Abstract
Transcriptional regulator PcaU from Acinetobacter sp. strain ADP1 governs expression of genes for protocatechuate degradation (pca genes) as a repressor or an activator depending on the levels of the inducer protocatechuate and of its own gene. PcaU is a member of the IclR protein family. Here the DNA binding properties of the purified protein are described in terms of the location of the binding sites and the affinity to these sites. Native PcaU was purified after overexpression of the pcaU gene in Escherichia coli. It is a dimer in solution. The binding site in the pcaU-pcaI intergenic region is located between the two divergent promoters covering 45 bp, which includes three perfect 10-bp repetitions. A PcaU binding site downstream of pcaU is covered by PcaU across two palindromic sequence repetitions. The affinity of PcaU for the intergenic binding sites is 50-fold higher (dissociation constant [Kd], 0.16 nM) than the affinity for the site downstream of pcaU (Kd, 8 nM). The binding of PcaU was tested after modifications of the intergenic binding site. Removal of any external sequence repetition still allowed for specific binding of PcaU, but the affinity was significantly reduced, suggesting an important role for all three sequence repetitions in gene expression. The involvement of DNA bending in the regulatory process is suggested by the observed strong intrinsic curvature displayed by the pcaU-pcaI intergenic DNA.
The β-ketoadipate pathway is of central importance for growth of soil bacterium Acinetobacter sp. strain ADP1 on aromatic compounds, which comprise a considerable part of the biomass on earth in the form of lignin (21). In two separate sets of enzymatic reactions it catalyzes the successive breakdown of the starting metabolites protocatechuate and catechol into the common metabolites of the central carbon metabolism acetyl-coenzyme A (CoA) and succinyl-CoA, and the two branches of the pathway appear to underlie sophisticated specific regulation enabling efficient gene expression (5, 17, 35). Enzymes for the breakdown of protocatechuate are encoded by the pca genes, which form a cluster on the chromosome of Acinetobacter sp. strain ADP1. This cluster could be shown to form a supraoperonic chromosomal region by the discovery of other gene clusters adjacent to it encoding enzymes for pathways funneling more-complex aromatic compounds to protocatechuate (8, 9, 12, 13). The pca genes are likely to be expressed as a polycistronic transcript of at least 8 kbp from promoter pcaIp upstream of pcaI, the first structural gene as judged from the effect of a pcaIp mutation on pcaHG expression (16). This promoter is controlled in a complex manner by transcriptional regulator PcaU (17). Addition of protocatechuate or precursors thereof leads to a PcaU-dependent activation of gene expression, the level of which depends on the nature of the carbon source added (42). At the same promoter PcaU exerts a repressing function when no effector is added (42). In addition it is involved in regulation of its own gene expression. The pcaU gene is located upstream of the pca gene cluster and is transcribed in the opposite direction, forming a 282-bp intergenic region (Fig. 1). Promoter pcaUp is repressed 6- to 11-fold when cells contain functional PcaU (42).
FIG. 1.
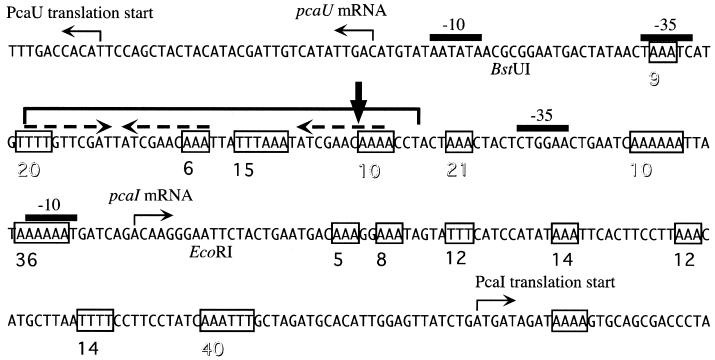
Intergenic region between pcaU and pcaI from Acinetobacter sp. strain ADP1. Dashed arrows, three 10-bp sequence repetitions located in the binding site of regulator protein PcaU; bracket, region protected from DNase I footprinting by PcaU; boxes, stretches of at least three successive adenosine residues. The distance from each stretch of As to the next one is given by a number underneath the sequence (printed open if the distance corresponds to one or multiple DNA helix turns). Vertical arrow, bending center for the 128-bp Bst UI-EcoRI fragment.
The PcaU protein has similarities in amino acid sequence to a number of proteins of wide distribution including proteins of members of the gram-positive bacteria, the Thermotogales, and the Archaea, indicating a well-conserved protein family. In all cases the function of these proteins as transcriptional regulators has either been shown or is predicted. The closest homologues are PobR from Acinetobacter sp. strain ADP1 (50% identical residues [8]) and PcaR from Rhodococcus opacus (33% identical residues [14]), from Pseudomonas aeruginosa (33% identical residues, GenBank accession no. AE004453), and from Pseudomonas putida (31% identical residues [36]). Specific binding of PcaU contained in an Escherichia coli cell extract to a DNA fragment consisting of part of the pcaU-pcaI intergenic region and to a DNA fragment containing DNA directly downstream of pcaU has been shown. Both fragments contain three sequence repetitions; it has been suggested that these are recognized and bound by PcaU (17). A first indication that full transcriptional activation requires the presence of all three sequence repetitions comes from the recent identification of spontaneous mutations in the pcaU-pcaI intergenic region (7). Nothing is known about the way in which PcaU interacts with this unusual potential binding site and how communication with RNA polymerase is accomplished. The only homologue that has been analyzed in more detail in this respect is PcaR from P. putida (18). Despite the high similarity of the two proteins and their potential (for PcaU) or proven (for PcaR) binding sites, they probably differ in activity, as judged from the location of the binding sites with respect to the regulated promoters. PcaR binding covers parts or all of the promoter signature (for the pcaR or pcaIJ promoter), whereas the suggested PcaU binding site is separated from the structural gene promoter (−35 signature) by 14 bp and from the pcaU gene promoter (−35 signature) by 4 bp (17, 18). Here, we describe the purification of the native PcaU protein and, qualitatively and quantitatively, the interaction with its DNA binding sites. Reducing the binding site by one of the external sequence repetitions reduced the affinity of PcaU for these modified sites but did not eliminate specific binding. The question of the biological function of the PcaU binding site downstream of the pcaU gene is addressed by determining the effect of its deletion on structural gene expression. Furthermore, the possible contribution to the process of gene expression by the topology of DNA within the pcaI-pcaU intergenic region is suggested by presenting data showing that this area is severely intrinsically bent.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
