purified over a 1% agarose gel in Tris-borate-EDTA buffer. The PCR fragments were cut with the appropriate restriction enzymes, purified again over a 1% agarose gel, isolated from the gel by electroelution, and cloned in an appropriate vector.
To clone Rd and alkane hydroxylase genes, easy-to-clone restriction fragments were identified by Southern blotting. Probes were prepared as follows. The cloned Alcanivorax borkumensis Rd PCR fragment obtained with DegRUBfw and DegRUBrv, and the cloned Prauserella rugosa NRRL B-2295 and Rhodococcus erythropolis NRRL B-16531 alkane hydroxylase gene fragments obtained earlier (34), were cut from pGEM7-Zf(+). The DNA fragments were purified over a 1% agarose gel or a 3% NuSieve gel, electroeluted, and labeled by the random priming method using the digoxigenin (DIG) labeling kit (Roche Diagnostics). Southern and colony blots were carried out by using the Roche Diagnostics DIG kit. Hybridizations were carried out by using standard hybridization buffer without formamide at 68°C. Alkaline phosphatase coupled to anti-DIG antibodies was used with CSPD as the chemiluminescent substrate.
Restriction fragments around the desired size were cut out from a preparative agarose gel, isolated by electroelution, ligated between the appropriate sites of pGEM7-Zf(+) (Promega) or pZErO-2.1 (Invitrogen), and transformed into E. coli DH10B (Life Technologies) by electroporation (6). E. coli transformants were selected with ampicillin at 200 μg/ml or kanamycin at 50 μg/ml. Plasmid DNA was isolated using the High Pure Plasmid Isolation kit (Roche Diagnostics). Both strands of the inserts were sequenced on a Li-Cor 4000L sequencer using IRD800-labeled −40 forward (AGGGTTTTCCCAGTCACGACGTT) and −40 reverse (GAGCGGATAACAATTTCACACAGG) primers and the Thermosequenase cycle sequencing kit (Amersham Pharmacia Biotech). Nucleotide and amino acid sequences were analyzed and compared using LASERGENE Navigator from DNASTAR. Nucleotide and amino acid sequences were compared with the EMBL, SwissProt, and GenBank databases using BLAST (1). BLAST searches were carried out at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Construction of pGEc47ΔG.
Plasmid pGEc283 (14) was used to construct an alkG deletion derivative of pGEc47. It has a 125-bp deletion in the N-terminal part of alkG, which places the rest of the gene out of frame with respect to the start codon of the AlkG gene. Plasmid pGEc283 has a pMB1 origin and an ampicillin resistance marker. To transfer the alkG deletion from pGEc283 to pGEc47, E. coli SF800 containing pGEc47 was transformed with pGEc283. SF800 lacks the PolA polymerase which is required for replication of pMB1 plasmids. The ampicillin marker can only survive when the plasmid integrates, usually in pGEc47 through homologous recombination. A second recombination, in which the ampicillin marker is lost, effectively results in an exchange of homologous DNA between pGEc47 and the pMB1 plasmid, sometimes including the deletion. Plasmid derivatives of pGEc47 were checked by gel electrophoresis for loss of the 125-bp fragment, by DNA sequencing, and by the inability to confer growth on n-octane to E. coli host strains.
Construction of expression plasmids encoding P. putida GPo1 Rds and Rd domains.
For expression of the N- and C-terminal domains of AlkG from the PalkB promoter, and further mutagenesis of the domains, the corresponding DNA fragments were amplified with primers alkG1-Eco-Nde (ATAAGAATTCCATATGGTGATGAGTATGGCT) and alkG1-RV-Bam (GCCGGATCCTCATACGCCGCTCTC) for AlkG1 and primers alkG2-Eco-Fw (CACAGGAAAGAATTCATGTTGAAGTGGATATG) and alkG-Rv-Bam (CCTGAGGATCCTAGAATGAC) for AlkG2. Both fragments were cloned between the EcoRI and BamHI sites of pKKPalk (35), resulting in pKK-G1 and pKK-G2. Cloned and expressed as described here, the amino acid sequence of AlkG1 starts and ends as shown (see Fig. 2) while the AlkG2 sequence starts with MLKW. Mutations were introduced in pKK-G1 and pKK-G2 using the QuikChange kit from Stratagene and primers G1-N10G-FW (CTATAAATGCCCGGATTGTGGCTATGTTTATGATGAGAGTG), G1-H21A-FW (AGTGCGGGTAATGTGGCCGAGGGGTTTTCTCCA), G1-PWHL29-32RWED-FW (GGGTTTTCTCCAGGTACGCGCTGGGAGGATATTCCTGAGGATTGG), G1-AVRD44-47GAT (TGCTGCCCCGATTGCGGGGCTACGAAGCTTGACTTCATG), G1-R45A-FW (GCCCCGATTGCGCAGTTGCAGACAAGCTTGACTTCAT), G2-G10N-FW (TGGATATGTATTACCTGCAACCATATATATGATGAGG), G2-A21H-FW(GCGTTGGGCGATGAGCACGAGGGTTTTACT), G2-RFED29-32PFHL-FW (TTTTACTCCAGGTACCCCCTTTCACCTTATTCCTGATGACTG), G2-D41L-FW (GACTGGTGCTGTCCGCTTTGCGGCGCCACGAAAGAAGAC), G2-D49A-FW (GGGGCTACGAAAGAGGCCTATGTGCTCTACGAG), and G2-GAT44-47AVRD-FW (GCTGTCCGGATTGCGCAGTTCGCGACAAAGAAGACTATGTG). The reverse complement primers are not shown. All cloned and mutated AlkG domains were sequenced using primers pAlkFw3 (GCCAGCTCGTGTTTTTCCAGCAGACG) and pKK-RV (GAGTTCGGCATGGGGTCAGGTG).
