Abstract
Loss of tonB1 adversely affects iron acquisition and intrinsic multidrug resistance in Pseudomonas aeruginosa. Several mutations in tonB1 compromised the protein's contribution to both processes, although TonB1 derivatives altered in residues C35, Q268, R287, Q292, R300, and R304 were compromised vis-à-vis their contribution to drug resistance only.
Energy-dependent, receptor-mediated ligand uptake across the outer membranes of gram-negative bacteria is dependent upon the function of the product of the tonB gene (22). In Escherichia coli, this proline-rich protein is anchored to the cytoplasmic membrane via its N terminus and extends into the periplasm (6, 23, 24), where it interacts with receptor proteins in the outer membrane (2, 5, 7, 25, 26). This disposition of the protein reflects its apparent role in coupling the energized state of the cytoplasmic membrane to the outer membrane receptors (17). Such interaction appears to be mediated by the C-terminal region of the TonB protein (10, 12) at or near residue Q160 (2, 7). Indeed, a recent cross-linking study confirmed that the interaction of TonB and the TonB box region of the TonB-dependent vitamin B12 receptor, BtuB, is mediated by residues in TonB near Q160 (3).
Two tonB genes in Pseudomonas aeruginosa, dubbed tonB1 (21) and tonB2 (30), have been described. Although TonB1 displays significant homology, e.g., to E. coli TonB (TonBEc), this protein is distinguished by the presence of additional sequences at its N terminus, making it larger than all other examples of TonB (21). Still, disruption of TonB1 (but not TonB2) abolishes siderophore-mediated iron uptake (21) and heme uptake (30), consistent with the idea that TonB1 is involved in iron acquisition. Interestingly, tonB1 (but not tonB2 [Q. Zhao, unpublished data]) mutants of P. aeruginosa are also drug hypersusceptible, apparently owing to a requirement for TonB1 for the operation of multidrug efflux systems in this organism (29). Indeed, the operation of the MexAB-OprM multidrug efflux system, responsible for intrinsic (11, 14, 20, 27) and acquired (9, 15, 16, 20, 32) resistance to several antimicrobials, appears in particular to be compromised in tonB1 mutants (29). In this report, we assess the functional importance of defined regions of the sequence of TonB1 for intrinsic multidrug resistance in P. aeruginosa and note the differential effects of several mutations on resistance versus iron acquisition.
Methods.
P. aeruginosa PAO6609 (met9011 amiE200 rpsL pvd9) (8) is the parent strain of the ΔtonB1 mutant strain K1040 (29). The chloramphenicol-resistant broad-host-range plasmid pMMB206 (18) was used to clone tonB1 (29) and its mutated and chimeric variants (31). Construction of chimeras and point mutations involved the use of PCR, and all mutations and constructions were confirmed by nucleotide sequencing (31). The plasmid-borne tonB1 constructs were introduced into P. aeruginosa K1040 via a triparental mating procedure (29). Expression of the various mutant and chimeric TonB1 proteins in P. aeruginosa K1040 was confirmed by using Western immunoblotting as described previously, following induction of the cloned genes with IPTG (isopropyl-β-d-thiogalactopyranoside; 20 μM) (31). TonB1 proteins were detected by use of a chicken polyclonal anti-MalE-TonB1 fusion antibody (31) or monoclonal antibody 1C3 to TonBEc (11a). Susceptibility testing was carried out with L broth with and without IPTG (20 μM) by the twofold serial broth dilution method (13). To alleviate the possible influence of iron limitation on the MIC results (tonB1 mutants are iron limited in L broth [30]), FeCl3 (200 μM) was included in the growth medium in some experiments (29).