Bacteria and plasmids used in this study are listed in Table 1. Strains of E. coli were grown in Luria-Bertani medium (37) with efficient aeration at 37°C. Antibiotics were added as required (ampicillin at 100 μg/ml, rifampin at 200 μg/ml, kanamycin at 50 μg/ml). Acinetobacter strains were grown at 30°C on mineral medium as described earlier and in the presence of antibiotics if required (ampicillin at 200 μg/ml, kanamycin at 12 μg/ml) (42).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Acinetobacter strains | ||
| ADP1 | Wild type (strain BD413, ATCC 33305) | 26 |
| ADPU6 | PcaU binding site downstream of pcaU replaced by Kmr cassette | This study |
| E. coli strains | ||
| DH5α | General-purpose cloning strain | 20 |
| BL21(DE3) | F−ompT hsdSB(rB−mB−) gal dcm (DE3); gene for T7 RNA polymerase under control of lacUV5 integrated into the chromosome | 39 |
| Plasmids | ||
| pBend5 | Vector designed for circular permutation assay based on pBluescript SK(−) | 46 |
| pET-21(+) | Transcription vector with T7 promoter | 39 |
| pKOK6 | Contains promoterless lacZ-Kmr cassette | 28 |
| pZR15 | 7-kbp HpaII fragment containing pcaU inserted into pUC19 | 17 |
| pZR17 | 2,792-bp EcoRI fragment from pZR15 cloned into pUC19 | 17 |
| pAC7 | 2,792-bp EcoRI fragment from pZR15 cloned into pET-21(+), T7 promoter upstream of pcaU | This study |
| pZR18 | 2,618-bp HindIII-HpaII fragment containing pcaU inserted into pUC19 | 17 |
| pAC25 | Corresponds to pZR18 after removal of 218-bp SwaI-Eco47III fragment | This study |
| p5/128 | 128-bp EcoRI-BstUI fragment of the pcaU-pcaI intergenic region in HpaI site of pBend5 | This study |
| pAC29 | pZR17 with 130-bp BstBI fragment replaced by Kmr cassette from pKOK6 | This study |
| pAC48 | Contains pcaI′-pcaU, lacZ-Kmr cassette from pKOK6 fused to pcaI′ on vector pRK415 | P. Patz and U. Gerischer, unpublished results |
| pAC52 | 130-bp BstBI fragment deleted from pAC48 | This study |
| p5DR | Intergenic PcaU binding site without the external inverted 10-bp sequence repetition on vector pBend5 | This study |
Gel retardation assay, determination of Kd for the protein-DNA interaction.
The DNA probes (a 131-bp BstBI fragment from plasmid pZR17 for the binding site downstream of the pcaU gene and a 214-bp XmnI-EcoRI fragment from plasmid pZR17 for the intergenic binding site) were generated by restriction digestion and radiolabeled at 3′ ends with the Klenow fragment of DNA polymerase or at 5′ ends with T4 DNA polymerase. The labeled fragments were purified with the NucleoSpin extract kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany). The probe, at 10,000 to 15,000 cpm (0.3 to 0.6 nM in the assay) was incubated with various amounts of protein in a mixture containing 50 mM Tris-HCl, pH 8.0, 50 mM potassium chloride, 5 mM magnesium chloride, 1 mM EDTA, 0.01% (wt/vol) bovine serum albumin, 10% (vol/vol) glycerol, 1 mM dithiothreitol, and 2 μg of poly(dI-dC) with a total volume of 20 μl for 15 min at 30°C. For all interactions tested, controls for specificity were included. Addition of 1 μg of nonspecific DNA did not alter the retardation, whereas addition of 0.1 μg of unlabeled probe prevented the shift. The samples were separated on a 1.2 or 1.5% (wt/vol) agarose gel in 0.5× TBE buffer (45 mM Tris base, 45 mM boric acid, 1 mM EDTA, pH 8). After electrophoresis, the gel was dried and radioactivity was detected with a Bio Imager Fujix BAS (Fuji Photo Film Co., Ltd., Tokyo, Japan). The software MacBAS (Fuji Photo Film Co., Ltd.) served to quantify the signals. For the determination of the dissociation constant (Kd) the unretarded part of the probe as a percentage of the total probe employed in one assay was plotted against the concentration of PcaU protein in the assay. The Kd was the PcaU concentration where 50% of the probe was retarded. This value was visually determined from the curves. The concentration of the probe was significantly lower than the PcaU concentration (4). Each Kd determination was repeated at least three times, and the standard deviation was 50% at maximum.
DNase I footprinting.
Radiolabeled DNA probes for the intergenic PcaU binding site were prepared as described above. For the binding site downstream of pcaU a 127-bp SspI-XbaI fragment was isolated after restriction enzyme cleavage of plasmid pZR17. Footprinting reactions were carried out in a total volume of 200 μl with 10,000 to 15,000 cpm of probe and 5 to 500 ng of protein in a mixture containing 25 mM Tris-HCl, pH 8.0, 25 mM potassium chloride, 15 mM magnesium chloride, 5 mM calcium chloride, 1 mM EDTA, 0.01% (wt/vol) bovine serum albumin, 1 mM dithiothreitol, and 2 μg of poly(dI-dC). This mixture was incubated for 30 min at 30°C before the addition of 5 μl of a solution of 5 to 10 ng of DNase I in 50 mM Tris-HCl, pH 7.2-10 mM magnesium sulfate-1 mM dithiothreitol-25% (vol/vol) glycerol. After 2 min at room temperature digestion was stopped by adding 700 μl of 90% (vol/vol) ethanol, 350 mM ammonium acetate, and 5 μg of tRNA. DNA was precipitated for 20 min at −20°C, centrifuged, washed twice with 70% (vol/vol) ethanol, and dried. The pellet was dissolved in loading buffer (95% [vol/vol] formamide, 10 mM EDTA, 0.1% [wt/vol] bromophenol blue, 0.1% [wt/vol] xylene cyanol) and denatured for 10 min at 90°C prior to electrophoresis. Samples were run alongside a dideoxy sequencing ladder of the same fragment on a 6 or 8% polyacrylamide gel containing 8 M urea with 1× TBE buffer (90 mM Tris base, 90 mM boric acid, 2 mM EDTA). The gel was soaked for 10 min in 10% (vol/vol) acetic acid and dried, and radioactivity was detected with a Bio Imager Fujix BAS (Fuji Photo Film Co., Ltd.).
Overexpression of native PcaU.
