The family of nucleic acid (NA) strand separation enzymes known as helicases are found in all organisms and participate in a wide variety of cellular processes. The central reaction catalyzed is always the same: hydrolysis of a nucleoside triphosphate (NTP; usually ATP) is coupled to the separation of an NA duplex, be it DNA-DNA, DNA-RNA, or RNA-RNA. This central process is required in almost every aspect of NA metabolism in the cell, including chromosomal and plasmid replication, transcription, translation, RNA processing, and DNA recombination and repair (30, 46). This widespread usage may be seen by examining the cellular complement of helicases; for example, at least 12 putative DNA helicases have been identified in the genome of Escherichia coli, while it has been estimated that more than 2% of the Saccharomyces cerevisiae genome encodes helicase-related proteins (48).
Helicases may be classified in various ways. A common functional distinction is to group them by directionality. A given length of NA duplex is essentially symmetric, possessing dyad symmetry (ignoring base composition); thus, helicase polarity was originally specified by referring to the direction of the flanking single-strand (ss) region of NA usually required to initiate unwinding. Thus, 3′-5′ helicases require a 3′ tail on the substrate duplex while a 5′ tail is required for the 5′-3′ enzymes. It should be noted, however, that some helicases are capable of initiating unwinding from a blunt-ended duplex, such as the RecBCD system in E. coli (45, 57). In view of this, it is more appropriate to describe the directionality in terms of the strand upon which the enzyme translocates, a definition that is consistent with, but expands upon, the previously described system.
At the sequence level, helicases have been divided into five main groups (16). The largest of these groups are the superfamily I and II (SF1 and SF2) helicases, most of which have a 3′-5′ directionality. These all contain seven so-called helicase motifs, I, Ia, II, III, IV, V, and VI. In both superfamilies, motifs I and II are the highly conserved Walker A and B sequences characteristic of ATPases (63). The other motifs are generally less highly conserved and differ between the SF1 and SF2 proteins. SF3 enzymes, usually from DNA or RNA viruses, only contain three conserved motifs, I, II, and III, while other helicases, such as the E. coli DnaB-like hexameric helicases, form another, smaller, distinct group. A very small fifth group contains enzymes such as the bacterial transcription termination factor Rho. As the primary chromosomal replicative helicases in prokaryotes and (probably) eukaryotes, the hexameric helicases have been extensively studied but are beyond the scope of this review (see, for example, reference 37) and will only be briefly mentioned.
Recent work (39, 67) has shown that some helicases, such as the RuvB branch migration enzyme and MCM proteins, belong to the AAA+ family (33) rather than any of the previously described helicase superfamilies.
Analysis of genomic sequence data has identified considerable numbers of open reading frames containing some or all of the characteristic helicase motifs and allowed classification of the respective gene product into one of the above classes. A small number of these proteins have also been studied biochemically, and it is becoming increasingly clear that not all (in fact, perhaps only a minority) of these enzymes actually possess true helicase activity. One explanation for this is that the helicases are only functional as part of a multiprotein complex or require activation, for example, phosphorylation, as part of a control mechanism. Another reason for this limited activity is that the helicase motifs are actually characteristic of NTP-dependent NA translocases, that is, enzymes that are capable of moving unidirectionally along a piece of ss or double-strand (ds) NA. The molecular motor of the translocase itself may not necessarily be able to unwind any duplex encountered, but the helicase activity is conferred by additional protein domains (which do not contain the helicase motifs) peripheral to this central core. Moreover, the extra domains do not necessarily have to provide a strand separation functionality and may be utilized for other purposes. In this review, we will discuss this principle of modularity and show how a general-purpose molecular motor may be coupled to a variety of molecular activities, including NA unwinding.