As a measure of mutant-TonB1 function in iron acquisition, the growth of tonB1 plasmid-carrying P. aeruginosa K1040 under iron-restricted conditions upon pyoverdine supplementation was assessed. The K1040 strain lacks tonB1 and is unable to synthesize pyoverdine. In the presence of a cloned, functional tonB1 gene, however, addition of exogenous pyoverdine can promote growth under conditions of iron restriction. Thus, P. aeruginosa K1040 carrying one of several mutant tonB1 plasmids was cultured overnight in succinate minimal medium (19) supplemented with methionine (100 μg/ml), FeCl3 (200 μM), and chloramphenicol (16 μg/ml) and used to inoculate an iron-deficient succinate minimal plate containing 180 μg of ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDHA) per ml and 20 μM IPTG (to induce the tonB1 genes cloned into pMMB206) as described previously (31). Filter disks impregnated with 8 μl of pyoverdine (50 mg/ml of stock) were then placed on the plates. Following incubation at 37°C for ca. 40 h, plates were examined for evidence of growth in the region surrounding the filter disks and the diameter of any zone of growth was measured. Strain K1040 expresses a chromosomal copy of the tonB2 gene but is not able to grow on the aforementioned EDDHA- and pyoverdine-supplemented medium. Thus, any growth afforded by the cloned tonB1 genes is a measure of TonB1 function only.
Influence of mutations at the N terminus on TonB1 function.
Recently, several mutations were constructed in tonB1 in an attempt to assess the functional importance of, e.g., the novel N-terminal extension as well as of the conserved C-terminal region of the TonB1 protein (31). To assess the impact of these mutations on intrinsic resistance compared to their effects on iron acquisition, the antibiotic susceptibility and siderophore-dependent growth capability (under iron-restricted conditions) of P. aeruginosa K1040 expressing the mutant TonB1 proteins were determined. Introduction of the wild-type tonB1 gene (on pQZ-6) into P. aeruginosa K1040 fully restored the antibiotic resistance of this mutant strain to the level of the PAO6609 parent strain (Table 1). Similarly, while the mutant is unable to grow on EDDHA-supplemented minimal medium upon pyoverdine supplementation, the plasmid with wild-type tonB1 permitted pyoverdine-dependent growth of K1040 (Table 1). Since this strain is unable to make its own pyoverdine and lacks tonB1, growth under iron-restricted (i.e., EDDHA-supplemented) conditions requires exogenously added pyoverdine as well as a functional TonB1 protein (pyoverdine-mediated iron acquisition is TonB1 dependent [21]). Thus, any growth of tonB1 plasmid-containing K1040 on EDDHA- and pyoverdine-supplemented minimal medium is an indication of a TonB1 contribution to pyoverdine-mediated iron uptake.
TABLE 1.
Influence of tonB1 mutations on the antibiotic susceptibility and pyoverdine-stimulated growth of P. aeruginosaa
| Plasmid | TonB or TonB chimerab | MIC (μg/ml) ofc:
|
Pyoverdine-stimulated growth (%)d | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAR | CFP | CPM | CIP | NOR | NOV | ERY | TET | IPM | |||
| None | Wild typee | 64 | 4 | 2 | 0.2 | 1 | 256 | 256 | 8 | 0.5 | NDf |
| pMMB206 | None | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 0 |
| pQZ-06 | Wild type | 64 | 4 | 2 | 0.2 | 1 | 256 | 256 | 8 | 0.5 | 100 |
| pQZ-N2 | ΔS6-P83 | 2 | 0.25 | 0.06 | 0.02 | 0.12 | 8 | 8 | 2 | 0.5 | 0 |
| pQZ-CY | C36G | 2 | 0.25 | 0.06 | 0.25 | 0.12 | 4 | 8 | 2 | 0.5 | 89 |
| pQZ-T | ΔP82-T111 | 2 | 2 | 0.25 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 0 |