A 2,792-bp EcoRI fragment containing the pcaU gene and 175 bp of upstream DNA was isolated from plasmid pZR15 and cloned into vector pET-21(+). In the resulting plasmid, pAC7, the T7 promoter of the vector was located upstream of the pcaU gene. Transformation of plasmid pAC7 into E. coli BL21(DE3) enabled expression of the chromosomally encoded T7 RNA polymerase, which is under the control of lacUV5, and thereby specific expression of PcaU. In initial purification attempts copurification of DNase activity was observed. To suppress the simultaneous expression of E. coli proteins, the bacterial RNA polymerase was inhibited by addition of rifampin to the medium. The cells were grown in Erlenmeyer flasks in Luria-Bertani medium. IPTG (isopropyl-β-d-thiogalactopyranoside) was added at an optical density at 600 nm of 0.8; addition of rifampin was 30 min after the induction. The cultures were incubated for another 3 h, cooled on ice for 10 min, and then harvested by centrifugation. The cells were washed with 50 mM Tris-Cl, pH 8.0-2 mM EDTA-5% (wt/vol) glycerol and stored at −20°C if not processed immediately.
Purification of PcaU.
Cells were suspended in 50 mM Tris-HCl, pH 8.0-2 mM EDTA-5% (wt/vol) glycerol-0.5 mM phenylmethylsulfonyl fluoride and disrupted by ultrasonication. The first two chromatography steps were performed on a BioCAD perfusion chromatography workstation (Perseptive Biosystems, Framingham, Mass.). The extract was cleared by centrifugation (50,000 × g, 15 min, 4°C) and injected onto an anion exchange column (POROS 20 QE; Perseptive Biosystems) that had been equilibrated with 50 mM Tris-HCl, pH 8.0. The column was washed with the same buffer, and the proteins were eluted by using a gradient of 0 to 1 M NaCl within 50 column volumes. PcaU-containing fractions were identified by performing the gel retardation assay using a 216-bp XmnI-EcoRI DNA fragment with the presumptive pcaU-pcaI intergenic PcaU binding site. They were desalted by dialysis and then subjected to chromatography on immobilized heparin (POROS 20 HE), which had been equilibrated with 50 mM Tris-HCl, pH 8.0. After a washing with 10 column volumes, elution was done in two steps at 0.2 and at 0.5 M NaCl. After being identified, PcaU-containing fractions were desalted and concentrated by dialysis against a mixture containing 10% (wt/vol) polyethylene glycol 20000, 50 mM Tris-HCl, pH 8.0, 2 mM EDTA, and 10% (vol/vol) glycerol and stored at −20°C. Finally the PcaU-enriched preparation was subjected to gel filtration by fast-protein liquid chromatography on Superdex 75 (Amersham Pharmacia Biotech Europe GmbH, Freiburg, Germany) using 50 mM Tris-HCl, pH 8.0-150 mM KCl-5% (vol/vol) glycerol for elution. That the PcaU protein was the major peak was confirmed by a gel retardation assay. The protein solution was concentrated using Microcon centrifugal filter devices (YM-10; Millipore, Eschborn, Germany) and was stored at −20°C after being adjusted to 50% (vol/vol) glycerol-2 mM dithiothreitol-1 mg of glycine/ml.
Protein determination, SDS-PAGE, and size determination of the native protein in solution.
Protein concentrations were determined by using the methods of Markwell et al. (32) or Bradford (3). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (30). Gel filtration on Superdex 75 (Amersham Pharmacia Biotech Europe GmbH) was used for size determination of the native PcaU protein. The system was calibrated by determining the elution volume for globular standard proteins RNase (molecular weight [MW], 13,700), chymotrypsinogen A (MW, 25,000), ovalbumin (MW, 43,000), and albumin (MW, 67,000) (Amersham Pharmacia Biotech Europe GmbH) in 50 mM Tris-HCl, pH 8.0. Kav values were calculated according to equation Kav = Ve − V0/Vt − V0, where Ve is the elution volume for the protein, V0 is the column void volume (equal to the elution volume for blue dextran 2000), and Vt is the total bed volume.
Deletion of the PcaU binding site downstream of the pcaU gene.
Acinetobacter sp. strain ADPU6 with a deletion of the PcaU binding site downstream of the pcaU gene was constructed as follows. Plasmid pZR17 was incubated with restriction endonuclease BstBI, and the biggest fragment was isolated. A DNA fragment carrying a Kmr gene and a terminator from phage fd was prepared by treating plasmid pKOK6 with restriction endonucleases BamHI and EcoRI and was gel purified. Both fragments were treated with the Klenow fragment of DNA polymerase I to produce blunt ends and incubated together with T4 DNA ligase. After transformation in E. coli DH5α, selection for ampicillin and kanamycin resistance led to identification of plasmid pAC29, which was verified by restriction analysis. pAC29 was cleaved with SnaBI and MscI, releasing a fragment containing the modified site with flanking ADP1 wild-type sequences (169 and 724 bp, respectively). Acinetobacter sp. strain ADP1 was transformed with the fragment mixture, and transformants were selected on plates with succinate mineral agar containing kanamycin. Sensitivity to ampicillin was tested to exclude cointegration of the vector. PCR with oligonucleotides 5′-TGAATCAGATCGTATGGC-3′ and 5′-AAACCACCAATCAGGATG-3′ was used to verify new strain ADPU6. In a second approach, the PcaU binding site downstream of the pcaU gene was deleted from a plasmid (pAC48) that contained the complete pcaU gene plus 1,410 bp downstream of pcaU, the pcaU-pcaI intergenic region, the first 92 bp of pcaI, and a lacZ-Kmr cassette for determination of the activity of pcaIp. pAC48 was treated with BstBI, and the resulting fragment mixture was exposed to T4 DNA ligase. After transformation of the ligation products into E. coli, strains with the desired plasmid, pAC52, were identified by the absence of the XbaI restriction site, which was contained on the 130-bp BstBI fragment to be deleted.
Modification of the pcaI-pcaU intergenic PcaU binding site.
Plasmid pAC25 containing a PcaU binding site without the direct repetition of the 10-bp sequence found three times within the PcaU binding site was created by removal of a 218-bp SwaI-Eco47III fragment from plasmid pZR18. Plasmid pBend5DR was created by hybridizing oligonucleotide 5′-TATCGAACAAATTATTTAAATATCGAACAAAACCTACTAAACTAC-3′ to the respective complementary oligonucleotide and cloning the product into the HpaI cloning site of vector pBend5 after phosphorylation using T4 polynucleotide kinase. The resulting plasmids were confirmed by restriction analysis or sequence analysis.
Determination of DNA curvature.