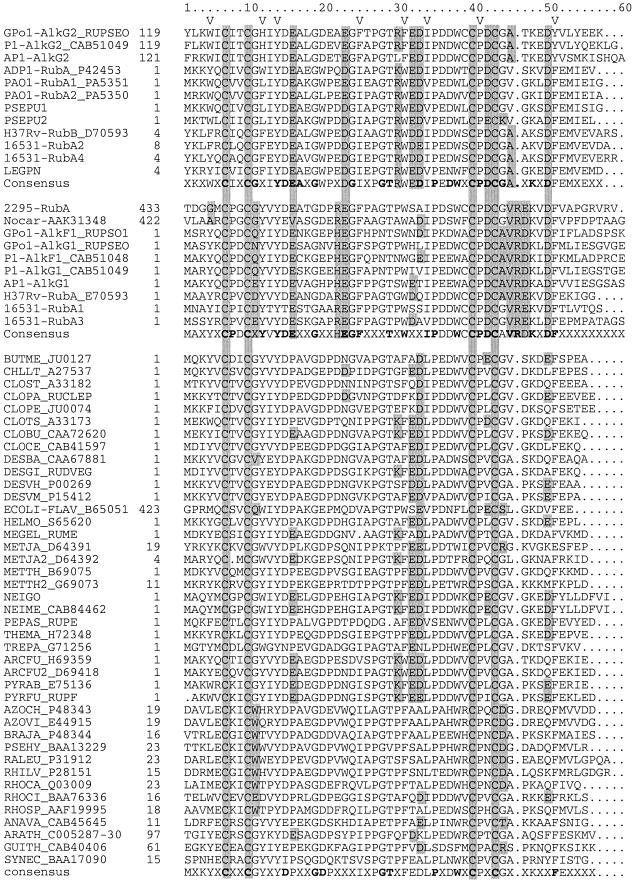
FIG. 2.
Alignment of Rd sequences. The names of the peptides are explained in the text (Alk related) or below. In most cases the accession number is shown with the strain abbreviation. The first group of sequences includes the AlkG2-type Rds, the second group includes the AlkG1-type Rds, and the third group contains the remaining Rds. In several cases the sequence was truncated for the alignment. The position of the first amino acid in the full-length protein is indicated. A V in the second line marks the hydrophobic core residues. Features discussed in the text are shaded. ANAVA, Anabaena variabilis; ARCFU, Archaeoglobus fulgidus; AZOCH, Azotobacter chroococcum (strain mcd 1); AZOVI, Azotobacter vinelandii; BRAJA, Bradyrhizobium japonicum; BUTME, Butyribacterium methylotrophicum; CHLLT, Chlorobium limicola f. sp. thiosulfatophilum; CLOBU, Clostridium butyricum; CLOCE, Clostridium cellulolyticum; CLOPA, C. pasteurianum; CLOPE, Clostridium perfringens; CLOST, Clostridium sticklandii; CLOTS, Clostridium thermosaccharolyticum; DESBA, Desulfoarculus baarsii; DESGI, Desulfovibrio gigas; DESVM, Desulfovibrio vulgaris (strain Miyazaki); DESVU, Desulfovibrio vulgaris; ECOLI, E. coli K-12; GUITH, Guillardia theta; HELMO, Heliobacillus mobilis; MEGEL, Megasphaera elsdenii; METJA, Methanococcus jannaschii; METTH, Methanobacterium thermoautotrophicum; NEIME, Neisseria meningitidis; PEPAS, Peptostreptococcus asaccharolyticus; PSEHY, Pseudomonas hydrogenovora; PYRAB, Pyrococcus abyssi (strain Orsay); PYRFU, P. furiosus; RALEU, Ralstonia eutropha; RHILV, Rhizobium leguminosarum bv. viciae; RHOCA, Rhodobacter capsulatus; RHOSP, Rhodobacter sphaeroides; RHOCI, Rhodococcus sp. CIR2; SYNEC, Synechocystis sp.; THEMA, Thermotoga maritima; TREPA, Treponema pallidum. A number of sequences are from unfinished genomes and have no accession numbers yet: PSEPU1 and PSEPU2, P. putida KT2440; LEGPN, Legionella pneumophila; NEIGO, Neisseria gonorrhoeae.
As a part of an earlier study (J. B. van Beilen, unpublished data), AlkG1 was also expressed separately from the trc promoter in pKK233-2 (2). First, a PCR was carried out to introduce an EcoRI site upstream of the ATG (alkG1-Eco-FW, GACATGGTGAATTCTATGGCTAGCTATAAATGC) while the reverse primer changed the glycine at position 56 to a stop codon and introduced an ApaI site immediately downstream (alkG1-Apa-RV, GCCCTTTTCGGGCCCTCAGCTCTCAATTAAC). The PCR product was cloned in pGEM-7Zf(+) cut with EcoRI and ApaI. The N10G and A43G mutations were introduced in alkG1 by site-directed mutagenesis (16) by using primers G1-N10G (CATCATAAACATAACCACAATCTGGGC) and G1-A43G (CAAGTTTGTCTCGAACGCCGCAATCGGGGC). This plasmid and pGEM-7Zf(+) containing the wild-type alkG1 gene were digested with EcoRI and PvuII, and the alkG1 fragments were isolated. Plasmid pKK233-2 was cut with HindIII, treated with Klenow, and cut with EcoRI. The purified alkG1 fragments were ligated to pKK233-2 and transformed into E. coli DH10B, resulting in plasmids pCR14 (alkG1) and pCR15 (alkG1 N10G A43G).
Expression constructs for Rds from other bacteria.
To express Rd genes from other bacteria in E. coli GEc137(pGEc47ΔG), forward primers containing an EcoRI or, in the case of A. borkumensis AP1 alkG, an NdeI site and reverse primers containing a HindIII site were used to amplify the Rd genes from chromosomal DNA (A. borkumensis AP1 alkG, AP1-FW [AGGTGCTTCATATGGCTAAATATC] and AP1-RV [ATTAAAGCTTGCCGAATGAGCCTA]; Mycobacterium tuberculosis RubA and RubB, MT-rubA-FW [CCGGAATTCATATGATGGCCGCCTAC], MT-rubA-RV [CGTAAGCTTAGTTACCCTCCTATC], MT-rubB-FW [CCGGAATTCATATGAACGACTACAAACTG], and MT-rubB-RV [CAGTAAAGCTTGGAGTGCTCACG]; R. erythropolis RubA1, RubA2, RubA3, and RubA4, rubA1FW [GGGGAATTCGCATGAACGCC], rubA1Rv [GGACCAAGCTTCGGGAATC], rubA2FW [GGTGGAATTCTTATGACAGC], rubA2RV [CGGGAAGCTTTCCATCGCC], rubA3FW [CGGAGAATTCTGATGAGCAGC], rubA3Rv [GGCAACTCCAAGCTTCGGG], rubA4FW [GGGGAGAATTCCATGAACGACTACAAG], and rubA4RV [CCGGCAAGCTTGGCCAGAACC]). The PCR fragments were cloned between the EcoRI or NdeI and HindIII sites of pKKPalk, a pKK223-3 derivative in which the Rd genes are under control of the GPo1 alkB promoter (35).