| pQZ-H | H98G | 32 | 4 | 1 | 0.2 | 1 | 256 | 256 | 8 | 0.5 | 96 |
| pQZ-C | ΔD260-A310 | 2 | 0.12 | 0.06 | 0.12 | 0.12 | 4 | 8 | 2 | 0.5 | 0 |
| pQZ-Y1 | Y264A | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 0 |
| pQZ-Y2 | Y264F | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 0 |
| pQZ-P1 | P265A | 32 | 4 | 1 | 0.2 | 1 | 256 | 256 | 8 | 0.5 | 100 |
| pQZ-P2 | P265G | 32 | 4 | 1 | 0.2 | 1 | 256 | 256 | 8 | 0.5 | 96 |
| pQZ-Q1 | Q269A | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 93 |
| pQZ-E1 | E274A | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 0 |
| pQZ-E3 | E274A/D304N | 4 | 0.25 | 0.12 | 0.02 | 0.12 | 8 | 16 | 2 | 0.5 | 93 |
| pQZ-E2 | E274K | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 0 |
| pQZ-K1 | K278A | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 37 |
| pQZ-K2 | K278E | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 41 |
| pQZ-R1 | R288A | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 81 |
| pQZ-R2 | R288E | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 85 |
| pQZ-Q2 | Q293A | 2 | 0.12 | 0.06 | 0.02 | 0.12 | 4 | 8 | 2 | 0.5 | 96 |
| pQZ-V1 | V294A | 8 | 0.5 | 0.25 | 0.1 | 0.25 | 64 | 64 | 4 | 0.5 | 100 |
| pQZ-V2 | V294T | 32 | 4 | 1 | 0.1 | 0.5 | 256 | 256 | 8 | 0.5 | 100 |
| pQZ-R3 | R301A | 2 | 0.12 | 0.06 | 0.02 | 0.02 | 4 | 8 | 2 | 0.5 | 93 |
| pQZ-R4 | R301E | 2 | 0.12 | 0.06 | 0.02 | 0.02 | 4 | 8 | 2 | 0.5 | 97 |
| pQZ-D1 | D304A | 2 | 0.12 | 0.06 | 0.02 | 0.02 | 4 | 8 | 2 | 0.5 | 0 |
| pQZ-D2 | D304K | 2 | 0.12 | 0.06 | 0.02 | 0.02 | 4 | 8 | 2 | 0.5 | 0 |
| pQZ-R5 | R305A | 2 | 0.12 | 0.06 | 0.02 | 0.02 | 4 | 8 | 2 | 0.5 | 93 |
| pQZ-R6 | R305E | 2 | 0.12 | 0.06 | 0.02 | 0.02 | 4 | 8 | 2 | 0.5 | 100 |
| pQZ-CEc1 | TonB1 (1-81) + TonBEc (1-239) | 2 | 0.12 | 0.06 | 0.02 | 0.02 | 4 | 8 | 2 | 0.5 | 37g |
| pQZ-CPa1 | TonBEc (1-152) + TonB1 (254-342) | 4 | 0.25 | 0.12 | 0.05 | 0.02 | 8 | 32 | 4 | 0.5 | 96 |
| pQZ-NPa1 | TonB1 (1-254) + TonBEc (154-239) | 2 | 0.12 | 0.06 | 0.02 | 0.02 | 4 | 8 | 2 | 0.5 | 0 |
The antibiotic susceptibility of P. aeruginosa K1040 (ΔtonB1) expressing the indicated plasmid-encoded mutant TonB1 proteins was assessed as described in the text. Identical results were obtained when assays were carried out in FeCl3-supplemented growth medium.
The mutations present in the TonB1 proteins encoded by the indicated plasmids are listed. The amino acid numbers in parentheses refer to the positions in the protein inclusive of the initiation Met. Results for only three of the TonB1-TonBEc chimeras are shown. The remaining chimeras yielded results indistinguishable from those with TonB of K1040 harboring pMMB206.
CAR, carbenicillin; CFP, cefoperazone; CPM, cefpirome; CIP, ciprofloxacin; NOR, norfloxacin; NOV, novobiocin; ERY, erythromycin; TET, tetracycline; IPM, imipenem. Comparable results were obtained with or without induction of the cloned tonB1 genes with IPTG.
P. aeruginosa K1040 harboring the indicated plasmids was streaked onto an iron-deficient succinate minimal plate supplemented with EDDHA, and a pyoverdine-impregnated disk was placed on the surface of the plate. The diameter (in millimeters) of any resultant zone of growth was measured and reported relative to (i.e., as a percentage of) the diameter of the zone observed for pQZ-06-carrying K1040. Results are representative of three independent experiments.
Data for the plasmid-free TonB+ parent strain PAO6609 are shown for comparison purposes. The same results were obtained for K201 carrying pMMB206.
ND, not determined.
The observed growth was very weak.
