Abstract
The biosynthesis of the thaxtomin cyclic dipeptide phytotoxins proceeds nonribosomally via the thiotemplate mechanism. Acyladenylation, thioesterification, N-methylation, and cyclization of two amino acid substrates are catalyzed by the txtAB-encoded thaxtomin synthetase. Nucleotide sequence analysis of the region 3′ of txtAB in Streptomyces acidiscabies 84.104 identified an open reading frame (ORF) encoding a homolog of the P450 monooxygenase gene family. It was proposed that thaxtomin A phenylalanyl hydroxylation was catalyzed by the monooxygenase homolog. The ORF was mutated in S. acidiscabies 84.104 by using an integrative gene disruption construct, and culture filtrate extracts of the mutant were assayed for the presence of dehydroxy derivatives of thaxtomin A. Reversed-phase high-performance liquid chromatography (HPLC) and HPLC-mass spectrometry indicated that the major component in culture filtrate extracts of the mutant was less polar and smaller than thaxtomin A. Comparisons of electrospray mass spectra as well as 1H- and 13C-nuclear magnetic resonance spectra of the purified compound with those previously reported for thaxtomins confirmed the structure of the compound as 12,15-N-dimethylcyclo-(l-4-nitrotryptophyl-l-phenylalanyl), the didehydroxy analog of thaxtomin A. The ORF, designated txtC, was cloned and the recombinant six-His-tagged fusion protein produced in Escherichia coli and purified from cell extracts. TxtC produced in E. coli exhibited spectral properties similar to those of cytochrome P450-type hemoproteins that have undergone conversion to the catalytically inactive P420 form. Based on these properties and the high similarity of TxtC to other well-characterized P450 enzymes, we conclude that txtC encodes a cytochrome P450-type monooxygenase required for postcyclization hydroxylation of the cyclic dipeptide.
Thaxtomins are highly phytotoxic cyclic dipeptides produced by plant-pathogenic members of the genus Streptomyces (22). All of the thaxtomins that have been identified from extracts of infected plant tissue and from filtrates of culture medium have the basic structure cyclo-(l-4-nitrotryptophyl-l-phenylalanyl) (19). The diketopiperazine moiety may be N-methylated, and most thaxtomin congeners also carry phenylalanyl α- and often ring-carbon hydroxyl groups. The predominant thaxtomin produced by Streptomyces scabies, Streptomyces acidiscabies, and Streptomyces turgidiscabies is thaxtomin A, with phenylalanyl m-ring and α-C hydroxyl additions.
Using PCR-amplified peptide synthetase gene fragments as probes, we previously cloned the thaxtomin synthetase genes, txtAB, from cosmid libraries of S. acidiscabies 84.104 and demonstrated that thaxtomin A production was abolished in txtA mutants, confirming the role of txtAB in thaxtomin biosynthesis (14). The genes encode two similar peptide synthetases (exhibiting 44% identity to one another at the amino acid level), and each consists of requisite acyladenylation, thiolation, and condensation domains. Both synthetases also have integratedS-adenosylmethionine-dependent N-methyltransferase do-mains, consistent with the occurrence of N-methyl groups on each of the amino acid substrates in thaxtomin A. The origins of the C-4 nitro addition to l-tryptophan as well as the l-phenylalanyl hydroxyl groups in most of the described thaxtomins (19), the enzymes catalyzing these additions, and their place in the thaxtomin biosynthetic pathway, however, have remained unclear.
Enzymes belonging to the class I b-type cytochrome P450 monooxygenase family and the genes encoding them have been characterized in many streptomycetes (10, 23, 25, 26, 29, 30, 38, 46, 49). These enzymes typically catalyze the addition of hydroxyl groups, arising from protons and the cleavage of dioxygen, to a wide variety of secondary metabolite and xenobiotic substrates. Reducing equivalents are provided by NAD(P)H, and electron pair transfer is mediated by a flavoprotein reductase and Fe-S ferredoxin (29). Structural studies of P450s have also demonstrated a critical role for an invariant cysteinyl residue involved in the coordination of a prosthetic heme group ferrous iron atom in the monooxygenase substrate binding pocket (8, 34). The complete reaction pathway of the best-studied bacterial P450, the P450cam of Pseudomonas putida, has recently been described (40).
Our analysis of the nucleotide sequence beyond the 3′ end of txtB in S. acidiscabies 84.104 identified an open reading frame (ORF), transcribed in the same direction as txtAB, the predicted gene product of which was similar to cytochrome P450 enzymes. The occurrence of hydroxyl groups on the phenylalanyl constituent of most described thaxtomins (19) and the location of the monooxygenase gene homolog in the vicinity of txtAB prompted us to investigate the possible role of this homolog in thaxtomin biosynthesis. This work presents evidence that this P450 monooxygenase homolog, designated txtC, encodes a cytochrome P450-type monooxygenase required for thaxtomin phenylalanyl hydroxylation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Streptomyces acidiscabies strain 84.104 is a wild-type strain that produces thaxtomins (20). Escherichia coli strain DH5αMCR (Gibco-BRL) was used for routine subcloning of recombinant plasmids. E. coli strain S17-1, carrying chromosomally integrated conjugal transfer functions for RK2/RP4-type broad-host-range plasmids (43), was used as a donor for conjugal transfer of recombinant plasmids to Streptomyces recipients. E. coli strain BL21(λDE3) (Novagen) was used as a host for expression of recombinant txtC. Plasmid pUC19 was used for the construction of cosmid subclones; plasmid pOJ260 (3) was used for monooxygenase gene disruption via Campbell-type integration. txtC was cloned into plasmid vector pET15b (Novagen) for production of a six-His [(His)6]-TxtC fusion protein.
Culture conditions.
The S. acidiscabies 84.104 parent strain and the monooxygenase mutant txtC strain were cultured on ISP2 agar medium for spore production (42); spore suspensions were maintained as 20% glycerol stocks at −80°C. txtC mutants were selected on AS-1 medium with apramycin soft agar overlays (25-μg/ml final concentration) following conjugation with E. coli S17-1 as previously described (14); S17-1 donors were counterselected with nalidixic acid (25-μg/ml final concentration). S. acidiscabies 84.104 and txtC mutants were cultured in oatmeal broth medium (OMB) at 28°C to evaluate toxin production as previously described (14). OMB cultures of the txtC mutant were amended with apramycin sulfate (25 μg/ml) to eliminate the possibility of revertants arising during growth of the culture. E. coli strains were grown in Luria-Bertani (LB) medium or on LB agar containing ampicillin or apramycin sulfate (Sigma Chemical Co.), 100 μg/ml, where appropriate.
Cloning, sequencing, and analysis of txtC.
The thaxtomin synthetase genes (txtAB) were originally isolated from S. acidiscabies 84.104 genomic libraries constructed in cosmid vector pOJ446 (14). One such cosmid, SACOS1, carries the txtAB genes as well as DNA sequence downstream of the 3′ end of txtB. Plasmid pFGH202 was constructed by subcloning a 9-kb KpnI fragment from SACOS1 containing the 3′ end of txtB in KpnI-digested pUC19. Plasmid pFGH204 was constructed by subcloning the 5-kb txtB-proximal KpnI-EcoRI fragment from pFGH202 in KpnI-EcoRI-digested pUC19 (Fig. 1). Nucleotide sequencing was done with an ABI 377XL automated sequencer at the Bioresource Center, Cornell University. Both strands were sequenced with M13 forward/reverse and custom oligonucleotide primers, synthesized by the Bioresource Center or Integrated DNA Technologies (Coralville, Iowa). The deduced amino acid sequence of the txtC gene product was compared with those of P450 enzymes by using the NCBI BLASTP server, and protein alignments were constructed with CLUSTAL W (45). Three-dimensional homology modeling was done with the automated comparative protein modeling server SWISS-MODEL (11). Stereochemical quality of TxtC models was assessed by using the PROCHECK V3.5 (21) and WHAT IF (39) utilities.