GENERAL HELICASE MECHANISMS
The physical mechanism by which helicases move along an ss or ds stretch of NA has been extensively studied by both structural and biochemical methods. Although various models for this translocation have been proposed (1), the so-called inchworm (69) mechanism has emerged as the most plausible, especially in the light of recent work (reviewed in reference 29). This mechanism has been elucidated mainly through work on the SF1 and SF2 enzymes, which are generally monomeric, but it is possible that a modified version is applicable to the hexameric ring helicases as well (34, 49).
The overall action of the inchworm model may be likened to that of a snowplow pushed or pulled along the NA duplex in an NTP hydrolysis-coupled manner, mechanically forcing apart the duplex as it does so. The exact means by which the duplex is forced apart seem to differ, even among enzymes of the same superfamily, and some alternative strategies the enzymes use will be considered later in this review.
Whatever the exact details of the mechanism, one topic of much debate is whether the duplex unwinding is active or passive with respect to NTP hydrolysis. In other words, is the free energy of hydrolysis used for translocation and duplex separation or for translocation alone? The free energy of ATP hydrolysis under physiological conditions, ΔG0, is approximately −10 kcal/mol. Averaged over G · C and A · T pairs, the energy required to open a single nucleotide base pair has been estimated to be about +1.6 kcal/mol (9), demonstrating the favorable energetics of the coupled hydrolysis-base-opening reaction. Other estimates have suggested that from 2 to 4 (45) to 9 to 12 (27) bases could be opened by a single NTP hydrolysis event. However, is this actually necessary? At room temperature, the thermal fraying of a DNA duplex is significant, with some estimates suggesting that a single base at an ss-ds junction may open up at a rate of 1,000 times per second (7). It could be argued that it would be sufficient for a duplex-unwinding enzyme to wait for such fraying to occur and simply advance along the transiently denatured strand to prevent reannealing and so trap the unwound state. On a macroscopic scale, this kinetic trap model appears to enhance the rate of thermal fraying by reducing the rate of the reverse (reannealing) reaction while hydrolyzing NTPs purely for the purpose of translocation. However, recent biochemical (52) and structural (61) studies have shown that at least some helicases actively destabilize the NA duplex at the ss-ds junction and that the free energy of NTP hydrolysis powers both translocation and unwinding. This does not preclude the possibility that some helicases utilize a passive process, but intuition suggests that even a so-called passive process would require the helicase to sense and therefore contact a junction prior to thermal fraying and that this contact would probably affect the equilibrium of the ds-ss melting process. Moreover, some helicases (e.g., RecBCD) are capable of translocation at greater than 1 kb/s, a rate likely to be faster than the (passive) rate of duplex fraying.
TRANSLOCATION—A STRUCTURAL PERSPECTIVE
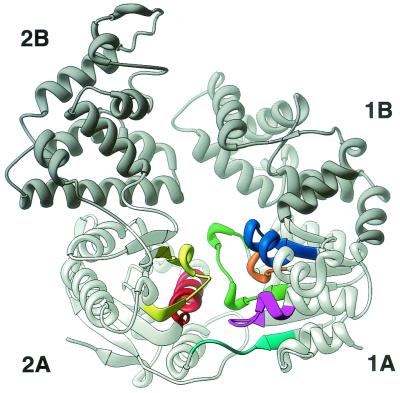
The first helicase crystal structure solved was that of the SF1 helicase PcrA from Bacillus stearothermophilus, both as the apo form and in complex with ADP (56). The structure revealed a monomeric enzyme with four prominent domains, designated 1A, 1B, 2A, and 2B (Fig. 1 and 2A). A striking feature was the structural similarity of domains 1A and 2A to the recombination strand exchange enzyme RecA from E. coli (55). These tandemly repeated domains were also found to contain all seven of the conserved SF1 helicase motifs, as well as the nucleotide-binding site. These findings led the authors to propose that all helicases would contain a RecA-like domain at some position in their structure (3). This has proved to be the case in all of the helicase structures examined thus far, although it appears that in the SF2 helicases, the fold of this domain shows a connectivity more like that of the enzyme adenylate kinase than like that of RecA. The folds are extremely similar though. For the sake of brevity, the term RecA like will be used in this review to refer to both folds. Helicase motifs I, Ia, II, V, and VI occupy roughly the same positions in the three-dimensional structures of both superfamilies, but major differences have been noted in the position, and presumably function, of motifs III and IV (25).