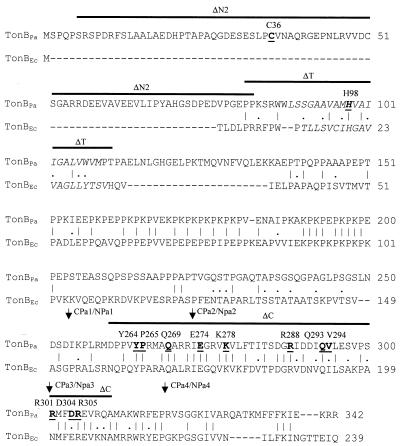
Several mutations in tonB1 compromised the TonB1 contribution to both intrinsic antimicrobial resistance and pyoverdine-mediated iron acquisition, including one carried by plasmid pQZ-N2 that resulted in a product with a deletion of 78 amino acids (from S6 to P83) (see ΔN2, Fig. 1) from the novel N-terminal extension. Deletion of the predicted transmembrane domain (P82 to T111) (pQZ-T; see ΔT, Fig. 1) or a highly conserved C-terminal region (D260 to A310) (plasmid pQZ-C; see ΔC, Fig. 1) also failed to promote either intrinsic antibiotic resistance or pyoverdine-mediated iron acquisition (Table 1). The lack of activity of the TonB1 derivative lacking the transmembrane domain was expected, given the importance of this region in TonBEc for proper membrane localization. Still, a conserved His residue (H98 in TonB1) present within the transmembrane domain of several TonB proteins (H20 in TonBEc) (28) and required for TonB function was dispensable for function in P. aeruginosa (see the H98G mutation [pQZ-H], Table 1), although mutation of H98 did abolish TonB1 function in E. coli (31). The C terminus of TonBEc is apparently involved in receptor interaction (3, 10, 12) (and possibly TonBEc dimerization [4]), and the C terminus of TonB1 in P. aeruginosa likely plays a similar role. Whether its contribution to intrinsic resistance is related to interaction with, e.g., efflux components remains to be examined.
FIG. 1.
Alignment of P. aeruginosa TonB1 (TonBPa) and TonBEc, and identification of mutations constructed in TonB1. Identical (|) and conserved (.) residues are indicated. The putative transmembrane domain is in italics. Deletions are indicated by a solid bar above the TonB1 sequence. Point mutations are underlined, in bold text, and identified above the TonB1 sequence. The crossover points of TonB1-TonBEc (NPa series) and TonBEc-TonB1 (CPa series) chimeras are indicated by arrows. Numbers at the right represent the position of the right-most amino acid within the sequence of the respective TonB proteins. Alignment was carried out using the PALIGN program of the PCGene software package (Intelligenetics, Inc.).
Influence of mutations at the C terminus on TonB1 function.
The highest degree of similarity between TonB1 and other TonB proteins, including that of E. coli, occurs within the C termini of these proteins. To assess the functional importance of a number of residues found within the TonB1 C terminus, including residues conserved in other TonB proteins (e.g., TonBEc), several of these were mutated (Fig. 1), with the resulting TonB1 proteins being expressed in P. aeruginosa K1040 (Fig. 2B), and the effects on the pyoverdine-dependent growth (i.e., iron acquisition) and antibiotic resistance of this strain were determined. Of 11 residues mutated, 6, including Q269 (pQZ-Q1), R288 (pQZ-R1 and pQZ-R2), Q293 (pQZ-Q2), V294 (pQZ-V1 and pQZ-V2), and R305 (pQZ-R5 and pQZ-R6), were dispensable for TonB1 function as regards pyoverdine-mediated iron acquisition (Table 1). Substitutions at Y264 (pQZ-Y1 and pQZ-Y2), E274 (pQZ-E1 and pQZ-E2), and D304 (pQZ-D1 and pQZ-D2), on the other hand, inactivated TonB1, and K1040 expressing these TonB1 derivatives failed to grow on a pyoverdine-supplemented iron-restricted medium (these were also compromised vis-à-vis antibiotic resistance) (Table 1). Mutations at K278 (pQZ-K1 and pQZ-K2) reduced but did not eliminate the TonB1 contribution to iron acquisition (Table 1). Intriguingly, many of the tonB1 mutations that had no or only a modest impact on the TonB1 contribution to the pyoverdine-promoted growth of K1040 completely eliminated its contribution to the antibiotic resistance of this strain (Table 1). Indeed, only substitutions P265 and V294 failed to abolish a TonB1 contribution to antibiotic resistance. Thus, substitutions at Q269, R288, Q293, R301, and R305 preferentially compromised the TonB1 contribution to intrinsic antibiotic resistance. A D304N intragenic suppressor of the original E274A mutation (31), which restored the growth of P. aeruginosa K1040 in a pyoverdine-supplemented iron-restricted medium (Table 1), had a very modest impact on resistance, providing a slight increase in resistance to only some of the antibiotics tested (Table 1). The specificity of the intragenic suppression with respect to iron acquisition clearly indicates that TonB1 operates differently in these two processes.