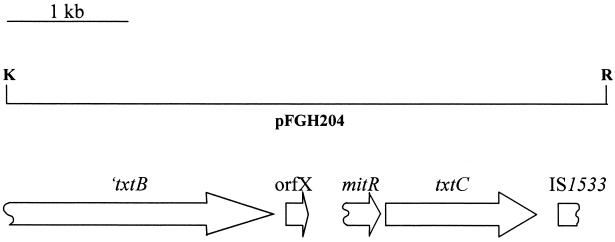
FIG. 1.
Genetic organization of the txtC region (plasmid pFGH204) in S. acidiscabies 84.104. ORFs other than ′txtB and txtC are denoted by the reported genes to which they are most similar (see text). Restriction endonuclease recognition sites are abbreviated as follows: K, KpnI; R, EcoRI.
txtC gene disruption.
Based on the nucleotide sequence of txtC, PCR oligonucleotide primers were designed to amplify an internal 744-bp fragment of the gene. EcoRI adapters were included on the 5′ ends of the primers to facilitate cloning in the gene disruption vector pOJ260 (3). The oligonucleotide primer sequences were as follows: P450F, 5′-GGAATTCCGCGCCGACCCAACTTCACC; and P450R, 5′-GGAATTCGGAGCGCCGCCACCACCATCTG. The monooxygenase gene fragment was amplified from plasmid pFGH204 in a 100-μl reaction volume containing 10 μl of 10× Taq DNA polymerase reaction buffer (Perkin-Elmer), 10 μl of 25 mM MgCl2, 2 μl of dimethyl sulfoxide (DMSO), 20 μM each deoxynucleoside triphosphate, 1 μM each oligonucleotide primer, 0.3 U of Taq DNA polymerase, and 10 ng of pFGH204 template DNA. The 758-bp amplification product was digested with EcoRI and ligated to EcoRI-digested plasmid pOJ260 with T4 DNA ligase. The resulting integrative plasmid, pFGH308, was used to disrupt the monooxygenase gene by using intergeneric conjugal transfer techniques described previously (14). Southern hybridization analysis of KpnI-digested genomic DNA of the parent and mutant strains with radiolabeled pFGH308 was done as previously described (14).
Isolation of thaxtomins.
OMB culture filtrates of parent strain S. acidiscabies 84.104 and txtC mutant were extracted in ethyl acetate, and the extracts were dried in vacuo. To purify thaxtomins, extracts were dissolved in CH2Cl2-CH3OH and flash chromatographed on silica gel (Baker 40-μm diameter), eluted with a mobile phase consisting of binary mixtures of CH2Cl2 and CH3OH at a flow rate of 5 ml/min. Extracts of the parent strain were eluted with 93% CH2Cl2; txtC extracts were eluted with 95% CH2Cl2. Fractions were monitored by UV A254 on an ISCO UA-6 detector. Thaxtomins were crystallized by slow evaporation from methanol.
HPLC analysis of thaxtomins.
OMB culture filtrates of the parent and monooxygenase mutant were extracted with ethyl acetate and dissolved in methanol. Thaxtomin production was estimated from samples by reversed-phase high-performance liquid chromatography (HPLC) with a Waters model 600 series pump (dwell volume= 7 ml) and a Phenomenex Prodigy ODS3 column (RP-C18, 250 by 4.6 mm, 5-μm particle size, 100-Å pore size). A two-component mobile phase consisting of (i) 0.1% (vol/vol) trifluoroacetic acid (TFA) in acetonitrile and (ii) 0.1% (vol/vol) TFA (aqueous) was ramped in a linear fashion from 25 to 100% 0.1% (vol/vol) TFA in acetonitrile over 10 min. Samples were eluted at a flow rate of 1 ml/min, and A370 was monitored with a Waters 2687 UV detector. Thaxtomins were quantified by comparing peak areas in extract chromatograms with areas from standard curves generated for purified thaxtomins A and D.
ESI-MS and HPLC-ESI-MS.
Electrospray (ESI) mass spectra were obtained by direct infusion of methanolic solutions into a Micromass ZMD mass detector at a flow rate of 5 or 10 μl/min with a model 55-1199 syringe pump (Harvard Apparatus, Holliston, Mass.). Spectra were obtained for thaxtomins in the negative ion mode with a cone voltage of 60 V and a capillary voltage of 1.82 or 4 kV and in the positive ion mode with a cone voltage of 90 V and a capillary voltage of 3 kV. Source block temperature was held at 100°C. Desolvation gas (N2) was heated to 400°C for liquid chromatography-mass spectrometry (MS) and 150°C for direct infusion of methanolic samples. HPLC-MS was carried out with a Waters 2690 Alliance HPLC pump with a Phenomenex Prodigy ODS-3 column at a flow rate of 0.25 ml/min. A mobile phase consisting of (i) 0.1% (vol/vol) TFA in acetonitrile and (ii) 0.1% (vol/vol) TFA (aqueous) was ramped in a linear fashion from 25 to 75% 0.1% (vol/vol) TFA in acetonitrile over 10 min.
NMR analyses.
1H-nuclear magnetic resonance (NMR) spectra were obtained from samples dissolved in CD3OD with Varian Unity 500 and XL-200 spectrometers. The spectra thus obtained were compared with published spectral data for purified thaxtomins (19). 13C-NMR spectra were obtained on a Varian Inova VXRS-400 spectrometer.
Cloning and expression of txtC and purification of (His)6-TxtC.
txtC was amplified from plasmid pFGH204 template by PCR (described above) with the following nucleotide primer pair: txtCF (5′-GGAAACGACCATATGGAATCTCCGGCCACCCAGGTC) and txtCR (5′-CTGGTGGATCCCTCGCGGCAGGCCCGGTCACCAG). Purified PCR product was cut at NdeI and BamHI recognition sites incorporated into nucleotide primers (underlined) and then cloned into plasmid pET15b, which was linearized with the same restriction enzyme pair. The nucleotide sequence of txtC cloned in pET15b was verified on both strands by dideoxy chain termination sequencing methods. Recombinant plasmid was used to transform BL21(λDE3) to ampicillin resistance. Overnight cultures (approximately 5 ml) grown in LB medium containing ampicillin (100 μg/ml) were used to inoculate 1 liter of the same medium. This culture was grown at 30°C until the optical density of the culture at 420 nm (OD420) reached 0.5. At this time, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the culture was grown for another 10 h. All subsequent steps were carried out at 4°C. Cells were collected by centrifugation and resuspended in 50 mM Tris-Cl (pH 7.5). Cells were broken by passage through a French pressure cell at 20,000 psi. The cell extract was centrifuged at 100,000 × g for 1 h, and the resulting supernatant was loaded on a 5-ml HiTrap Chelating HP column (Amersham Pharmacia Biotech), which had been charged with 100 mM NiCl2 and equilibrated in 50 mM Tris-Cl (pH 7.5). Fractions were eluted stepwise with 50 mM Tris-Cl (pH 7.5) containing increasing amounts of imidazole. (His)6-TxtC was eluted from the column with 75 mM imidazole and was dialyzed extensively against 50 mM Tris-Cl (pH 7.5). The protein concentration was estimated with Bradford reagent with bovine serum albumin as a standard. (His)6-TxtC was also purified with 50 mM phosphate buffer (pH 7.2), containing 20% glycerol and 0.1 mM dithiothreitol (DTT). Thrombin protease treatment of (His)6-TxtC fusion protein was done according to instructions provided by the manufacturer (Amersham Pharmacia Biotech).