DNA curvature was determined in the circular permutation assay (43). A 128-bp EcoRI-BstUI fragment including most of the DNA between the two transcriptional start sites of pcaI and pcaU was cloned into the HpaI site of bending vector pBend5 after converting the single-stranded DNA resulting from the EcoRI cut into double-stranded DNA with the Klenow fragment of DNA polymerase I. The construct was verified and its orientation was determined by sequence analysis. Permutated fragments were produced by digestion with the restriction enzymes contained in the permutation element and analyzed on 8% acrylamide gels in 1× TBE buffer, pH 7.5, at 4°C and a voltage of 7 V/cm. The relative mobilities of the fragments (mobility of a fragment divided by mobility of an unbent control fragment of identical length) were plotted against the number of base pairs of permutation element DNA fused to the test DNA in the direction of the HindIII restriction site of pBend5. Polynomials of fourth order best described the plotted data, and tangents were laid into their turning points. The crossing of these tangents was at the distance of the fragment that had the bending center in the middle of the fragment. The bending angle (α) was determined according to the empirical relationship μM/μE = cos(α/2), where μM is the mobility of a fragment with a central curvature and μE is the mobility with the bend at the end (41). Since the mobility of a fragment with the bend at the end is comparable to the mobility of an unbent fragment, relative mobility was used for μM/μE. Computational analysis of the DNA topology was done using the program DIAMOD (11).
DNA sequence determination.
DNA sequence determination was performed with a SequiTherm Excel Long-Read DNA sequencing kit LC (Biozym Diagnostik GmbH, Oldendorf, Germany) by using primers with the fluorescent label IRD800 (MWG-Biotech GmbH, Ebersberg, Germany). Electrophoresis was done on a LI-COR DNA sequencer 4000L, and the accompanying software was used for annotation (MWG-Biotech GmbH). Sequence reactions used as a size standard for DNase I footprints were performed with a T7 sequencing kit (Amersham Pharmacia Biotech Europe) and 35S-dATP (15).
RESULTS
Overexpression and purification of native PcaU.
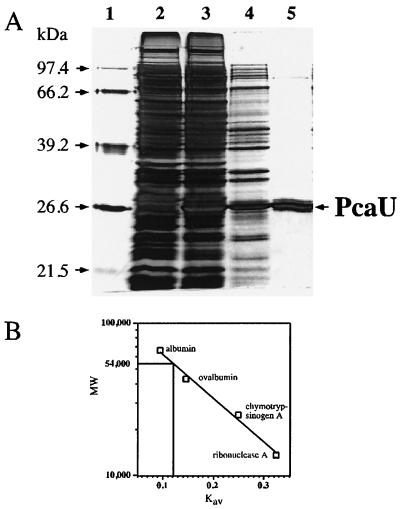
To obtain stable overexpression of PcaU, T7 polymerase-based expression was used (39, 40). The pcaU gene was cloned into transcription vector pET-21(+) under the control of the T7 promoter, creating plasmid pAC7. After transformation into host strain E. coli BL21(DE3) expression of PcaU could be directed by activation of the lacUV5 promoter controlling the gene for T7 RNA polymerase by addition of IPTG. Cell extracts prepared from such induced cultures contained significant amounts of an additional 28,000-Da protein (Fig. 2) and showed specific binding of a 216-bp XmnI-EcoRI DNA fragment containing the PcaU binding site (data not shown). The recombinant PcaU protein was purified to near homogeneity in a three-step procedure employing anion exchange chromatography, affinity chromatography on heparin, and gel filtration as detailed in Materials and Methods (Fig. 2). The native protein had a molecular mass of 54,000 Da as determined by using globular reference proteins for calibration (Fig. 2). From these data it is concluded that the oligomeric structure of the PcaU protein in solution is a homodimer.
FIG. 2.
(A) Denaturing SDS-PAGE from fractions of the purification of native PcaU after overexpression in E. coli. Lane 2, cell extract of the host strain with pET-21(+) without insert; lane 3, cell extract of the strain overproducing PcaU; lane 4, fraction after anion exchange chromatography; lane 5, fraction after affinity chromatography on heparin resin; lane 1, size standard. A low-molecular-weight protein still present in the preparation after chromatography on heparin was separated from PcaU by gel filtration (not shown). (B) Size determination of the native PcaU protein by gel filtration on Superdex 75. Squares, Kav values for reference proteins of known size; vertical line, respective value for PcaU.
Determination of the binding sites of PcaU.
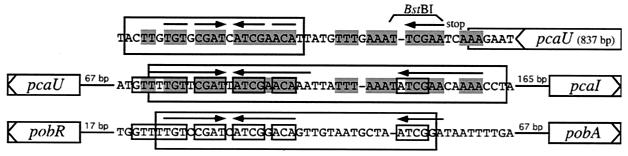
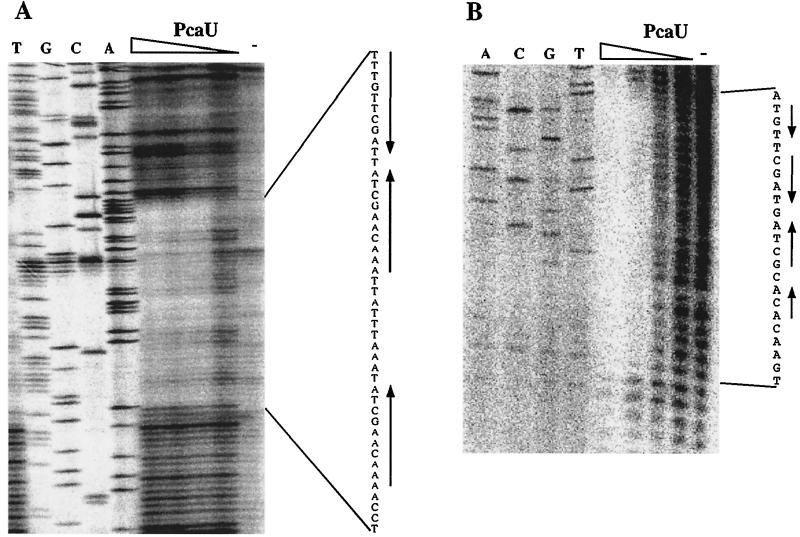
A previous investigation had revealed two PcaU binding sites, one in the 282-bp intergenic region between pcaI and pcaU (detected by retardation of a 216-bp XmnI-EcoRI DNA fragment) and a second one downstream of the pcaU gene (detected by retardation of a 300-bp PCR fragment) (17). Identification of a sequence motif within the intergenic DNA, which consisted of three perfect 10-bp sequence repetitions two of which formed a palindrome, led to the hypothetical formulation of a PcaU binding site. The presumption was strengthened by the finding that DNA directly downstream of the pcaU gene contained a motif which showed high similarity to the intergenic motif (33 out of 43 nucleotides identical after introduction of one gap corresponding to 1 nucleotide). In addition, DNA bound by the PobR protein, necessary for expression of the pobA gene, also contained a similar motif (22 out of 37 nucleotides identical after introduction of one gap corresponding to 1 nucleotide) (Fig. 3). The purified PcaU protein was used to reveal its binding site in the pcaU-pcaI intergenic region as well as downstream of pcaU. In both cases the presumed sites could be verified (Fig. 4). Bound PcaU covered 45 bp within the pcaU-pcaI intergenic DNA including the three 10-bp sequence repetitions (Fig. 1 and 3). For the binding site downstream of pcaU the footprint of PcaU was found to include the palindrome but did not extend to include the direct sequence repetition (Fig. 3 and 4).