To clone the Rd domain of the P. rugosa AlkB, two forward primers introducing an EcoRI site and a start codon at position 432 of the complete AlkB-AlkG fusion protein (2295-GGG, GTGAATTCGCCATGGACGGCGGGATGTGCCCC; 2295-TGG, GTGAATTCGCCATGGACGGCTGGATGTGCCCC) and a reverse primer introducing a BamHI site (2295-RV, CGGCGAGGATCCGGTCCAGCTC) were designed. Primer 2295-GGG amplifies the wild-type sequence while primer 2295-TGG introduces the mutation G436W (position 4) (see Fig. 2).
Alkane hydroxylase activity assay.
In vivo alkane hydroxylase activity was determined by measuring the conversion of 1-nonene to 1,2-epoxynonane by induced cells. LB medium (10 ml) was inoculated with 0.5 ml of an overnight culture in LB medium and incubated at 150 rpm at 30°C. After 3 h, alkane hydroxylase was induced by adding dicyclopropylketone to a concentration of 0.02%. Three hours after induction, 5 μl of 1-nonene (99.9% purity; Wiley Organics) was added and the incubation was continued for 30 min at 200 rpm. Subsequently, the cultures were extracted with 1 ml of n-hexane containing 0.01% n-dodecane as the internal standard. The hexane phase was analyzed for 1,2-epoxynonane on a Fisons GC8000 gas chromatograph, equipped with an Optima 5 column (Macherey Nagel, Oensingen, Switzerland), isotherm at 80°C. All experiments were carried out in duplicate. Activity is expressed as units per gram (1 U = 1 μmol of product formed per min). The minimum activity that can be detected in this assay is less than 0.01 U g of cells−1.
3-D structure modeling.
Three-dimensional (3-D) structure predictions were carried out by using Swiss Model (31). The first approach mode was used for AlkG2-type Rd sequences. The AlkG1-type Rds were modeled in the optimize mode after manual alignment of the sequences with the Clostridium pasteurianum and Pyrococcus furiosus Rds (see Fig. 2 and 4).
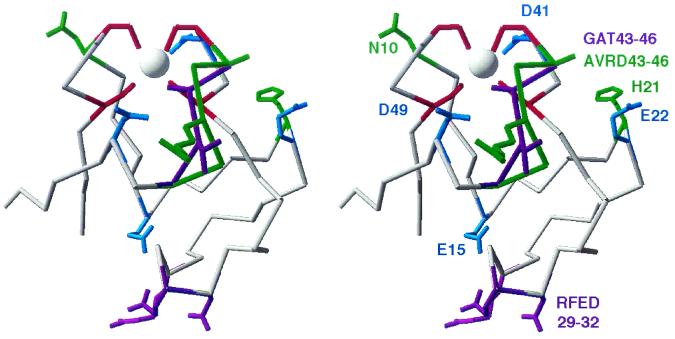
FIG. 4.
3-D structure model of Rd domains AlkG1 and AlkG2. The 3-D models were generated by Swiss Model by using the P. furiosus and C. pasteurianum Rd structures as templates. The modeled structure of AlkG2 is shown in grey. Residues that are conserved in AlkG1- and AlkG2-type Rds, but not in other Rds, are shown in light blue (E15, E22, D41, and D49). Residues that are conserved in the AlkG2-type Rds, but not in the AlkG1-type Rds, are shown in purple (GAT43-46 and RFED29-32) while sequence features conserved in the AlkG1-type Rds are shown in green (N10, H21, and AVRD43-46). The iron-binding cysteines of AlkG2 are shown in red. Light blue, green, red, and purple residues are shown with side groups (with the exception of AlkG1 V44 and D46).
Sequences.
Sequences used in this study for comparisons are available in the sequence databases (see Fig. 2 for accession numbers).
Nucleotide sequence accession number.
Novel sequences determined in this study were deposited in GenBank and are available under the following accession numbers: AJ295164 (A. borkumensis AP1 alkS alkBGHJ), AJ009587 (P. rugosa NRRL B-2295 alkB), AJ009586 (R. erythropolis NRRL B-16531 alkB1 rubA1 rubA2 rubB), and AJ297269 (R. erythropolis NRRL B-16531 alkB2 rubA3 rubA4).
RESULTS AND DISCUSSION
Rds involved in alkane degradation.
The sequence databases contain several Rd sequences that are likely to be involved in alkane oxidation, as they are more closely related to the P. putida GPo1 and Acinetobacter sp. ADP1 Rds than to other Rds, and are associated with alkane hydroxylase or Rd reductase genes. The Pseudomonas aeruginosa PAO1 genome (37) encodes two alkane hydroxylase homologs, two Rds, and an Rd reductase. In M. tuberculosis H37Rv (5), two Rd genes are located immediately downstream of an alkane hydroxylase homolog (40% identity to the GPo1 alkane hydroxylase) while other mycobacteria have similar gene arrangements. Several unfinished genome sequences encode Rds that are quite closely related to Rds involved in alkane hydroxylation. Some of these were included in the sequence comparisons, but they were not included in the functional studies.
Cloning of novel Rd genes.
Recently, four Rd genes and four alkane hydroxylase homologs from R. erythropolis NRRL B-16531 have been cloned by using highly degenerate primers that amplify internal fragments of alkane hydroxylase genes (Whyte et al., submitted for publication). In short, an alkane hydroxylase gene fragment obtained earlier from strain 16531 (34) was used to clone flanking DNA. This DNA was found to encode two Rds, RubA1 and RubA2, and an Rd reductase, RubB. A DNA fragment encoding a second alkane hydroxylase gene was cloned from the same strain by using an alkane hydroxylase PCR fragment, obtained from a related R. erythropolis strain, as a probe. This fragment encoded another two Rds, RubA3 and RubA4.
The alkane hydroxylase of P. rugosa NRRL B-2295 (previously named Amycolatopsis rugosa) was also cloned by using an alkane hydroxylase gene fragment as a probe (34). A 3.0-kb SacI-BamHI fragment was selected by Southern blotting, cloned in pGEM7-Zf(+), and sequenced. A 1,470-bp alkane hydroxylase open reading frame was identified, the C-terminal portion of which was homologous to Rds. A closely related alkane hydroxylase sequence from Nocardioides sp. CF8, which also contains a C-terminal Rd domain, was included in the sequence comparisons (11).