Spectral analysis of TxtC.
Absorption spectra for purified TxtC were obtained from 400 to 600 nm with a Beckman DU 640 spectrophotometer. For oxidized samples, solutions of TxtC in 50 mM Tris-Cl (pH 7.5) were placed in a cuvette, and spectra were recorded directly. For reduced samples, a few grains of sodium dithionite were added to samples prior to recording spectra. CO-reduced samples were prepared by first adding dithionite to protein samples, followed by gentle bubbling of CO into sample cuvettes for approximately 1 min prior to spectral analysis. Heme content of TxtC was calculated by measuring the dithionite-reduced minus oxidized difference spectrum of pyridine-hemochromogen (31), with ɛ = 34.7 mM−1 cm−1 for the reduced pyridine-hemochromogen (33).
Nucleotide sequence accession number.
The nucleotide sequence of the txtC region has been deposited in the GenBank database under accession no. AF393159.
RESULTS
Nucleotide sequence analysis of the region 3′ of txtB surrounding txtC.
Analysis of the nucleotide sequence 3′ of the txtAB region revealed the presence of two complete ORFs and two truncated ORFs (Fig. 1). ORFX begins 95 nucleotides (nt) from the txtB stop codon and is predicted to encode a 52-amino-acid product. The predicted product is similar to several conserved small hypothetical proteins found adjacent to the 3′ ends of other bacterial peptide synthetase genes. These include the calcium-dependent antibiotic (CDA)-associated ORFX of Streptomyces coelicolor (50% identity, 69% similarity) (6, 37), mbtH of Mycobacterium tuberculosis (65% identity, 77% similarity) (7), and cumB of Streptomyces rishiriensis (56% identity, 77% similarity) (48).
Downstream of ORFX, a region of approximately 200 bp was identified, the deduced amino acid sequence of which exhibits strong similarity to the C termini of a putative S. coelicolor lipoprotein, SCD72A.20 (43% identity, 54% similarity)(http://www.sanger.ac.uk/Projects/S_coelicolor/blast_server.shtml) (37), and MitR, a component of the mitomycin C biosynthetic cluster in Streptomyces lavendulae (43% identity, 54% similarity) (23). This region also exhibits strong codon bias typical of streptomycete coding regions, suggesting that this may represent a truncated portion of a once functional gene. Similarly, a nucleotide sequence was identified at the txtB-distal end of pFGH204, the deduced amino acid sequence of which exhibits high similarity to the N termini of several transposases. This sequence exhibits the highest similarity to IS 1533 of M. tuberculosis (67% identity, 80% similarity) (7), and is 51% identical (63% similar) to a transposase sequence from Yersinia pestis virulence-associated plasmid pPCP1 (15). The transposase sequence homology ends approximately 300 nt from the EcoRI site.
The deduced amino sequence of the txtC gene product is homologous with cytochrome P450 enzymes.
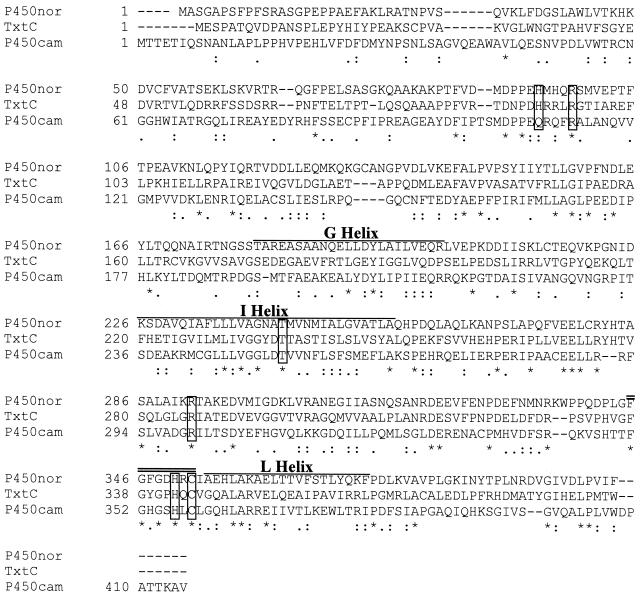
Protein sequence database searches revealed that the deduced amino acid sequence of txtC was similar to those of many previously characterized P450 enzymes. A CLUSTAL W alignment (45) of the TxtC sequence with the well-characterized Pseudomonas putida P450cam (NCBI accession no. P00183) and Fusarium oxysporum P450nor (NCBI accession no. JC5150) is shown in Fig. 2. Over a 426-position alignment, the three proteins shared 12% identity (51 of 426 positions), and the extent of amino acid similarity was 27.7% (118 of 426 positions).
FIG. 2.
Clustal W alignment of S. acidiscabies TxtC with Fusarium oxysporum P450nor and Pseudomonas putida P450cam. Residues that are identical in both sequences are marked with asterisks, and those that are strongly or weakly similar are marked with colons and single dots, respectively. Conserved propionate H-bond donor residues are boxed, along with conserved I-helix Thr and thiolate Cys. Regions corresponding to G, I, and L helices are overlined. The heme pocket motif is doubly overlined.
Closer inspection of the alignment and a comparison with published crystallographic structures of P450cam (Protein Data Bank ID 5CP4) (34) and P450nor (Protein Data Bank ID CL6A) (41) revealed the conservation in TxtC of individual residues and regions that play critical structural and functional roles for these well-characterized enzymes. Residues R112, R299, and H355 in P450cam, which serve as H-bond donors to propionate groups of the heme prosthetic group, are conserved in TxtC (R95, R286, and H342, respectively). A high degree of sequence conservation was also found among the I and L helices of P450cam, P450nor, and the corresponding regions of TxtC. These secondary structural features serve to bracket the distal and proximal faces of the heme group, respectively, in P450cam (34). The oxygen-binding sites of helix I in particular, which correspond to residues 248 to 252 in P450cam and residues 233 to 237 in TxtC, is highly conserved in TxtC. The adjacent Thr residue in P450cam, which is also conserved in TxtC (T237), is thought to play a role in proton translocation during the monooxygenase reaction (35). Additionally, the region in TxtC corresponding to the heme binding pocket, including the cysteinyl residue that forms the proximal heme iron thiolate ligand (C358 in P450cam, C353 in P450nor, and C344 in TxtC), is conserved and conforms to the consensus sequence found in all cytochrome P450 enzymes, FXXGXXXCXG.