FIG. 2.
Expression of plasmid-encoded TonB1 in the tonB1 deletion strain P. aeruginosa K1040. Cell extracts of K1040 harboring various mutant tonB1 genes (A and B) and tonB1-tonBEc chimeras (C) were immunoblotted and probed with antibodies to TonB1 (with the exception of the CPa and CEc series of chimeras, which were probed with monoclonal anti-TonBEc antibody 1C3). The nature of the mutation in the sequence of TonB1 is indicated above each lane in panels A and B, with the exceptions of lanes ΔC (deletion of D260 to A310), ΔN2 (deletion of S6 to P83), and ΔT (deletion of P82 to T111). Lanes: WT, wild-type TonB1; pMMB, pMMB206, vector control without a cloned tonB1 gene. Variations seen in the levels of the various mutant TonB1 proteins in panel B were within the natural variation seen for any given mutant TonB1 as observed over three separate experiments. Chimeras shown in panel C are described in the legend to Fig. 1. E.c.tonB, tonBEc.
Activities of TonB-TonBEc chimeras.
The earlier observation that TonB1 could complement an E. coli tonBEc mutation (21) but that TonBEc could not return the favor in P. aeruginosa (Q. Zhao, data not shown) indicated that sequences unique to TonB1 were crucial for TonB function in P. aeruginosa. Initially, it was surmised that the novel N-terminal TonB1 extension that is lacking in TonBEc might be important for this. Still, the addition of the TonB1 extension to the N terminus of TonBEc (see pQZ-CEc1, Table 1) failed to promote significant pyoverdine-dependent growth or antibiotic resistance (Table 1). It was reasoned, then, that sequences present within the TonB1 C terminus but lacking in TonBEc might be involved. To test this, a number of TonB1-TonBEc chimeras were constructed by swapping various portions of the C termini of these proteins (31) and the impact on the iron acquisition (i.e., pyoverdine-dependent growth under conditions of iron restriction) and intrinsic antibiotic resistance of this strain was assessed. Of four TonB derivatives carrying the N terminus of TonBEc and the C terminus of TonB1 (CPa-1 through CPa-4, Fig. 1), only the chimera encoded by pQZ-CPa1 in which the N-terminal 152 amino acid residues of TonBEc were fused to the C-terminal 79 amino acids of TonB1 (Fig. 1) promoted both the pyoverdine-dependent growth of K1040 under iron-restricted conditions and antibiotic resistance (Table 1), although the impact on resistance was very modest. Chimeras carrying N-terminal sequences of TonB1 and C-terminal sequences of TonBEc (NPa-1 through NPa-4, Fig. 1) were all inactive as regards antibiotic resistance and pyoverdine-mediated iron acquisition (Table 1). Thus, the C terminus indeed carries a sequence important for the operation of TonB1 in P. aeruginosa.
Expression of TonB1 derivatives.