In the case of A. borkumensis AP1, we were unable to amplify internal fragments of alkane hydroxylase genes. As an alternative, we developed degenerate primers based on conserved sequence elements (CGXXYDEA and CPDCG) in previously sequenced Alk-Rds (see Fig. 2). Using these primers, a fragment of the expected size was amplified from AP1, sequenced, and subsequently used as a probe in colony blots to obtain a larger genomic DNA clone. A 3.5-kb EcoRI-BamHI fragment was selected, cloned in pZErO-2.1, and sequenced (pAF1). The fragment was found to contain a 525-bp open reading frame, very similar to the GPo1 AlkG. It consists of an N-terminal and a C-terminal Rd domain connected by a linker. The linker is of the same length as the GPo1 and P1 AlkG linkers but shows no homology, even though the N- and C-terminal domains are closely related to AlkG1 and AlkG2, respectively (see Fig. 2 and 3). AP1 does not contain an alkF gene. The degenerate Rd gene primers were also tested on other alkane-degrading strains known or expected to contain Rds but only yielded PCR products in the case of GPo1 and R. erythropolis NRRL B-16531. The resulting sequences were identical to known Rd sequences (data not shown). The organization of the Rd genes is shown in Fig. 1A.
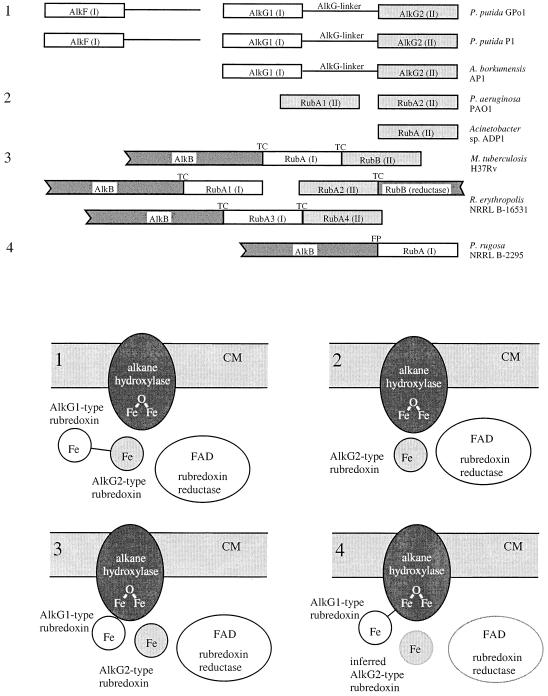
FIG. 1.
Rd genes in gram-positive and gram-negative alkane-degrading strains and general architecture of alkane hydroxylase systems. (A) Organization of Rd genes in alkane-degrading strains. (B) General architecture of alkane hydroxylase systems. CM, cytoplasmic membrane; FAD, flavin adenine dinucleotide. AlkG1- and AlkG2-type Rds and Rd genes are shown in white and grey, respectively. Rd reductase is shown in white. The alkane hydroxylases are shown in dark grey. The P. putida GPo1 and P1 and A. borkumensis AP1 AlkG molecules consist of an AlkG1- and an AlkG2-type Rd connected by a linker (1). Acinetobacter sp. ADP1 and P. aeruginosa PAO1 possess only an AlkG2-type Rd (2). The gram-positive R. erythropolis NRRL B-16531 and M. tuberculosis H37Rv possess separate AlkG1- and AlkG2-type Rds (one or two of each) (3). The P. rugosa NRRL B-2295 AlkB and an AlkG1-type RubA form a natural fusion protein (FP) (4). Genes linked by translational coupling are marked TC.
Functional studies.
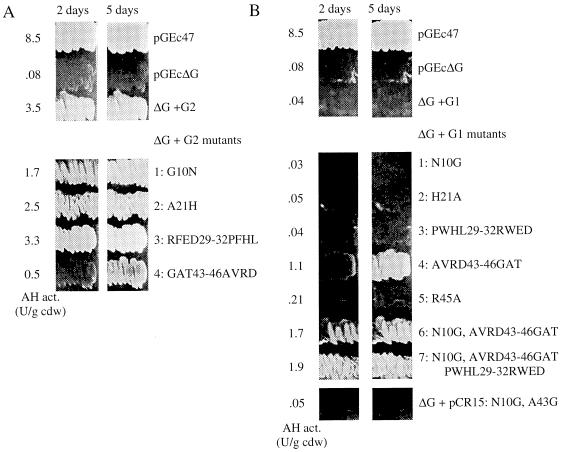
Earlier biochemical studies have shown that spinach ferredoxin and ferredoxin reductase can replace P. putida GPo1 Rd and Rd reductase in vitro (4). To investigate whether heterologous Rds or Rd domains are able to transfer electrons from the GPo1 Rd reductase to alkane hydroxylase in vivo, we constructed an alkG deletion derivative of pGEc47, a plasmid containing all genes (alkBFGHJKL and alkST) necessary for growth of E. coli recombinants on n-octane vapor (7). Plasmid pGEc47ΔG has a 125-bp HindIII deletion in alkG, which removes a small part of the AlkG1 domain and most of the linker, and puts the remainder of the protein out of frame. E. coli GEc137 containing this plasmid has 1 to 2% alkane hydroxylase activity relative to GEc137 containing pGEc47. This residual activity is not due to AlkF and/or the remaining part of AlkG1 or to expression of the AlkG2 domain from an internal start codon, as an E. coli recombinant containing only the alkB and alkST genes showed the same residual activity (40). Plasmids encoding the GPo1 Rd AlkG, such as pBG219 (pKK233-2 containing alkG), restore growth of the E. coli GEc137(pGEc47ΔG) recombinant on n-octane. The GPo1 AlkG1 and AlkG2 Rd domains were also cloned separately and expressed from the trc promoter in pKK233-2 (2) and the alkB promoter in pKKPalk (35). As expected, in view of the earlier biochemical and deletion studies, a plasmid encoding the AlkG2 domain (pKK-G2) fully restored growth on n-octane while the AlkG1 domain of GPo1 (pCR14 and pKK-G1) was unable to complement the alkG deletion (Table 2) (see Fig. 5).
FIG. 5.
Growth on n-octane and alkane hydroxylase activity of E. coli GEc137 recombinants containing pGEc47, pGEc47ΔG, or pGEc47ΔG combined with wild-type and mutagenized AlkG2 (A) and AlkG1 (B) Rd domains. Growth on n-octane was documented after 2 and 5 days. Alkane hydroxylase activities are shown to the left of the growth zones. The mutations are shown to the right of the growth zones. For AlkG1, 1 to 7 correspond to plasmids pKK-G1mut1 to pKK-G1mut7. For AlkG2, 1 to 4 correspond to plasmids pKK-G2mut1 to pKK-G2mut4 in Table 1.