FIG. 3.
Comparison of the two PcaU binding sites characterized here and the binding site of the closest homologue of PcaU, PobR from Acinetobacter sp. strain ADP1. Genes are represented as boxes with arrowheads inside indicating the direction of transcription. Arrows above sequence, sequence repetitions. Sequence identity between the two PcaU binding sites is highlighted by shaded boxes; identity between the pca and pob intergenic binding sites is shown by small open boxes. Large open boxes indicate the areas protected by the respective regulators in DNase I footprinting experiments.
FIG. 4.
Determination of the PcaU binding sites by DNase I footprinting in the pcaU-pcaI intergenic region (A) and downstream of pcaU (B). PcaU was added in various amounts (20, 10, 5, and 2.5 ng)
Different affinities of PcaU for the two binding sites upstream and downstream of its gene.
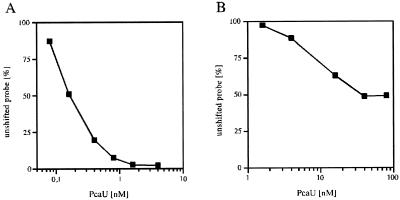
Despite the high similarity between the two recognized binding sites, they probably differ in their biological functions. Here, the affinities of PcaU for the sites were compared by employing a DNA retardation assay. The Kd of PcaU for the pcaU-pcaI intergenic binding site was 0.16 nM, and the Kd for the PcaU binding site downstream of its gene was 8 nM (Fig. 5). Thus the affinity of PcaU for the downstream binding site was 50 times lower than that for the pcaU-pcaI intergenic binding site.
FIG. 5.
Determination of the Kd for PcaU binding to the pcaU-pcaI intergenic region (A) and to the binding site downstream of pcaU (B). The Kd was the PcaU concentration at which 50% of the DNA was bound. For the downstream binding site, a maximum of 50% of the total probe was bound under any condition; thus the Kd was the PcaU concentration at which 25% of the DNA was bound.
The PcaU binding site downstream of the pcaU gene does not significantly influence the expression of the structural gene cluster.
Identification of a second functional binding site downstream of the pcaU gene poses the question of its biological function. Most likely would be an involvement of PcaU binding to this site in the regulation of the structural gene cluster or of the pcaU gene itself. To explore the first possibility, a strain that differed from the wild type in that the downstream binding site was removed on a 130-bp fragment and replaced with a kanamycin resistance cassette was constructed (see Materials and Methods). pca gene expression by that strain was compared with that by the wild type on a medium that required the respective gene products (mineral medium with quinate as the carbon source). There was no significant difference in growth between the two strains (data not shown). To get a more detailed picture of the expression level itself, we used a heterologous system of monitoring PcaU-dependent pca gene expression. PcaU expressed in E. coli in small amounts functions in concert with E. coli RNA polymerase at pcaIp in the same way it does in Acinetobacter sp. strain ADP1 as shown by using a transcriptionally fused lacZ cassette (P. Patz and U. Gerischer, unpublished results). For these studies a fragment of Acinetobacter DNA which contained the complete pcaU gene and 1,785 bp downstream of it, the complete pcaU-pcaI intergenic region, and the first 92 bp of the first structural gene, pcaI, was used. The pcaI gene was transcriptionally fused with reporter gene lacZ. A 130-bp fragment of DNA containing the downstream PcaU binding site was removed from the reporter construct. The expression pattern of this construct (pAC52) was indistinguishable from that of the respective wild-type construct (data not shown).
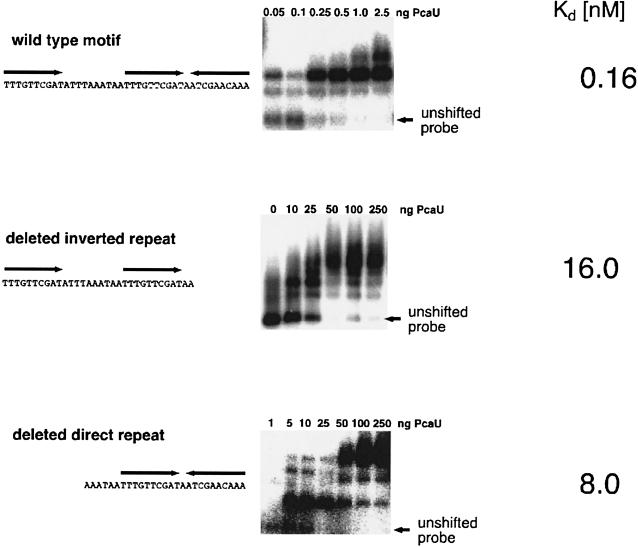
Removal of the external sequence repetitions does not eliminate the specific interaction of PcaU with the binding site but decreases its affinity.
The PcaU binding sites contain three 10-bp sequence repetitions, two in a palindromic order and, separated by 10 bp, a direct repetition. To investigate the influence of the removal of one of these repeated sequences on PcaU binding, modified DNA fragments were constructed and tested for PcaU binding. A 212-bp BamHI-XmnI fragment containing only the palindrome was isolated from plasmid pAC25. A 287-bp HindIII-EcoRI fragment from plasmid pBend5DR contained the PcaU binding site without the external palindromic sequence repetition. Gel retardation assays with the purified PcaU protein were performed. In both cases PcaU still specifically bound the modified binding sites. For both modifications the affinity of PcaU for the DNA site decreased considerably (Fig. 6). Kd for the direct sequence repetition was 16 nM, and that for the palindrome was 8 nM. Thus Kds of both shortened PcaU binding sites are in a range comparable to the Kd for the binding site downstream of the pcaU gene.
FIG. 6.
Qualities of purified PcaU binding to modified intergenic binding sites. The relevant part of the respective DNA fragment used for the gel retardation assay is shown on the left. The wild-type motif was contained on a 214-bp EcoRI-XmnI fragment, the motif containing only the direct sequence repetitions was contained on a 45-bp fragment cloned into plasmid pBend5, and the motif containing only the palindrome was contained on a derivative of pZR18 after removal of the 218-bp SwaI-Eco47III fragment containing the direct sequence repetition. Results of the respective gel retardation assays are shown on the right. The amount of unshifted probe was plotted against the respective PcaU concentration and served for determination of Kd.