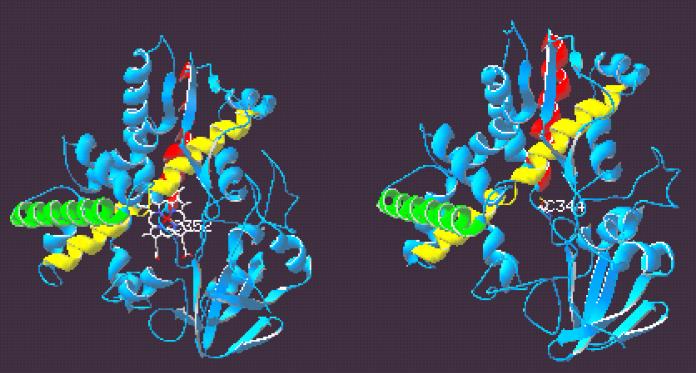
A more thorough investigation of potential structural similarities between TxtC and the P450 enzymes was carried out by constructing a three-dimensional model of TxtC by using the Swiss-Model homology modeling server and the coordinates of P450cam and P450nor as structural templates. The results of quality assessments of the TxtC model obtained revealed that 91% of residues were in most favored regions of the Ramachandran plot, and no residues were found in disallowed regions. Comparison of the TxtC model with P450cam and P450nor revealed an overall conservation of the major secondary structural features common to P450 enzymes, as shown in Fig. 3. The side-by-side comparison between the P450nor structure and the TxtC model reveals the conservation of prominent α-helical structures (e.g., the G, I, J, K, and L helices). A “vacant” heme pocket can be seen in the TxtC model; the conserved C344 is also positioned in such a way as to provide a putative fifth axial ligand within the heme pocket region. Moreover, the Cys ligand loop following the signature heme consensus sequence is conserved in the model.
FIG. 3.
Ribbon diagrams of P450nor (left) and modeled three-dimensional structure of TxtC (right). Heme prosthetic group and cysteinyl thiolate ligand Cys352 are shown in P450nor, and the conserved putative ligand Cys344 is shown in TxtC. G, I, and L helices are depicted in green, yellow, and red, respectively. The figure was prepared with Swiss-PdbViewer v3.7b2.
txtC gene disruption.
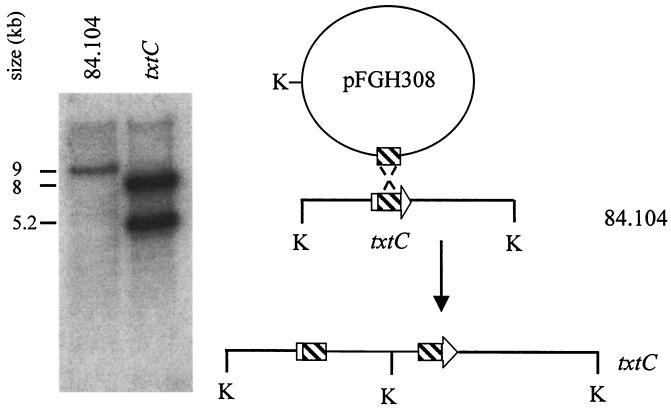
The primary sequence and modeled secondary structural similarities of TxtC with other well-characterized P450 enzymes led us to address whether a txtC mutation would result in changes in thaxtomin product formation. Repeated attempts to create a txtC-null mutant by double crossover recombination failed, and for this reason, the integrative plasmid pFGH308 was constructed to create an S. acidiscabies txtC mutant by Campbell-type integration. This approach was successful, and Southern hybridization analysis of KpnI-digested genomic DNA samples of the parent and mutant strain is shown in Fig. 4. Using a radiolabeled pFGH308 probe, the expected 9-kb KpnI fragment hybridized to DNA from the parent strain, while the presence of a single KpnI site in plasmid pOJ260 introduced a KpnI site within the parental 9-kb fragment in the genome of the monooxygenase mutant. Consequently, the probe hybridized to 8- and 5.2-kb fragments.
FIG. 4.
Southern hybridization analysis of KpnI-digested (K) genomic DNA of S. acidiscabies 84.104 and txtC mutant, probed with radiolabeled pFGH308 DNA. A diagram illustrating txtC gene disruption is shown to the right.
Production of thaxtomin A didehydroxy derivative (thaxtomin D) by S. acidiscabies txtC mutants.
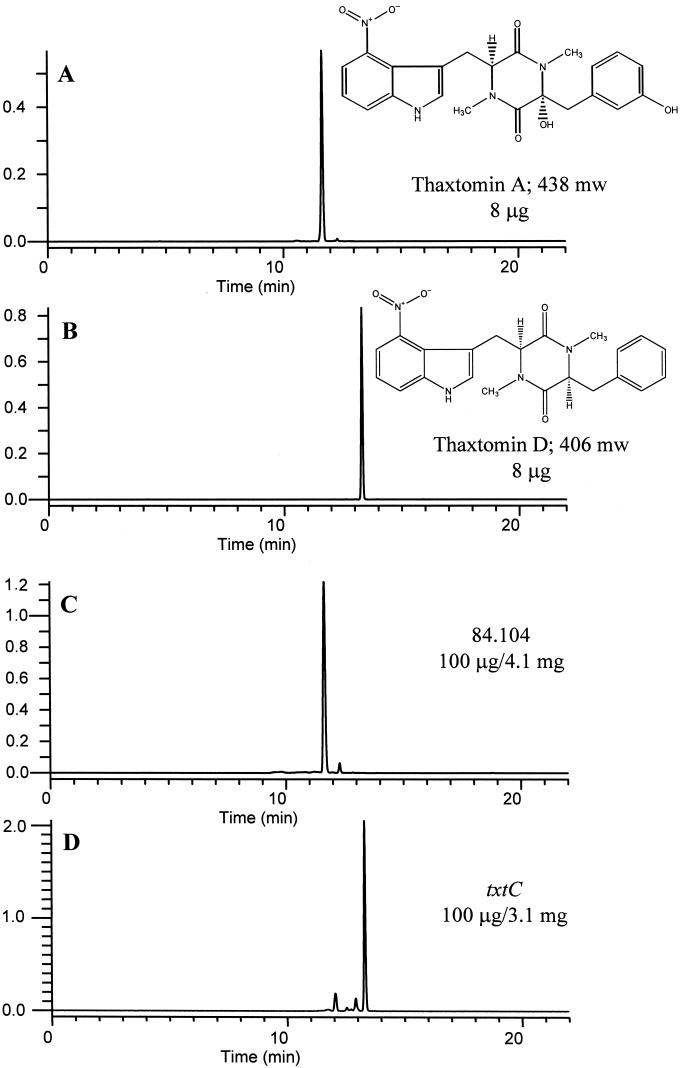
Extraction of 3 liters of S. acidiscabies 84.104 OMB culture filtrate with ethyl acetate yielded 120 mg of crude extract; silica gel flash chromatography of this extract yielded 28 mg of yellow orange rosettes. Crystals dissolved in methanol exhibited a quasimolecular ion, [M−H+]−, at 437 amu (negative-ion-mode ESI-MS) and a sodium adduct, [M+Na]+, at 461 amu (positive-ion-mode ESI-MS). The 1H-NMR spectrum (CD3OD) agreed with that previously reported for thaxtomin A (19) (not shown). Extraction of 3 liters of the txtC mutant OMB culture filtrate yielded 84 mg of crude extract; silica gel flash chromatography of this extract yielded 55 mg of lemon-yellow rosettes. Crystals dissolved in methanol exhibited a quasimolecular ion, [M−H+]−, at 405 amu (negative-ion-mode ESI-MS) and a sodium adduct, [M+Na]+, at 429 amu (positive-ion-mode ESI-MS). The 1H-NMR spectrum (CD3OD) was identical to that previously reported for 12,15-N-dimethylcyclo-(l-4-nitrotryptophyl-l-phenylalanyl) (thaxtomin compound 5 [19], here referred to as thaxtomin D) (not shown). The 13C-NMR spectrum (CD3OD) indicated the presence of 20 nonidentical carbon atoms, which further confirmed our structural determination. HPLC chromatograms of representative extracts and purified compounds are shown in Fig. 5.