The Acinetobacter sp. ADP1 Rd, RubA (10), was cloned in pKKPalk. The recombinant E. coli GEc137[pGEc47ΔG, pKKrubA(ADP1)] was able to grow as well on n-octane as a similar recombinant containing the GPo1 Rd gene in trans. RubA1 and RubA2 from P. aeruginosa PAO1, RubA2 and RubA4 from R. erythropolis NRRL B-16531, and RubB from M. tuberculosis H37Rv, cloned and expressed in the same way, also complemented the deletion. RubA from the latter strain showed very weak complementation while RubA1 and RubA3 from R. erythropolis did not show any activity. Because the P. putida P1 and A. borkumensis AlkG proteins are so similar to the P. putida GPo1 AlkG, only the complete alkG genes were tested (Table 2).
Comparison of the P. rugosa Rd domain with the other Rd sequences (Fig. 2 and see below) showed that the first hydrophobic core residue (position 4 in the alignment, usually W, Y, or F) is replaced by a glycine, whereas in the Nocardioides sequence, an alanine is found at this position. As these are the only Rds that are fused to the alkane hydroxylase, it is possible that the bulky side group of the tryptophan residue is replaced by an amino acid from the fusion partner. Therefore, we expressed the P. rugosa Rd domain separately in two forms, one with the original glycine residue, and the other with a tryptophan at this position. However, neither the wild-type Rd domain nor the G4W mutant complemented the deletion in pGEc47ΔG, even though the complete fusion protein shows alkane hydroxylase activity (33).
The finding that all AlkG2-type Rds were able to replace the GPo1 Rd 2 means that strains expressing any AlkG2-type Rd and an Rd reductase can be used as hosts to test whether newly cloned alkane hydroxylase homologs are indeed functional (33).
Comparison of Rd and Rd domain sequences.
The sequence databases contain about 60 Rd or Rd-like sequences, many of which are between 52 and 55 amino acids in length. Several Rd-like sequences that are associated with hydrogenase genes have a 15- to 25-amino-acid N-terminal extension while Rd-like sequences from algae and plants have variable length extensions (between 60 and 250 amino acids). A 479-amino-acid hypothetical protein, b2710, from E. coli has an Rd-like domain at its C terminus (ECOLI_FLAV in Fig. 2 and 3,) which is, with 37% sequence identity, the Rd-like sequence most distantly related to GPo1 AlkG2. For sequence comparisons, all Rds were trimmed to approximately the same length. The P. putida P1 and A. borkumensis AP1 Alk-Rd domains were named according to the GPo1 terminology (AlkG1 and AlkG2 for P. putida P1 and A. borkumensis AP1 AlkG and AlkF1 for the P. putida P1 AlkF) while the names of the other Rds are derived from the organism or strain name (Fig. 2). Some Rd-like sequences from unfinished genomes that were similar to Rds involved in alkane hydroxylation (PSEPU1, PSEPU2, and LEGPN) were included in this analysis.
Alignment of the Rds and Rd domains shows that all sequences contain four iron-binding cysteines in two CXXC motifs. Y13 and P40 are absolutely conserved while the hydrophobic core residues (Fig. 2) and a few other residues are also very well conserved. Rds involved in alkane oxidation (Alk-Rds) are on three deep-rooted branches in the tree of identity shown in Fig. 3. The percentage of sequence identity within the Alk-Rd branches (>37%) varies almost as widely as similarity levels between Alk-Rds and other Rds (usually over 40%, with some exceptions under 30%). A conspicuous difference between the two groups is that the Alk-Rds have conserved negatively charged residues at positions 15 (E), 22 (D/E), 41 (D), and 49 (D), whereas the other Rds usually have uncharged, nonconserved residues at these positions. Inspection of a 3-D model of AlkG2 (see below) shows that residues D41 and D49 are located on opposite sides of the redox center while the two other negative charges are further away from the iron (Fig. 4).
Comparison of Alk-Rds.
Rds or Rd domains that restore growth on n-octane of E. coli recombinants lacking AlkG are separated in two branches that are as distantly related to each other as they are to the other Rds (down to 40% sequence identity). All Alk-Rds that were not effective in the complementation assay are on one branch of the tree of identity (>52% sequence identity) together with a few sequences that were not tested but can be expected to be ineffective as well because they are closely related to tested Alk-Rds (e.g., the AlkG1 domains of P. putida P1 and A. borkumensis AP1) (Fig. 3). Several sequence features distinguish these Alk-Rds (henceforth named AlkG1 type) from the active Alk-Rds (AlkG2 type). Whereas in AlkG2-type Rds, the CXXC motifs are followed by a glycine (positions 10 and 43 in the alignment), the AlkG1-type Rds have a D, E, Q, or N at position 10, and an A, or less frequently a G, at position 43 (Fig. 2).
In all AlkG1-type Rds, the sequence immediately downstream of cysteine 42 ([A/G]VR[D/E]K) is well conserved, is one amino acid longer, and contains much bulkier amino acids than the corresponding sequence in the AlkG2-type Rds (G[A/V][G/A/S/T]K). In the AlkG2-type Rds, amino acids 29 to 32 constitute a well-conserved polar patch ([K/R][W/F][E/D][E/D]) opposite to the redox center (Fig. 4 and see below). This stretch is not conserved in the AlkG1-type Rds, except for W30, which is part of the hydrophobic core. At position 21, the AlkG1-type Rds have a positively charged residue (R/H) that is not conserved in AlkG2-type Rds. The P. rugosa Rd sequence shows all features of the AlkG1-type Rds, except that both CXXC motifs are followed by a glycine.
Modeling of AlkG1- and AlkG2-type Rds.
All Alk-Rd sequences were modeled with Swiss Model, which calculates a 3-D model based on sequence alignments and X-ray crystallography data of related proteins (31). The models of AlkG2-type (active) Rds are hard to distinguish from published Rd structures, as the hydrophobic core residues of Rd are unchanged or similar in size, and no insertions or deletions have to be accommodated (Fig. 4).