The pcaI-pcaU intergenic region displays strong intrinsic bending.
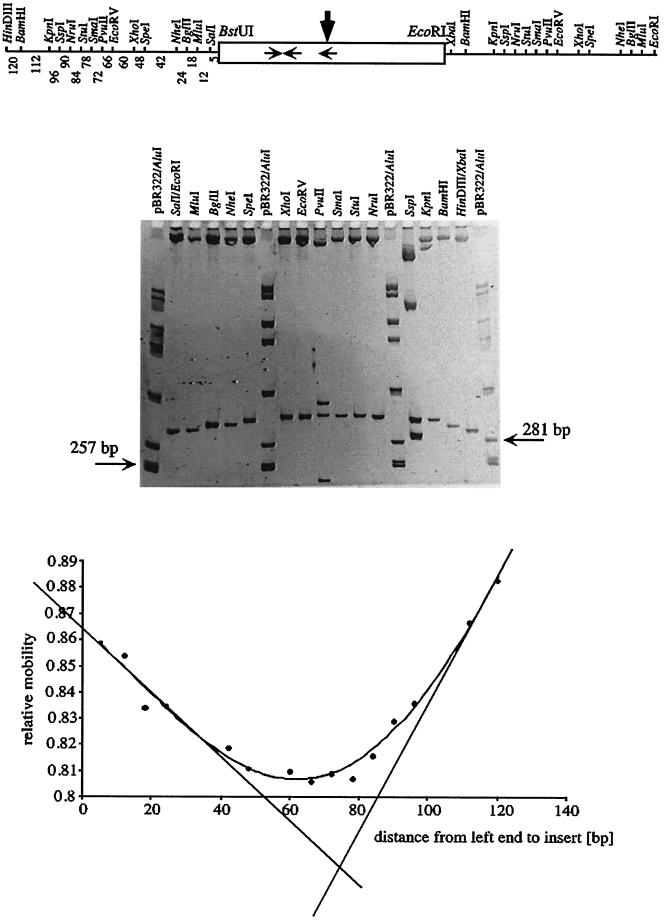
The intergenic DNA between the pcaU gene and the pcaI gene contains multiple polyadenosine nucleotides (Fig. 1). Such sequences are known to cause intrinsic bending of DNA, particularly when multiple poly(A) stretches (with each individual one at least 4 nucleotides long) are in phase with the turns of the DNA helix (10.5 bp) (19). Previous data indicated that there may be physical deviation from straight topology in this region (A. Segura and L. N. Ornston, unpublished results). Therefore the pcaI-pcaU intergenic region was investigated in more detail in this respect by employing the circular permutation assay (43). This assay is based on the observation of retardation of a bent DNA fragment through a native polyacrylamide gel in comparison with that of an unbent fragment of identical length (31). The extent of retardation depends on the position of the bend, and this can be exploited to determine the bending angle and the bending center (41, 43). Positioning the DNA sequence under investigation at different positions on a series of fragments of identical lengths is simplified by the use of special vectors (46). Here a 128-bp fragment encompassing roughly the region between promoters pcaIp and pcaUp was cloned into vector pBend5 and used for the determination of DNA curvature as detailed in Materials and Methods. This DNA displayed strong intrinsic bending centered within the direct sequence repetition of the PcaU binding site with a bending angle of 72° (Fig. 7). Computational calculation of the topology of the respective 128-bp fragment also resulted in the prediction of strongly bent DNA with multiple bending centers (data not shown).
FIG. 7.
Determination of DNA bending within the 128-bp BstUI-EcoRI fragment from the pcaU-pcaI intergenic region (Fig. 1) by a circular permutation assay using bending vector pBend5. (Top) The relevant part of plasmid p5/128 is shown with the restriction sites of the permutation element and the test DNA as a box in the middle. Horizontal arrows, locations of the PcaU binding sites; vertical arrow, bending center revealed in this experiment. Numbers under the left half of the permutation element, distances in base pairs between the BstUI site of the test DNA and the left end of the fragment resulting from restriction digestion with the respective enzymes. These numbers were used as the distances from left end to insert in the graph (bottom). (Middle) Example of a gel containing samples of plasmid p5/128 after restriction cleavage with the enzymes indicated. The permutated fragments had a length of 255 bp; for comparison two fragments of the standard are indicated. (Bottom) Plot of the relative mobilities of the fragments against the distance between the left end of the permutated fragments and the test DNA enables the determination of the bending center. Relative mobilities are the averages of six gels; the error was between 0.7 and 1.5%.
DISCUSSION
As a prerequisite for a study of the interaction of transcriptional regulator PcaU from Acinetobacter sp. strain ADP1 with its DNA targets the protein was purified in its native state after overexpression in E. coli. On the basis of the appearance of the protein in denaturing polyacrylamide gels and upon gel filtration, it is suggested that the protein is a dimer in solution. This is an observation that is made frequently for regulatory proteins, such as PcaR from P. putida and the Gal repressor from E. coli (18, 25). Both binding sites for PcaU on DNA upstream and downstream of the pcaU gene, suggested earlier based on the threefold repetition of a sequence motif which is also found in the binding sites of other regulatory proteins, could be verified by DNase I footprinting experiments. In the pcaU-pcaI intergenic region, all three sequence repetitions are included in the DNA covered by PcaU whereas the motif directly downstream of the pcaU gene was protected from DNase I digestion only in the palindromic area (Fig. 3). The fact that the sequence repetitions are not as well preserved as in the intergenic DNA may be the reason for this observation and may also explain the lower affinity of PcaU for the downstream motif than for the intergenic PcaU binding motif. No specific binding of PcaU to the binding site for the closely related PobR protein located in the pobR-pobA intergenic region from the same organism could be detected (data not shown). Taking into consideration that the downstream PcaU binding site binds PcaU specifically without involvement of a third direct sequence repetition, specificity for PcaU binding must be determined by the palindromic structure alone. The sequences of the palindrome in the downstream PcaU binding site differ in 3 out of 20 positions from that of the pcaU-pcaI intergenic binding site; the palindrome of the PobR binding site displays the same number of mismatches, two of which are in the same positions as mismatches between the two PcaU binding sites (Fig. 3). Thus it is suggested that DNA sequence requirements are pronounced. In this context the discovery of a protein that binds to both the intergenic PcaU binding site and to the PobR binding site appears remarkable in that the protein is a derivative of PobR with the single-amino-acid substitution T57A producing a stretch of six residues that are identical within the presumed helix-turn-helix motifs of PcaU and PobR (27).