FIG. 5.
Reversed-phase HPLC chromatograms of purified thaxtomin A (A), purified thaxtomin D (B), and extracts of OMB culture filtrates of S. acidiscabies 84.104 (C) and the txtC mutant (D).
Several minor components were also found in extracts of culture filtrates from both strains. Based on LC-MS analyses, these components exhibited masses and retention times that were similar to those reported for some of the minor thaxtomin compounds produced by S. scabies (19). However, these compounds were not present in quantities large enough for preparative isolation and structural determination.
TxtC produced in E. coli is a heme-containing protein with spectral properties similar to those of catalytically inactive P450 enzymes (P420 form).
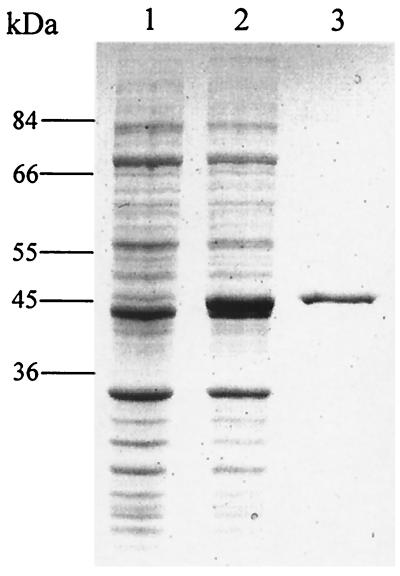
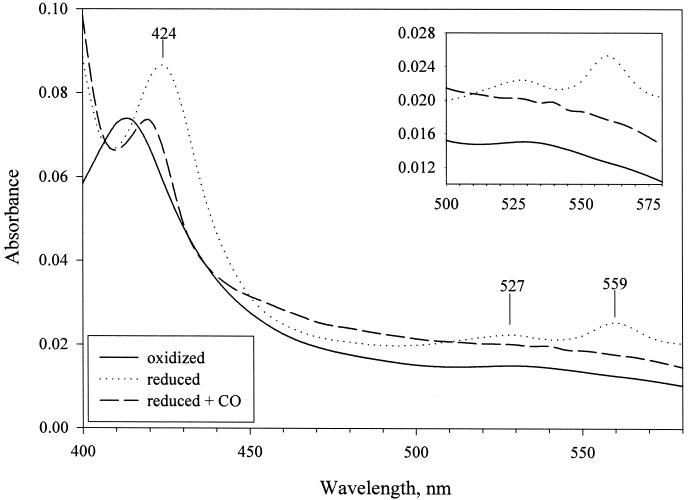
In order to examine the spectral properties of TxtC, the plasmid-encoded txtC gene was expressed in E. coli from an IPTG-inducible promoter. (His)6-TxtC (predicted molecular mass, 45.6 kDa) was purified to apparent homogeneity by metal chelation chromatography (Fig. 6). Ferric protoporphyrin IX (heme) content of purified TxtC was estimated at 3.5 nmol/mg of protein. Absorbance spectra for the ferric-oxidized, dithionite-reduced, and dithionite-reduced CO forms of TxtC are shown in Fig. 7. TxtCox exhibited a Soret absorption band at 413 nm; the reduced form exhibited the Soret absorption peak at 424 nm, with α and β bands at 559 and 527 nm, respectively. Upon formation of the CO adduct with TxtCred, a shift in the Soret absorption was observed—from 424 nm to 420 nm.
FIG. 6.
SDS-PAGE (8.5% polyacrylamide) analysis of TxtC expression in E. coli BL21(λDE3). Lanes: 1, total soluble protein from uninduced cultures; 2, total soluble protein from IPTG-induced cultures; 3, TxtC (3 μg) purified by nickel chelation chromatography.
FIG. 7.
Absorption spectra of oxidized, reduced, and reduced-plus-CO forms of TxtC at 25°C (6 μM in 50 mM Tris-Cl [pH 7.5]). An inset shows α and β bands in the 500- to 575-nm region.
The CO-reduced minus reduced difference spectra were also recorded, although we were unable to detect a 450-nm Soret absorption peak (data not shown). Similar results were obtained when protein was treated with CO prior to dithionite reduction. We were also unsuccessful in detecting type I spectral shifts associated with binding of the presumed substrate, thaxtomin D, with substrate concentrations ranging from 0.05 to 1.0 mM (data not shown). A thrombin protease cleavage recognition sequence is present between the N-terminal (His)6 sequence and the N terminus of TxtC. Treatment with thrombin protease releases the (His)6 used for purification purposes and leaves an N-terminal Gly-Ser-His preceding the TxtC-initiating Met residue. Spectral absorption data were similar with the protease-treated and untreated proteins. Similar results were also obtained when TxtC was purified from cell extracts by using phosphate buffer (pH 7.2), containing 20% glycerol and 0.1 mM DTT. Treatment of catalytically inactive P450s (P420 form) with sulfhydryl reagents has been shown, in some instances, to restore native P450 substrate binding, catalytic, and spectral characteristics (50). Neither substrate binding spectra nor 450-nm CO-reduced difference spectra were observed after treatment of TxtC preparations with cysteine, β-mercaptoethanol, or dithioerythritol.
DISCUSSION
Nucleotide sequence analysis of the region surrounding the previously characterized thaxtomin synthetase genes txtAB from S. acidiscabies 84.104 identified an ORF homologous to other small conserved hypothetical peptide synthetase-associated proteins and a putative P450 monooxygenase gene, txtC, adjacent to the 3′ end of txtB. Our analysis of the region also identified truncated homologs of other genes, including a pathogenicity-associated transposase. The chimeric architecture of the txt region in particular serves to further support our model of the recent pathogenicity island-mediated emergence of taxonomically diverse Streptomyces pathogens (4, 5, 13). Our analysis of the 5-kb region did not reveal the presence of any ORFs resembling the class I P450 redox partner proteins, ferredoxin or the FAD-containing NAD(P)H ferredoxin oxidoreductase. The genes for these components may reside further than 5 kb from txtC; alternatively, the TxtC monooxygenase reaction may not involve “dedicated” ferredoxin/NAD(P)H reductase enzymes, but rather the appropriation of enzymes, which also serve other physiological functions in the cell, as has been discussed previously (29).