Modeling of the AlkG1-type Rds suggests that the hydrophobic core and most of the backbone are very similar as well. Based on the results of site-directed mutagenesis experiments with the C. pasteurianum Rd, replacement of G10 and G43 can be expected to cause relatively minor shifts in the overall structure (15, 23). The side groups of the residues replacing G10 and G43 are oriented towards the outside of the protein and make the protein more bulky at the redox site. The arginine inserted in position 45 is more difficult to accommodate. In the initial approach mode of Swiss Model, the downstream amino acids were shifted by one position in the 3-D model, which removed the Y/F at position 50 from the hydrophobic core and replaced it with the conserved aspartate D49. As this is unlikely, the stretch between the CXXC motif and Y50 probably forms a small loop. Modeling of this loop in the optimize mode of Swiss Model resulted in two different loops for the eight AlkG1-type Rds, one involving the Cα atoms of four amino acids (positions 43 to 47) and the other involving only the two amino acids at positions 44 and 46. In this model, the Cα atoms of amino acids 43 and 47 stay at the same position, but the side chains move (data not shown). The short-loop model could be used as a template to model the other AlkG1-type Rds without affecting the hydrophobic core or the iron-binding cysteine residues (Fig. 4). As this model shows relatively minor changes in the predicted structure relative to the available 3-D structures, we consider this a plausible solution. However, 3-D structure determination will have to show whether the structure of the predicted loop comes close to the real structure.
Mutagenesis of AlkG1 and AlkG2.
To determine which features of the AlkG1 and AlkG2 consensus sequences are important for the inability of the AlkG1-type Rd domains to replace AlkG, we exchanged the amino acids at positions 10, 21, 29 to 32, and 43 to 46 in AlkG1 and AlkG2 by site-directed mutagenesis. The mutated AlkG domains were subsequently tested in the E. coli complementation system by using growth and alkane hydroxylase activity as criteria for the ability of the mutated Rd domains to function as electron transfer proteins (Fig. 5). In all cases, the alkane hydroxylase activities correspond well to the observed growth density on minimal medium plates. Both for AlkG1 and AlkG2, the results were unequivocal. In AlkG2, only the GAT43-46AVRD mutation caused significantly slower growth on n-octane, while the G10N mutation slightly affected growth. The opposite mutation AVRD43-46GAT in AlkG1 enabled this Rd to replace the intact AlkG while the other mutations (N10G, H21A, PWHL29-32RWFD, R45A, or the combination of N10G and A43G) had no effect. The AlkG1 domain was further improved by the combination of AVRD43-46GAT and N10G. The reduced electron transfer ability of AlkG1 might be due to steric hindrance at the site of electron transfer, altered midpoint redox potentials, or reduced binding of iron to the cysteines.
Steric hindrance.
Figure 4 shows that the residues replacing G10 and G43 in AlkG1 make the Rd more bulky close to the iron atom. Actually, the AlkG2 G10N mutation clearly reduces alkane hydroxylase activity. Therefore, we constructed a double mutant of AlkG1 in which both N10 and A43 are changed to glycine. However, this mutant could not replace AlkG2, like the P. rugosa Rd, which shows all sequence features of the AlkG1-type Rds except for the glycines at positions 10 and 43. Steric effects could also be due to bulky residues in the AVRD43-46 loop. In fact, the AlkG1 mutant R45A, which was constructed because R45 seems to shield the conserved residue D49 (Fig. 4), is clearly more active than the wild-type AlkG1, even if the activity is not sufficient to allow growth on n-octane.
Redox potential.
Mutations G10E, G43A, and A44V in the C. pasteurianum Rd all shift the FeIII-FeII midpoint redox potential downward by about 50 to 100 mV, resulting in increased stability of the oxidized [FeIII(S-Cys)4] center relative to the reduced form (23). However, this redox potential is still significantly higher than the midpoint potential of the flavin in Rd reductase. Moreover, the measured midpoint redox potential of reconstituted AlkG1 is similar to that of AlkG2 (20), and the reconstituted AlkG1 domain is reduced by Rd reductase as efficiently as that of AlkG2 (25), suggesting that the midpoint redox potential is not a major factor.
Iron binding.
It is possible that the AVRD43-46 loop affects metal binding. As AlkG is usually isolated from GPo1 and E. coli as the one-iron form (20), it is not clear whether AlkG binds two-iron atoms in vivo. A large proportion of AlkG, isolated from recombinant E. coli strains, binds zinc instead of iron, probably due to subtle differences in the metal housekeeping of this bacterium (level of free metal ions and redox state of the cell). Overexpression of the AlkG1 and AlkG2 domains in E. coli using the T7 promoter of pET11a resulted in dark-red cell extracts for AlkG2 but completely colorless cell extracts for AlkG1 even though the expression levels were similar (data not shown), indicating that most of the AlkG1 in E. coli does not bind iron.
To summarize, the loop introduced in the AlkG1-type domains at positions 43 to 46 prevents this type of Rd from donating electrons to the alkane hydroxylase, probably due to a combination of steric effects and a reduced affinity for iron.
The identity of residue 44 is correlated with the presence or absence of AlkG1-type Rds.
Correlation of specific positions with the presence or absence of AlkG1-type Rds in particular alkane-degrading strains shows that the three AlkG2-type Rds from organisms that only contain AlkG2-type Rds have a valine at position 44, while all AlkG2-type Rds from organisms that also contain AlkG1-type Rds have an alanine at this position. Swartz and coworkers have found that the identity of this residue correlates with the redox potential; V44 Rds have reduction potentials that are about −50 mV while A44 Rds have values around 0 mV (38). This was substantiated by Eidsness and coworkers, who have shown that replacement of A44 by V in the P. furiosus Rd reduces the redox potential by 95 mV while the reverse replacement in the C. pasteurianum Rd results in an 86-mV increase (8). The A/V44 correlation may mean that for optimal electron transfer in the absence of an AlkG1-type Rd, the redox potential of AlkG2-type Rds needs to be tuned to a value near −50 mV, the expected value for the Acinetobacter sp. ADP1 and P. aeruginosa PAO1 Rds. However, the identity of residue 44 is not critical because both A44 and V44 Rds are active in the complementation assay.
Composition and general architecture of alkane hydroxylase systems.