The Kd value determined for the intergenic PcaU binding site is comparable to the Kd found for PcaR binding to the pcaIJ promoter (0.16 and 0.13 nM, respectively), whereas the affinity of PcaU for the binding site downstream of the pcaU gene is 50-fold lower. The existence of an additional regulator binding site is reminiscent of what is found for other regulatory systems where multiple binding sites for the same regulator exist and contribute to gene expression, as in the well-characterized systems governed by AraC and LacI (33, 38) and the regulation brought about by DeoR from E. coli (6). In all these cases, simultaneous binding of two or more DNA binding sites by the respective regulator protein consisting of a dimer or oligomer and loop formation of the intervening DNA as a consequence have been described. Here we explored the possibility of a contribution of the downstream binding site to the regulated expression of the pca genes by deleting it. In two different approaches we could not detect any difference in pca gene expression or in growth between situations in which the downstream binding site was present and those in which it was absent. The nature of the downstream binding site is very unlikely to be random due to the site's complexity. The downstream binding site may have a function which affects pca gene expression but which is so subtle that it could not be detected in the two approaches we chose. Alternatively the downstream binding site may contribute to the strong autoregulatory repression observed for the pcaU gene, which has not been tested (42).
The intergenic PcaU binding site contains three perfect 10-bp sequence repetitions, which are all protected from DNase I digestion by PcaU as demonstrated here (Fig. 3 and 4). The PcaU binding site downstream of its gene displays a similar structure with less-well-preserved sequence repetitions, and there only binding of PcaU to the palindrome could be demonstrated. To determine the function of the individual sequence repetitions, we tested the binding of PcaU to modified intergenic PcaU binding sites missing the direct sequence repetition or the external inverted repetition. Sites with either modification still bound PcaU specifically but with a lower affinity. Indication for a physiological function of these external sequence repetitions comes from the observed growth phenotypes of two spontaneous mutant strains, one containing a C→T base substitution within the external sequence repetition and the second one with a deletion of the direct sequence repetition. The respective strains show a cold-enhanced block in protocatechuate catabolism (7, 42). Thus both PcaU binding and gene expression are impaired when one of the external sequence repetitions is modified or deleted, and possibly pca gene expression is not inducible at all. The arrangement of three sequence repetitions in one palindrome and one direct repetition spaced by 10 bp from the palindrome is unusual for regulator binding sites in that the most abundant motif is a twofold-repeated sequence arranged as a palindrome. A direct repetition of two sequences as observed for araO1 and araI is rare (22, 34). Binding sites structured similarly to the PcaU binding sites are found only for regulators with close homology to PcaU such as the PobR and PcaR binding sites (Fig. 3) (10, 18). We are aware of one other example of a similarly structured regulator binding site. The DeoR repressor from Bacillus subtilis, regulating genes required for deoxyribonucleoside and deoxyribose utilization, was shown to protect an area of 43 bp that contains three repetitions of a 6-bp sequence arranged in a palindrome and a direct sequence repetition from DNase I digestion (45). For the interaction of this regulator, which shows no homology to PcaU, with its binding site all three sequence repetitions are required (44, 45). This differs from the DNA binding quality of PcaU described here. Such data are not available for proteins closely related to PcaU (PobR and PcaR) (10, 18), but comparing the architectures of the respective regulatory regions reveals significant differences for each individual regulator (18). These differences may be an indication of differences in the way in which the regulators interact with RNA polymerase.
We show here that the stretch of DNA between the two promoters of the pcaU-pcaI intergenic DNA is intrinsically bent by 72° and that the center of this strong curvature is within the external sequence repetition of the PcaU binding motif. The assay used can only reveal the sum of the effects of multiple bends if there is more than one bend. The DNA fragment assayed was 128 bp long and contained multiple polyadenosine stretches spaced by roughly one (10 or 11 bp) or two helix turns (20 to 22 bp) (Fig. 1). Thus it is possible that this stretch of DNA contains multiple curved regions. This assumption is supported by the computational prediction, which revealed three bends, one close to the −10 site of pcaIp and two near the PcaU binding site. This observation is in agreement with the prediction of intrinsic bending for a substantial fraction of all promoters or the respective upstream regions of many mesophilic bacteria (2). The nonlinear DNA conformation may, for example, be necessary to correct for a nonoptimal spacing of the −10 and the −35 regions of pcaIp, a mechanism found to apply for SoxR and MerR and proposed to apply to PcaU homologue PcaR (1, 18, 23). Alternatively, the intrinsic DNA curvature may support a potential DNA looping brought about by the two PcaU binding sites as discussed above.
In addition to the potential function of curved DNA in the promoter area, PcaU-dependent regulation has other features in common with regulation by metalloregulator MerR and by redox-regulated SoxR of E. coli. These proteins act as repressors or as activators of the respective structural gene promoters, depending on the absence or presence of the respective inducing signal, an unusual quality of a regulatory protein shared by PcaU (24, 29, 42). In contrast to the binding of MerR and SoxR, which overlap the structural gene promoters creating an unusual situation for activator proteins, PcaU binding between positions −48 and −92 with respect to the transcriptional start of the structural genes is observed. The specific binding of PcaU to the intergenic binding site was observed in the absence or presence of the inducer protocatechuate (100 μM), thus allowing the speculation that both repression and activation at pcaIp are based on a specific interaction between PcaU and RNA polymerase. The binding of the inducer must change the nature of this interaction in a way that leads from a repression to a strong activation of RNA polymerase at this promoter. The mechanism leading to repression by PcaU at pcaUp may be similar to that leading to repression at pcaIp since the PcaU binding site (positions −41 to −85 with respect to the transcriptional start site of pcaUp) does not overlap with this promoter either.
Acknowledgments
This research was supported by grant GE 672/3-1 from the Deutsche Forschungsgemeinschaft.
We thank S. Adhya for plasmid pBend5. Iris Steiner contributed to this work with brilliant technical assistance. We thank P. Dürre for critical reading of the manuscript.