Based on our identification of the monooxygenase gene homolog txtC in the vicinity of the previously characterized txtAB genes, it was hypothesized that thaxtomin A phenylalanyl α- and/or ring carbon hydroxyl group additions were catalyzed by this monooxygenase homolog. In order to test this, a txtC mutant was constructed and assayed for the production of dehydroxy thaxtomin derivatives. S. acidiscabies 84.104 txtC mutants produced the didehydroxy thaxtomin A derivative, herein designated thaxtomin D, as the predominant thaxtomin compound. This finding was confirmed by HPLC, LC-MS, and 13C- and 1H-NMR spectroscopic methods.
The phenotypic characterization of the txtC mutation as a deficiency in thaxtomin D hydroxylation suggested that txtC indeed encodes a P450-type monooxygenase activity. Using the Swiss-Model homology-modeling server, we constructed a three-dimensional TxtC model by using P450 enzyme structural coordinates as templates. These results demonstrate that TxtC can adopt folds and structural features similar to those of the P450 template structures.
TxtC was purified from E. coli in order to provide biochemical evidence for the P450 hemoprotein nature of TxtC. The heme content of our purified TxtC preparation, based on pyridine-hemochromogen measurements, was 3.5 nmol/mg of protein, which amounts to only 16% occupancy of the heme pocket (3.5 nmol of heme/mg of protein·mg of protein/21.93 nmol = 0.159). It is possible that TxtC overexpression in E. coli overwhelms the cell's heme biosynthetic capabilities, resulting in poor incorporation of the heme into the P450 apoprotein. Nonetheless, these measurements as well as the occurrence of clear α, β, and Soret spectral absorbance bands clearly establish that TxtC is a hemoprotein. These data are consistent with three-dimensional modeling data that suggest TxtC could form a heme pocket capable of accommodating the prosthetic group and also adopt folding topologies common to P450-type enzymes.
The inability of purified TxtC to exhibit the 450-nm reduced-CO difference spectrum typical of P450 enzymes, but which instead displays a ferrous 420-nm shift, can be explained by the observation that cytochrome P450 enzymes commonly undergo, in certain instances, a transition to an inactive yet stable structural state referred to as P420. The P450→P420 conversion was first described by Omura and Sato (31), by using snake venom- or deoxycholate-treated rabbit liver microsomal P450 preparations. Subsequently, it has been shown that P450→P420 transitions may be induced by temperature or pressure perturbations (9, 16), as well as treatment with various chemicals (17) or pH extrema (28). Some of these treatments have been exploited to investigate P450 structural architecture and prosthetic group ligand coordination (24).
Various enzymatic structural changes as a result of the aforementioned treatments have been posited to account for the conversion to the inactive P420 state. These include proximal thiolate protonation (12), loss or substitution of, for example, His, Met, or water for the Cys thiolate ligand (32, 44), and lengthening of the thiolate Fe-S bond (18).
The absorbance spectra we obtained for our purified recombinant TxtC are consistent with the protein having undergone P450→P420 conversion. TxtC displayed a shift in the Soret band from 424 nm to 420 nm for the ferrous and ferrous-CO forms, respectively. Conversion from a native P450 to P420 state could also explain the failure to observe type I substrate binding spectra with thaxtomin D. Treatment of TxtC with a variety of sulfhydryl reagents had no effect on its absorbance profiles, although in at least one instance, treatment with sulfhydryl compounds restored P420cam to the P450 form (50). Our inability to obtain recombinant TxtC that exhibited reduced CO difference spectra with a 450-nm Soret absorbance or type I binding spectra may be caused by improper protein folding or incorporation of the heme group into the apoenzyme in E. coli, since crude cell extracts of BL21(λDE3) expressing txtC also failed to exhibit 450-nm reduced-CO difference spectra (data not shown). It is likely that the low heme content of the recombinant TxtC preparation would also contribute to these shortcomings. Future biochemical studies will require that TxtC be purified in its native form from S. acidiscabies.
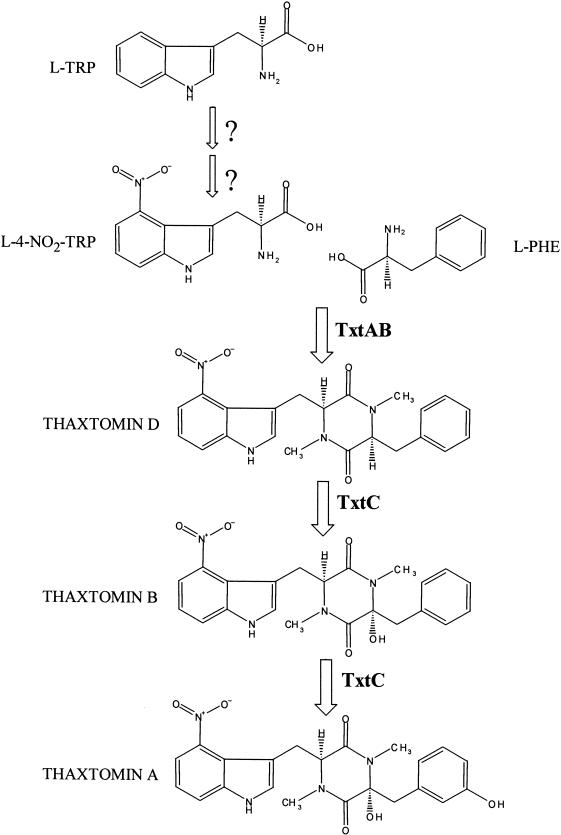
The observation that the txtC mutant produces thaxtomin D as virtually the sole identifiable thaxtomin derivative allows us to draw the following inferences regarding the most plausible biosynthetic pathway leading to production of thaxtomin A: (i) l-phenylalanyl hydroxylation follows diketopiperazine formation (amino acid cyclization), whether TxtC catalyzes one or both hydroxylations, and (ii) l-tryptophanyl nitration should also precede phenylalanyl hydroxylation. From this, it would also follow that l-Phe is one of the substrates for either TxtA or TxtB; acyladenylation assays using l-Trp and l-4-NO2-Trp should allow the substrate assignment for the other synthetase. Based on this argument, the most likely pathway for thaxtomin A biosynthesis, including the central reactions catalyzed by TxtAB and TxtC, is presented in Fig. 8.
FIG. 8.
Proposed pathway of thaxtomin A biosynthesis in S. acidiscabies 84.104. Central reactions catalyzed by TxtAB, and consecutive TxtC-catalyzed hydroxylations are shown. Cofactors and accessory proteins are not shown. Initial sequences catalyzing l-Trp nitration are not known. For simplicity, it is assumed that l-4-NO2-Trp is the substrate for the TxtAB peptide synthetase.
The accumulation of the didehydroxy derivative thaxtomin D in extracts of the txtC mutant indicates that, at the very least, the txtC-encoded monooxygenase catalyzes a single initial postcyclization hydroxyl addition to thaxtomin D. The identification of txtC adjacent to txtAB, coupled with the observation that secondary metabolite pathways are typically organized in a linear manner, would be consistent with this role for TxtC. If we assume the monooxygenase catalyzed a single initial postcyclization hydroxylation, then the most likely monooxygenase target would be the l-phenylalanyl α-carbon atom based on the logic of natural thaxtomin congener profiles found in extracts (1). This is because the only monohydroxylated thaxtomins that have been isolated from extracts and characterized are hydroxylated on the phenylalanyl α-carbon (thaxtomin B and thaxtomin 6) (19); thaxtomins carrying single hydroxyl groups on the phenylalanyl ring have not been identified in extracts. Thaxtomins containing multiple hydroxyl groups are hydroxylated on the phenylalanyl α-carbon in addition to carrying phenylalanyl ring hydroxyl groups. Therefore, it seems most likely that a first postcyclization hydroxyl addition by a monofunctional monooxygenase would occur on the phenylalanyl α-carbon atom, resulting in the accumulation of thaxtomin B, and subsequent additions would occur on the phenylalanyl ring carbon atoms. TxtC then would function in the conversion of thaxtomin D to thaxtomin B.