A perhaps surprising finding from this study was the differential effects of tonB1 mutations on the TonB1 contributions to intrinsic resistance and iron acquisition. Most mutations did in fact adversely affect the TonB1 contribution to intrinsic resistance without impacting its contribution to iron acquisition. To rule out lack of mutant TonB1 production as an explanation for these defects in activity, extracts of TonB1-expressing K1040 strains were probed with a TonB1-specific antiserum. As seen in Fig. 2, all TonB1 derivatives, with the exception of the pQZ-N2-encoded N-terminal deletion derivative ΔN2 (Fig. 1), were expressed during growth under conditions used for the MIC assay. No substantial differences were observed in the levels of the various mutant TonB1 proteins and wild-type TonB1, indicating that loss of activity was not attributable to decreased production of the mutant TonB1 constructs. Significantly, detection of the TonB1 derivatives required induction of the plasmid-borne genes with IPTG, although the drug susceptibility and pyoverdine-stimulated growth data were unchanged with or without IPTG (Table 1). Thus, enhanced production of the TonB1 derivatives does not enhance activity or compensate for loss of function, so differences in TonB1 activity vis-à-vis iron acquisition and intrinsic antimicrobial resistance cannot be attributed to, e.g., better expression of TonB1 in the iron-limited minimal medium used in assessing iron acquisition.
The lack of expression of a TonB1 with an N-terminal deletion may reflect the importance of this region for protein stability, which is interesting given that this N-terminal extension is unique to TonB1 among TonB proteins. Within the N-terminal region of TonB1 are two cysteine residues, C36 and C51 (Fig. 1), of which one, C36, was successfully mutated (C36G, pQZ-CY). Such a mutation specifically compromised the TonB1 contribution to intrinsic resistance inasmuch as pQZ-CY-harboring K1040 exhibited resistance levels reminiscent of those in the plasmid-free ΔtonB1 mutant (Table 1), whereas strain K1040 carrying this vector was capable of near wild-type levels of pyoverdine-dependent growth under iron-restricted conditions (Table 1). The observation that deletion of a substantial portion of the C-terminal region of TonB1 did not destabilize TonB1 in P. aeruginosa (Fig. 2A) was interesting, given that a similar C-terminal deletion destabilizes TonBEc in E. coli (1).
Conclusions.
In examining the impact of tonB1 mutations on intrinsic multidrug resistance, it was interesting that susceptibility to agents known not to be substrates for multidrug resistance efflux systems in P. aeruginosa (e.g., imipenem) was not affected by such mutations. This finding further supports our earlier suggestion that the enhanced multidrug susceptibility of tonB1 mutants results from attendant defects in efflux and not in the outer membrane barrier, since defects in the latter would be expected to influence imipenem susceptibility. Most importantly, these results also distinguish the effects of tonB1 mutations on iron acquisition from their effects on multidrug resistance and in so doing provide evidence for a direct contribution of this protein to intrinsic resistance.
Acknowledgments
We thank Kathleen Postle for her kind gift of monoclonal antibody 1C3.
This work was supported by an operating grant from the Canadian Institutes for Health Research (formerly the Medical Research Council of Canada) and the Canadian Cystic Fibrosis Foundation (CCFF). Q.Z. is the recipient of a CCFF studentship. K.P. is a CCFF Scholar.