Comparison of the alkane hydroxylase systems in the different alkane-degrading strains shows that four arrangements can be distinguished (Fig. 1). In P. putida GPo1 and P1, and in A. borkumensis, an AlkG1-type Rd domain is fused to an AlkG2-type Rd domain, thereby fixing this ratio to 1:1 (AlkG). The first two strains also contain an additional AlkG1-type Rd named AlkF. In the case of the GPo1 alkane hydroxylase system, the ratio of the three components in the wild-type strain and various E. coli recombinants was studied in detail and found to be approximately 1:0.2.01 for alkane hydroxylase:Rd 2 (AlkG):Rd reductase (36). The gram-negative strains Acinetobacter sp. ADP1 and P. aeruginosa PAO1 only contain AlkG2-type Rds. These Rds are not encoded close to the alkane hydroxylase gene on the chromosome. Here, the ratio between the different components is not known, but in Acinetobacter sp. ADP1, the Rd is constitutively expressed while the alkane hydroxylase is inducible (10). In the Mycobacterium and Rhodococcus strains, the start codons of several alkB and rub genes overlap with the stop codons of the preceding gene (alkB1-rubA1, rubA2-rubB, and alkB2-rubA3-rubA4 in R. erythropolis and alkB-rubA-rubB in M. tuberculosis). This phenomenon, which is known as translational coupling, is quite common to Rhodococcus genes involved in biodegradation (18) and probably fixes the ratio of the involved proteins to 1:1. Finally, in P. rugosa, an AlkG1-type Rd is fused to the alkane hydroxylase open reading frame, fixing this ratio at 1:1. In view of the results of the complementation assays and sequence analysis, it is likely that this strain also contains an AlkG2-type Rd, which must be encoded at some distance from the alkane hydroxylase gene.
Conclusions.
Cloning, sequencing, and functional analysis show that Rds involved in alkane degradation can be exchanged between alkane hydroxylase systems present in gram-positive and gram-negative strains because certain structural features that are necessary for electron transfer are highly conserved. Rds found in alkane-degrading bacteria can be divided into two groups. AlkG2-type Rds are able to restore growth on n-octane of a recombinant E. coli lacking an active Rd while AlkG1-type Rds are not functional in this assay. Sequence analysis shows that specific positions, such as the negative charges at positions 15, 22, 41, and 49 are common to both AlkG1- and AlkG2-type Rds while other sequence features are specific for either AlkG1- or AlkG2-type Rds. One of the latter features, the insertion of an arginine (R45), is responsible for the lack of activity of the AlkG1-type Rds in the complementation assay.
It is apparent from the sequence distances and the tree of identity that the AlkG1-type Rds are conserved across phylogenetic boundaries. The degree of conservation, and the fact that the GPo1 AlkG1 domain can be converted to an active Rd by changing the sequence downstream of the second iron-binding cysteine motif (CXXC), strongly suggests that these Rds are not just the product of gene duplications but play an as yet undefined role in alkane hydroxylation. Future studies will focus on the role of AlkG1-type Rds in alkane oxidation and the molecular basis of electron transfer between Rd reductases, AlkG1 and AlkG2 Rds, and the alkane hydroxylases.
Acknowledgments
This research was supported by the Swiss National Science Foundation through the Swiss Priority Program in Biotechnology, grant no. 5002-037023.
We thank Alessandro Franchini for assistance with the cloning of the A. borkumensis AP1 Rd gene and Martina Röthlisberger for DNA sequencing.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, E., and J. Brosius. 1985. “ATG vectors” for regulated high-level expression of cloned genes in Escherichia coli. Gene 40:183-190. [DOI] [PubMed] [Google Scholar]
- 3.Benson, A., K. Tomoda, J. Chang, G. Matsueda, E. T. Lode, M. J. Coon, and K. T. Yasunobu. 1971. Evolutionary and phylogenetic relationships of rubredoxin-containing microbes. Biochem. Biophys. Res. Commun. 42:640-646. [DOI] [PubMed] [Google Scholar]
- 4.Benson, S., M. Fennewald, J. Shapiro, and C. Huettner. 1977. Fractionation of inducible alkane hydroxylase activity in Pseudomonas putida and characterization of hydroxylase-negative plasmid mutations. J. Bacteriol. 132:614-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLeah, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Soeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrett. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 6.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eggink, G., R. G. Lageveen, B. Altenburg, and B. Witholt. 1987. Controlled and functional expression of Pseudomonas oleovorans alkane utilizing system in Pseudomonas putida and Escherichia coli. J. Biol. Chem. 262:17712-17718. [PubMed] [Google Scholar]
- 8.Eidsness, M. K., A. E. Burden, K. A. Richie, D. M. Kurtz, R. A. Scott, E. T. Smith, T. Ichiye, B. Beard, T. Min, and C. Kang. 1999. Modulation of the redox potential of the [Fe(SCys)4] site in rubredoxin by the orientation of a peptide dipole. Biochemistry 38:14803-14809. [DOI] [PubMed] [Google Scholar]
- 9.Fox, B. G., J. Shanklin, A. Jingyuan, T. M. Loehr, and J. Sanders-Loehr. 1994. Resonance Raman evidence for an Fe-O-Fe center in stearoyl-ACP desaturase. Primary sequence identity with other diiron-oxo proteins. Biochemistry 33:12776-12786. [DOI] [PubMed] [Google Scholar]
- 10.Geiβdörfer, W., S. C. Frosch, G. Haspel, S. Ehrt, and W. Hillen. 1995. Two genes encoding proteins with similarities to rubredoxin and rubredoxin reductase are required for conversion of dodecane to lauric acid in Acinetobacter calcoaceticus ADP1. Microbiology 141(Part 6):1425-1432. [DOI] [PubMed] [Google Scholar]
- 11.Hamamura, N., C. M. Yeager, and D. J. Arp. 2001. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl. Environ. Microbiol. 67:4992-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou, C. T., M. A. Jackson, M. O. Bagby, and L. A. Becker. 1994. Microbial oxidation of cumene by octane-grown cells. Appl. Microbiol. Biotechnol. 41:178-182. [Google Scholar]
- 13.Juni, E., and A. Janik. 1969. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J. Bacteriol. 98:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kok, M., R. Oldenhuis, M. P. G. van der Linden, C. H. C. Meulenberg, J. Kingma, and B. Witholt. 1989. The Pseudomonas oleovorans alkBAC operon encodes two structurally related rubredoxins and an aldehyde dehydrogenase. J. Biol. Chem. 264:5442-5451. [PubMed] [Google Scholar]