Our nucleotide sequence analysis beyond pFGH204 has also identified a homolog of the FAD-dependent monooxygenase family. It was our initial belief that thaxtomin ring carbon additions might be catalyzed by an activity encoded by this ORF, similar to those of the hydroxybenzoate hydroxylases (47). However, we have recently mutated the ORF by the techniques described in this report, and mutants exhibited the wild-type thaxtomin production phenotype (unpublished data). This does not rule out the possibility that a second unlinked monooxygenase exists for the conversion of thaxtomin B to thaxtomin A. However, the lack of any candidate genes in the vicinity of the txtABC region that could encode ring hydroxylation activities leads us to propose that TxtC catalyzes both the phenylalanyl α- and ring-carbon additions. P450 enzymes that catalyze successive hydroxylations to a single substrate are not uncommon. For instance, Newman and Wackett (27) have shown that toluene 2-monooxygenase from Burkholderia cepacia catalyzes both hydroxylation of toluene to o-cresol and hydroxylation of o-cresol to 3-methylcatechol. These reactions allow B. cepacia to utilize toluene as a sole carbon and energy source by activating the aromatic ring for subsequent ring scission and metabolism. In Nicotiana tabacum, the antimicrobial bicyclic sesquiterpene compound capsidiol is formed through the hydroxylation of 5-epi-aristolochene; the product of this reaction, 1-deoxycapsidiol, is further hydroxylated to form capsidiol. Both of these hydroxylation reactions are catalyzed by the CYP71D20-encoded cytochrome P450 monooxygenase gene (36). In addition, in human mitochondria, the active form of vitamin D, 1,25-(OH)2D3, is metabolized through two pathways. The predominant C-23-C-24 catabolic pathway involves C-24 hydroxylation and ketonization, followed by C-23 hydroxylation and cleavage of the C-23-C-24 C-C bond, yielding 1,23-(OH)2-24,25,26,27-tetranor-D3. Beckman et al. (2) have demonstrated that all of these steps are carried out by the multicatalytic P450cc24 monooxygenase. Further in vitro biochemical studies using the purified native TxtC monooxygenase should allow us to establish the precise role played by this enzyme in thaxtomin biosynthesis.
Acknowledgments
We thank K. T. Shanmugam for thoughtful discussions. We also acknowledge two anonymous reviewers, whose suggestions improved this manuscript.
This work was supported by USDA NRI grant 97-35303-4487. Financial support was also provided to F.G.H. through the USDA National Needs Fellowship Program.
REFERENCES
- 1.Babcock, M. J., E. C. Eckwall, and J. L. Schottel. 1993. Production and regulation of potato-scab-inducing phytotoxins by Streptomyces scabies. J. Gen. Microbiol. 139:1579-1586. [Google Scholar]
- 2.Beckman, M. J., P. Tadikonda, E. Werner, J. Prahl, S. Yamada, and H. F. DeLuca. 1996. Human 25-hydroxyvitamin D3-24-hydroxylase, a multicatalytic enzyme. Biochemistry 35:8465-8472. [DOI] [PubMed] [Google Scholar]
- 3.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. Nagaraja Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Bukhalid, R. A., S. Y. Chung, and R. Loria. 1998. nec1, a gene conferring a necrogenic phenotype, is conserved in plant-pathogenic Streptomyces spp. and linked to a transposase pseudogene. Mol. Plant-Microbe Interact. 11:960-967. [DOI] [PubMed] [Google Scholar]
- 5.Bukhalid, R. A., and R. Loria. 1997. Cloning and expression of a gene from Streptomyces scabies encoding a putative pathogenicity factor. J. Bacteriol. 179:7776-7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong, P. P., S. M. Podmore, H. M. Kieser, M. Redenbach, K. Turgay, M. Marahiel, D. A. Hopwood, and C. P. Smith. 1998. Physical identification of a chromosomal locus encoding biosynthetic genes for the lipopeptide calcium-dependent antiobiotic (CDA) of Streptomyces coelicolor A3(2). Microbiology 144:193-199. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, S. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, S. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Cupp-Vickery, J. R., and T. L. Poulos. 1995. Structure of the P450eryF involved in erythromycin biosynthesis. Nat. Struct. Biol. 2:144-153. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, M. T., S. F. Scarlata, and S. G. Sligar. 1985. High-pressure investigations of cytochrome P-450 spin and substrate binding equilibria. Arch. Biochem. Biophys. 240:456-463. [DOI] [PubMed] [Google Scholar]
- 10.Fouces, R., E. Mellado, B. Diez, and J. L. Barredo. 1999. The tylosin biosynthetic cluster from Streptomyces fradiae: genetic organization of the left region. Microbiology 145:855-868. [DOI] [PubMed] [Google Scholar]
- 11.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modelling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 12.Hanson, L. K., W. A. Eaton, S. G. Sligar, I. C. Gunsalus, M. Gouterman, and C. R. Connell. 1976. Origin of the anomalous Soret spectra of carboxycytochrome P-450. J. Am. Chem. Soc. 98:2672-2674. [DOI] [PubMed] [Google Scholar]
- 13.Healy, F. G., R. A. Bukhalid, and R. Loria. 1999. Characterization of an insertion sequence element associated with genetically diverse plant pathogenic Streptomyces spp. J. Bacteriol. 181:1562-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Healy, F. G., M. Wach, S. B. Krasnoff, D. M. Gibson, and R. Loria. 2000. The txtAB genes of the plant pathogen Streptomyces acidiscabies encode a peptide synthetase required for phytotoxin thaxtomin A production and pathogenicity. Mol. Microbiol. 38:794-804. [DOI] [PubMed] [Google Scholar]
- 15.Hu, P., J. Elliott, P. McCready, E. Skowronski, J. Garnes, A. Kobayashi, R. R. Brubaker, and E. Garcia. 1998. Structural organization of virulence-associated plasmids of Yersinia pestis. J. Bacteriol. 180:5192-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hui Bon Hoa, G., C. Di Primo, I. Dondaine, S. G. Sligar, I. C. Gunsalus, and P. Douzou. 1989. Conformational changes of cytochromes P-450cam and P-450lin induced by high pressure. Biochemistry 28:651-656. [DOI] [PubMed] [Google Scholar]
- 17.Hui Bon Hoa, G., C. Di Primo, M. Geze, P. Douzou, J. A. Kornblatt, and S. A. Sligar. 1990. The formation of cytochrome P-450 from cytochrome P-420 is promoted by spermine. Biochemistry 29:6810-6815. [DOI] [PubMed] [Google Scholar]
- 18.Jung, C. 1985. Quantum chemical explanation of the “hyper” spectrum of the carbon monoxide complex of cytochrome P-450. Chem. Phys. Lett. 113:589-596. [Google Scholar]