REFERENCES
- 1.Ansari, A. Z., J. E. Bradner, and T. V. O'Halloran. 1995. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374:371-375. [DOI] [PubMed] [Google Scholar]
- 2.Bolshoy, A., and E. Nevo. 2000. Ecologic genomics of DNA: upstream bending in prokaryotic promoters. Genome Res. 10:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Carey, J. 1991. Gel retardation. Methods Enzymol. 208:103-117. [DOI] [PubMed] [Google Scholar]
- 5.Collier, L. S., G. L. Gaines, and E. L. Neidle. 1998. Regulation of benzoate degradation in Acinetobacter sp. strain ADP1 by BenM, a LysR-type transcriptional activator. J. Bacteriol. 180:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dandanell, G., K. Norris, and K. Hammer. 1991. Long-distance deoR regulation of gene expression in Escherichia coli. Ann. N. Y. Acad. Sci. 646:19-30. [DOI] [PubMed] [Google Scholar]
- 7.D'Argenio, D. A., A. Segura, P. V. Bünz, and L. N. Ornston. 2001. Spontaneous mutations affecting transcriptional regulation by protocatechuate in Acinetobacter. FEMS Microbiol. Lett. 201:15-19. [DOI] [PubMed] [Google Scholar]
- 8.DiMarco, A. A., B. Averhoff, and L. N. Ornston. 1993. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J. Bacteriol. 175:4499-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiMarco, A. A., B. A. Averhoff, E. E. Kim, and L. N. Ornston. 1993. Evolutionary divergence of pobA, the structural gene encoding p-hydroxybenzoate hydroxylase in an Acinetobacter calcoaceticus strain well-suited for genetic analysis. Gene 125:25-33. [DOI] [PubMed] [Google Scholar]
- 10.DiMarco, A. A., and L. N. Ornston. 1994. Regulation of p-hydroxybenzoate hydroxylase synthesis by PobR bound to an operator in Acinetobacter calcoaceticus. J. Bacteriol. 176:4277-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dlakic, M., and R. E. Harrington. 1998. DIAMOD: display and modeling of DNA bending. Bioinformatics 14:326-331. [DOI] [PubMed] [Google Scholar]
- 12.Elsemore, D. A., and L. N. Ornston. 1994. The pca-pob supraoperonic cluster of Acinetobacter calcoaceticus contains quiA, the structural gene for quinate-shikimate dehydrogenase. J. Bacteriol. 176:7659-7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsemore, D. A., and L. N. Ornston. 1995. Unusual ancestry of dehydratases associated with quinate catabolism in Acinetobacter calcoaceticus. J. Bacteriol. 177:5971-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eulberg, D., S. Lakner, L. A. Golovleva, and M. Schlömann. 1998. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 180:1072-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerischer, U., and P. Dürre. 2001. Primer design and primer-directed sequencing, p. 39-51. In C. A. Graham and A. J. M. Hill (ed.), DNA sequencing protocols, 2nd ed. Humana Press Inc., Totowa, New Jersey. [DOI] [PubMed]
- 16.Gerischer, U., and L. N. Ornston. 1995. Spontaneous mutations in pcaH and -G, structural genes for protocatechuate 3,4-dioxygenase in Acinetobacter calcoaceticus. J. Bacteriol. 177:1336-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerischer, U., A. Segura, and L. N. Ornston. 1998. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J. Bacteriol. 180:1512-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, Z., and J. E. Houghton. 1999. PcaR-mediated activation and repression of pca genes from Pseudomonas putida are propagated by its binding to both the −35 and the −10 promoter elements. Mol. Microbiol. 32:253-263. [DOI] [PubMed] [Google Scholar]
- 19.Hagerman, P. J. 1990. Sequence-directed curvature of DNA. Annu. Rev. Biochem. 59:755-781. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 21.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 22.Hendrickson, W., and R. Schleif. 1985. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc. Natl. Acad. Sci. USA 82:3129-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hidalgo, E., and B. Demple. 1997. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 16:1056-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hidalgo, E., V. Leautaud, and B. Demple. 1998. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. 17:2629-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh, M., and M. Brenowitz. 1997. Comparison of the DNA association kinetics of the Lac repressor tetramer, its dimeric mutant LacIadi, and the native dimeric Gal repressor. J. Biol. Chem. 272:22092-22096. [DOI] [PubMed] [Google Scholar]
- 26.Juni, E., and A. Janik. 1969. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum). J. Bacteriol. 98:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kok, R. G., D. A. D'Argenio, and L. N. Ornston. 1998. Mutation analysis of PobR and PcaU, closely related transcriptional activators in Acinetobacter. J. Bacteriol. 180:5058-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kokotek, W., and W. Lotz. 1989. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene 84:467-471. [DOI] [PubMed] [Google Scholar]
- 29.Kulkarni, R. D., and A. O. Summers. 1999. MerR cross-links to the alpha, beta, and sigma 70 subunits of RNA polymerase in the preinitiation complex at the merTPCAD promoter. Biochemistry 38:3362-3368. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Marini, J. C., S. D. Levene, D. M. Crothers, and P. T. Englund. 1983. A bent helix in kinetoplast DNA. Cold Spring Harbor Symp. Quant. Biol. 47:279-283. [DOI] [PubMed] [Google Scholar]
- 32.Markwell, M. A., S. M. Haas, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 33.Müller, J., S. Oehler, and B. Müller-Hill. 1996. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J. Mol. Biol. 257:21-29. [DOI] [PubMed] [Google Scholar]
- 34.Ogden, S., D. Haggerty, C. M. Stoner, D. Kolodrubetz, and R. Schleif. 1980. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc. Natl. Acad. Sci. USA 77:3346-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romero-Arroyo, C. E., M. A. Schell, G. L. Gaines III, and E. L. Neidle. 1995. catM encodes a LysR-type transcriptional activator regulating catechol degradation in Acinetobacter calcoaceticus. J. Bacteriol. 177:5891-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero-Steiner, S., R. E. Parales, C. S. Harwood, and J. E. Houghton. 1994. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J. Bacteriol. 176:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schleif, R. 1992. Regulation of the L-arabinose catabolic operon araBAD, p. 643-665. In S. L. McKnight and K. R. Yamamoto (ed.), Transcriptional regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. Methods Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 40.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. F., and A. Landy. 1988. Empirical estimation of protein-induced DNA bending angles: applications to λ site-specific recombination complexes. Nucleic Acids Res. 16:9687-9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trautwein, G., and U. Gerischer. 2001. Effects exerted by transcriptional regulator PcaU from Acinetobacter sp. strain ADP1. J. Bacteriol. 183:873-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, H.-M., and D. M. Crothers. 1984. The locus of sequence-directed and protein-induced DNA bending. Nature 308:509-513. [DOI] [PubMed] [Google Scholar]
- 44.Zeng, X., and H. H. Saxild. 1999. Identification and characterization of a DeoR-specific operator sequence essential for induction of dra-nupC-pdp operon expression in Bacillus subtilis. J. Bacteriol. 181:1719-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng, X., H. H. Saxild, and R. L. Switzer. 2000. Purification and characterization of the DeoR repressor of Bacillus subtilis. J. Bacteriol. 182:1916-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zwieb, C., and S. Adhya. 1994. Improved plasmid vectors for the analysis of protein-induced DNA bending. Methods Mol. Biol. 30:281-294. [DOI] [PubMed] [Google Scholar]