- 15.Kümmerle, R., H. Zhuang-Jackson, J. Gaillard, and J.-M. Moulis. 1997. Site-directed mutagenesis of rubredoxin reveals the molecular basis of its electron transfer properties. Biochemistry 36:15983-15991. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 17.Lageveen, R. G., G. W. Huisman, H. Preusting, P. Ketelaar, G. Eggink, and B. Witholt. 1988. Formation of polyesters by Pseudomonas oleovorans: the effect of substrates on the formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl. Environ. Microbiol. 54:2924-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin, M. J., R. De Mot, L. A. Kulakov, and I. Nagy. 1998. Applied aspects of Rhodococcus genetics. Antonie Leeuwenhoek 74:133-153. [DOI] [PubMed] [Google Scholar]
- 19.Lee, H. J., J. Basran, and N. S. Scrutton. 1998. Electron transfer from flavin to iron in the Pseudomonas oleovorans rubredoxin reductase-rubredoxin electron transfer complex. Biochemistry 37:15513-15522. [DOI] [PubMed] [Google Scholar]
- 20.Lee, H. J., L.-Y. Lian, and N. Scrutton. 1997. Recombinant two-iron rubredoxin of Pseudomonas oleovorans: overexpression, purification and characterization by optical, CD and 113Cd NMR spectroscopies. Biochem. J. 328:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, H. J., L. Y. Lian, and N. S. Scrutton. 1996. Rubredoxin/rubredoxin reductase of Pseudomonas oleovorans: a model system for investigating interprotein in electron transfer. Biochem. Soc. Trans. 24:447S. [DOI] [PubMed]
- 22.Lode, E. T., and M. J. Coon. 1971. Enzymatic ω-oxidation. V. Forms of Pseudomonas oleovorans rubredoxin containing one or two iron atoms: structure and function in ω-hydroxylation. J. Biol. Chem. 246:791-802. [PubMed] [Google Scholar]
- 23.Maher, M. J., Z. Xiao, M. C. J. Wilce, J. M. Guss, and A. G. Wedd. 1999. Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert. Acta Crystallogr. Sect. D 55:962-968. [DOI] [PubMed] [Google Scholar]
- 24.McKenna, E. J., and M. J. Coon. 1970. Enzymatic ω-oxidation. IV. Purification and properties of the ω-hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 245:3882-3889. [PubMed] [Google Scholar]
- 25.Perry, A., L.-Y. Lian, and N. S. Scrutton. 2001. Two-iron rubredoxin of Pseudomonas oleovorans: production, stability and characterization of the individual iron-binding domains by optical, CD and NMR spectroscopies. Biochem. J. 354:89-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson, J. A., D. Basu, and M. J. Coon. 1966. Enzymatic ω-oxidation. I. Electron carriers in fatty acid and hydrocarbon hydroxylation. J. Biol. Chem. 241:5162-5164. [PubMed] [Google Scholar]
- 27.Peterson, J. A., and M. J. Coon. 1968. Enzymatic ω-oxidation. III. Purification and properties of rubredoxin, a component of the ω-hydroxylation system of Pseudomonas oleovorans. J. Biol. Chem. 243:329-334. [PubMed] [Google Scholar]
- 28.Peterson, J. A., M. Kusunose, E. Kusunose, and M. J. Coon. 1967. Enzymatic ω-oxidation. II. Function of rubredoxin as the electron-carrier in ω-hydroxylation. J. Biol. Chem. 242:4334-4340. [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schwartz, R. D., and C. J. McCoy. 1973. Pseudomonas oleovorans hydroxylation-epoxidation system: additional strain improvements. Appl. Microbiol. 26:217-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwede, T., A. Diemand, N. Guex, and M. C. Peitsch. 2000. Protein structure computing in the genomic era. Res. Microbiol. 151:107-112. [DOI] [PubMed] [Google Scholar]
- 32.Shanklin, J., C. Achim, H. Schmidt, B. G. Fox, and E. Münck. 1997. Mössbauer studies of alkane ω-hydroxylase: evidence for a diiron cluster in an integral-membrane protein. Proc. Natl. Acad. Sci. USA 94:2981-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smits, T. H. M., S. B. Balada, B. Witholt, and J. B. van Beilen. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184:1733-1742. [DOI] [PMC free article] [PubMed]
- 34.Smits, T. H. M., M. Röthlisberger, B. Witholt, and J. B. van Beilen. 1999. Molecular screening for alkane hydroxylase genes in gram-negative and gram-positive strains. Environ. Microbiol. 1:307-318. [DOI] [PubMed] [Google Scholar]
- 35.Smits, T. H. M., M. A. Seeger, B. Witholt, and J. B. van Beilen. 2001. New alkane-responsive expression vectors for E. coli and Pseudomonas. Plasmid 46:16-24. [DOI] [PubMed] [Google Scholar]
- 36.Staijen, I. E., J. B. van Beilen, and B. Witholt. 2000. Expression, stability and performance of the three-component alkane monooxygenase of Pseudomonas oleovorans in Escherichia coli. Eur. J. Biochem. 267:1957-1965. [DOI] [PubMed] [Google Scholar]
- 37.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 38.Swartz, P. D., B. W. Beck, and T. Ichiye. 1996. Structural origins of redox potentials in Fe-S proteins: electrostatic potentials of crystal structures. Biophys. J. 71:2958-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueda, T., E. T. Lode, and M. J. Coon. 1972. Enzymatic ω-oxidation. VI. Isolation of homogeneous reduced diphosphopyridine nucleotide-rubredoxin reductase. J. Biol. Chem. 247:2109-2116. [PubMed] [Google Scholar]
- 40.van Beilen, J. B. 1994. Alkane oxidation by Pseudomonas oleovorans: genes and proteins. Ph.D. thesis. University of Groningen, Groningen, The Netherlands.
- 41.van Beilen, J. B., S. Panke, S. Lucchini, A. G. Franchini, M. Röthlisberger, and B. Witholt. 2001. Analysis of Pseudomonas putida alkane degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk-genes. Microbiology 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 42.van Beilen, J. B., D. Penninga, and B. Witholt. 1992. Topology of the membrane-bound alkane hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 267:9194-9201. [PubMed] [Google Scholar]
- 43.van Beilen, J. B., M. G. Wubbolts, and B. Witholt. 1994. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5:161-174. [DOI] [PubMed] [Google Scholar]
- 44.Witholt, B., M. J. de Smet, J. Kingma, J. B. van Beilen, M. Kok, R. G. Lageveen, and G. Eggink. 1990. Bioconversions of aliphatic compounds by Pseudomonas oleovorans in multiphase bioreactors: background and economic potential. Trends Biotechnol. 8:46-52. [DOI] [PubMed] [Google Scholar]
- 45.Yakimov, M. M., P. N. Golyshin, S. Lang, E. R. B. Moore, W.-R. Abraham, H. Lünsdorf, and K. N. Timmis. 1998. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 48:339-348. [DOI] [PubMed] [Google Scholar]
- 46.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]