- 19.King, R. R., C. H. Lawrence, and L. A. Calhoun. 1992. Chemistry of phytotoxins associated with Streptomyces scabies, the causal organism of potato common scab. J. Agric. Food Chem. 40:834-837. [Google Scholar]
- 20.Lambert, D. H., and R. Loria. 1989. Streptomyces acidiscabies sp. nov. Int. J. Syst. Bacteriol. 39:393-396. [Google Scholar]
- 21.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26:283-291. [Google Scholar]
- 22.Loria, R., R. A. Bukhalid, B. A. Fry, and R. R. King. 1997. Plant pathogenicity in the genus Streptomyces. Plant Dis. 81:836-846. [DOI] [PubMed] [Google Scholar]
- 23.Mao, Y., M. Varoglu, and D. H. Sherman. 1999. Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chem. Biol. 6:251-263. [DOI] [PubMed] [Google Scholar]
- 24.Martinis, S. A., S. R. Blanke, L. P. Hager, S. G. Sligar, G. Hui Bon Hoa, J. J. Rux, and J. H. Dawson. 1996. Probing the heme iron coordination of pressure-induced cytochrome P420cam. Biochemistry 35:14530-14536. [DOI] [PubMed] [Google Scholar]
- 25.Molnar, I., J.-F. Aparicio, S.-F. Haydock, L.-E. Khaw, T. Schwecke, A. Konig, J. Staunton, and P.-F. Leadlay. 1996. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene 169:1-7. [DOI] [PubMed] [Google Scholar]
- 26.Motamedi, H., A. Shafiee, S.-J. Cai, S. L. Streicher, B. H. Arison, and R. R. Miller. 1996. Characterization of methyltransferase and hydroxylase genes involved in the biosynthesis of the immunosuppressants FK506 and FK520. J. Bacteriol. 178:5243-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman, L. M., and L. P. Wackett. 1995. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry 34:14066-14076. [DOI] [PubMed] [Google Scholar]
- 28.O'Keefe, D. H., R. E. Ebel, J. A. Peterson, J. C. Maxwell, and W. S. Caughey. 1978. An infrared spectroscopic study of carbon monoxide bonding to ferrous cytochrome P-450. Biochemistry 17:5845-5852. [DOI] [PubMed] [Google Scholar]
- 29.O'Keefe, D. P., and P. A. Harder. 1991. Occurrence and biological function of cytochrome P450 monooxygenases in the actinomycetes. Mol. Microbiol. 5:2099-2106. [DOI] [PubMed] [Google Scholar]
- 30.Omer, C. A., R. Lenstra, P. J. Little, C. Dean, J. M. Tepperman, K. J. Leto, J. A. Romesser, and D. P. O'Keefe. 1990. Genes for two herbicide-inducible cytochromes P-450 from Streptomyces griseolus. J. Bacteriol. 172:3335-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Omura, T., and R. Sato. 1964. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 239:2370-2378. [PubMed] [Google Scholar]
- 32.Omura, T., and R. Sato. 1967. Isolation of cytochromes P-450 and P-420. Methods Enzymol. 10:556-561. [Google Scholar]
- 33.Paul, K., H. Theorell, and A. Akeson. 1953. The molar light absorption of pyridine ferroprotoporphyrin (pyridine hemochromogen). Acta Chem. Scand. 7:1284-1287. [Google Scholar]
- 34.Poulos, T. L., B. C. Finzel, and A. J. Howard. 1987. High-resolution crystal structure of cytochrome P450cam. J. Mol. Biol. 195:687-700. [DOI] [PubMed] [Google Scholar]
- 35.Raag, R., S. A. Martinis, S. G. Sligar, and T. L. Poulos. 1991. Crystal structure of the cytochrome P-450CAM active site mutant Thr252Ala. Biochemistry 30:11420-11429. [DOI] [PubMed] [Google Scholar]
- 36.Ralston, L., S. T. Kwon, M. Schoenbeck, J. Ralston, D. J. Schenk, R. M. Coates, and J. Chappell. 2001. Cloning, heterologous expression, and functional characterization of 5-epi-aristolochene-1,3-dihydroxylase from tobacco (Nicotiana tabacum). Arch. Biochem. Biophys. 393:222-235. [DOI] [PubMed] [Google Scholar]
- 37.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez, A.-M., C. Olano, C. Mendez, C. R. Hutchinson, and J.-A. Salas. 1995. A cytochrome P450-like gene possibly involved in oleandomycin biosynthesis by Streptomyces antibioticus. FEMS Microbiol. Lett. 127:117-120. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez, R., N. Chinea, T. Lopez, G. Pons, and G. Vriend. 1998. Homology modeling, model and software evaluation: three related resources. Comput. Appl. Biol. Sci. 14:523-528. [DOI] [PubMed] [Google Scholar]
- 40.Schlichting, I., J. Berendzen, K. Chu, A. M. Stock, S. A. Maves, D. E. Benson, R. M. Sweet, D. Ringe, G. A. Petsko, and S. G. Sligar. 2000. The catalytic pathway of cytochrome P450cam at atomic resolution. Science 287:1615-1622. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu, H., E. Obayashi, Y. Gomi, H. Arakawa, S. Y. Park, H. Nakamura, S. Adachi, H. Shoun, and Y. Shiro. 2000. Proton delivery in NO reduction by fungal nitric-oxide reductase. Cryogenic crystallography, spectroscopy, and kinetics of ferric-NO complexes of wild-type and mutant enzymes. J. Biol. Chem. 275:4816-4826. [DOI] [PubMed] [Google Scholar]
- 42.Shirling, E. B., and D. Gottlieb. 1966. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16:313-340. [Google Scholar]
- 43.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 44.Stern, J. O., J. Peisach, W. E. Blumberg, A. Y. H. Lu, and W. Levin. 1973. A low-temperature EPR study of partially purified, soluble ferric cytochromes P-450 and P-448 from rat liver microsomes. Arch. Biochem. Biophys. 156:404-413. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trower, M. K., R. Lenstra, C. Omer, S. E. Buchholz, and F. S. Sariaslani. 1992. Cloning, nucleotide sequence determination and expression of the genes encoding cytochrome P-450soy (soyC) and ferredoxinsoy (soyB) from Streptomyces griseus. Mol. Microbiol. 6:2125-2134. [DOI] [PubMed] [Google Scholar]
- 47.van Berkel, W. J. H., and F. Muller. 1991. Flavin-dependent monooxygenases with special reference to p-hydroxybenzoate hydroxylase, p. 1-29. In F. Muller (ed.), Chemistry and biochemistry of flavoenzymes, vol. 2. CRC Press, Boca Raton, Fla. [Google Scholar]
- 48.Wang, Z.-X., S.-M. Li, and L. Heide. 2000. Identification of the coumermycin A1 biosynthetic gene cluster of Streptomyces rishiriensis DSM 40489. Antimicrob. Agents Chemother. 44:3040-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe, I., F. Nara, and N. Serizawa. 1995. Cloning, characterization and expression of the gene encoding cytochrome P-450sca-2 from Streptomyces carbophilus involved in the production of pravastatin, a specific HMG-CoA reductase inhibitor. Gene 163:81-85. [DOI] [PubMed] [Google Scholar]
- 50.Yu, C. A., and I. C. Gunsalus. 1974. Cytochrome P-450cam. II. Interconversion with P-420*. J. Biol. Chem. 249:102-106. [PubMed] [Google Scholar]