REFERENCES
- 1.Anton, M., and K. J. Heller. 1991. Functional analysis of a C-terminally altered TonB protein of Escherichia coli. Gene 105:23-29. [DOI] [PubMed] [Google Scholar]
- 2.Bell, P. E., C. D. Nau, J. T. Brown, J. Konisky, and R. J. Kadner. 1990. Genetic suppression demonstrates interaction of TonB protein with outer membrane transport proteins in Escherichia coli. J. Bacteriol. 172:3826-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 96:10673-10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, C., A. Mooser, A. Pluckthun, and A. Wlodawer. 2001. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 276:27535-27540. [DOI] [PubMed] [Google Scholar]
- 5.Günter, K., and V. Braun. 1990. In vivo evidence for FhuA outer membrane receptor interaction with the TonB inner membrane protein of Escherichia coli. FEBS Lett. 274:85-88. [DOI] [PubMed] [Google Scholar]
- 6.Hannavy, K., G. C. Barr, C. J. Dorman, J. Adamson, L. R. Mazengera, M. P. Gallagher, J. S. Evans, B. A. Levine, I. P. Trayer, and C. F. Higgins. 1990. TonB protein of Salmonella typhimurium: a model for signal transduction between membranes. J. Mol. Biol. 216:897-910. [DOI] [PubMed] [Google Scholar]
- 7.Heller, K. J., R. J. Kadner, and K. Günther. 1988. Suppression of the btuB451 mutation by mutations in the tonB gene suggests a direct interaction between TonB and TonB-dependent receptor proteins in the outer membrane of Escherichia coli. Gene 64:147-153. [DOI] [PubMed] [Google Scholar]
- 8.Hohnadel, D., D. Haas, and J.-M. Meyer. 1986. Mapping of mutations affecting pyoverdine production in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 36:195-199. [Google Scholar]
- 9.Jalal, S., O. Ciofu, N. Høiby, N. Gotoh, and B. Wretlind. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 44:710-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaskula, J. C., T. E. Letain, S. K. Roof, J. T. Skare, and K. Postle. 1994. Role of the TonB amino terminus in energy transduction between membranes. J. Bacteriol. 176:2326-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köhler, T., M. Kok, M. Michea-Hamzehpour, P. Plesiat, N. Gotoh, T. Nishino, L. K. Curty, and J.-C. Pechere. 1996. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:2288-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Larsen, R. A., P. S. Myers, J. T. Skare, C. L. Seachord, R. P. Darveau, and K. Postle. 1996. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 178:1363-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen, R. A., D. Foster-Hartnett, M. A. McIntosh, and K. Postle. 1997. Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J. Bacteriol. 179:3213-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, X.-Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, X.-Z., L. Zhang, R. Srikumar, and K. Poole. 1998. β-Lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda, N., and S. Ohya. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuda, N., E. Sakagawa, and S. Ohya. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 18.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 19.Poole, K., S. Neshat, and D. Heinrichs. 1991. Pyoverdine-mediated iron transport in Pseudomonas aeruginosa: involvement of a high-molecular-mass outer membrane protein. FEMS Microbiol. Lett. 78:1-5. [PubMed] [Google Scholar]
- 20.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. E. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole, K., Q. Zhao, S. Neshat, D. E. Heinrichs, and C. R. Dean. 1996. The tonB gene of Pseudomonas aeruginosa encodes a novel TonB protein. Microbiology 142:1449-1458. [DOI] [PubMed] [Google Scholar]
- 22.Postle, K. 1990. TonB and the gram-negative dilemma. Mol. Microbiol. 4:2019-2025. [DOI] [PubMed] [Google Scholar]
- 23.Postle, K., and J. T. Skare. 1988. Escherichia coli TonB protein is exported from the cytoplasm without proteolytic cleavage of its amino terminus. J. Biol. Chem. 263:11000-11007. [PubMed] [Google Scholar]
- 24.Roof, S. K., J. D. Allard, K. P. Bertrand, and K. Postle. 1991. Analysis of Escherichia coli TonB membrane topology by use of PhoA fusions. J. Bacteriol. 173:5554-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schöffler, H., and V. Braun. 1989. Transport across the outer membrane of Escherichia coli K12 via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol. Gen. Genet. 217:378-383. [DOI] [PubMed] [Google Scholar]
- 26.Skare, J. T., B. M. M. Ahmer, C. L. Seachord, R. P. Darveau, and K. Postle. 1993. Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268:16302-16308. [PubMed] [Google Scholar]
- 27.Srikumar, R., X.-Z. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Traub, I., S. Gaisser, and V. Braun. 1993. Activity domains of the TonB protein. Mol. Microbiol. 8:409-423. [DOI] [PubMed] [Google Scholar]
- 29.Zhao, Q., X.-Z. Li, A. Mistry, R. Srikumar, L. Zhang, O. Lomovskaya, and K. Poole. 1998. Influence of the TonB energy-coupling protein on efflux-mediated multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:2225-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao, Q., and K. Poole. 2000. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol. Lett. 184:127-132. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, Q., and K. Poole. 2002. Mutational analysis of the TonB1 energy coupler of Pseudomonas aeruginosa. J. Bacteriol. 184:1503-1513. [DOI] [PMC free article] [PubMed]
- 32.Ziha-Zarifi, I., C. Llanes, T. Köhler, J.-C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]