FIG. 1.
Ribbon diagram of the PcrA apoenzyme. The RecA-like domains (1A/2A) are light grey, and the 1B/2B domains are dark grey. The colors of the seven helicase motifs are as follows: motif I, magenta; motif Ia, blue; motif II, orange; motif III, green; motif IV, cyan; motif V, yellow; motif VI, red. This and the subsequent figures were created with the RIBBONS program (5).
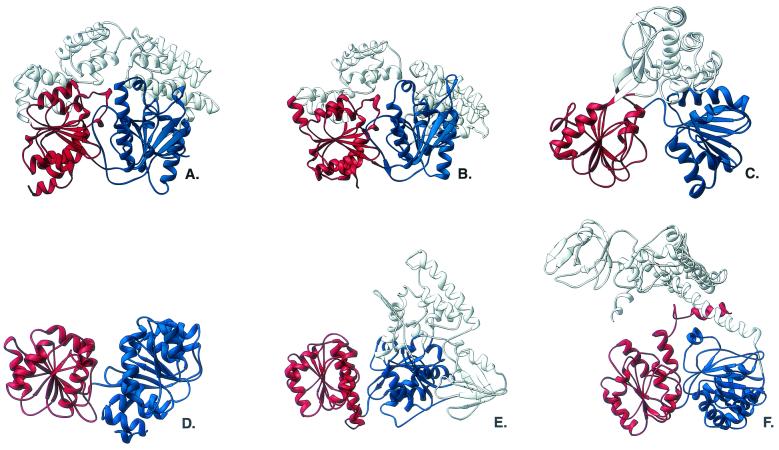
FIG. 2.
Ribbon diagrams of the SF1 and SF2 helicases. The RecA-like domains are in blue (equivalent to PcrA domain 1A) and red (equivalent to PcrA domain 2A). The structures are as follows: A, PcrA from B. stearothermophilus; B, Rep helicase from E. coli; C, NS3 protein helicase domain from hepatitis C virus; D, eIF4A-type RNA helicase from M. jannaschii; E, UvrB from Thermus thermophilus; F, RecG from Thermotoga maritima.
The cleft between the 1A and 2A domains forms the nucleotide-binding site, with residues from several of the conserved motifs in both domains forming protein-nucleotide interactions. Subsequent structures have shown that this cleft opens and closes in response to nucleotide binding and hydrolysis (61).
The 1B and 2B domains, in contrast, are almost entirely α-helical and show no significant similarity to each other. Recent analysis has shown that they belong to the helix-hairpin-helix motif that is commonly involved in nonspecific DNA binding (47). Sequence and structural analyses of other helicases have shown that these domains, which may be either insertions or extensions to the core RecA-like domains are highly variable or, in some cases, entirely absent.
More information about the role of these domains was gained with the solution of the structure of the homologous Rep helicase from E. coli (24) in complex with a 16-mer ssDNA (Fig. 2B). In the crystal structure, two forms of the enzyme were seen in differing conformations. The open conformation was rather similar to that of the apo-PcrA structure, but the closed conformation showed the 2B domain to have rotated by a large amount (∼120°), thus shutting the cleft between the domains. The structures also showed that the ssDNA bound along a groove on the top of the 1A and 2A domains. The authors proposed that the conformational change was related to the translocation action of the enzyme, but it was not until the structure of PcrA in complex with a tailed duplex in both ADPNP (a nonhydrolyzable analogue of ATP)- and ADP-bound forms was determined that the precise details of the process were elucidated (61).
These structures showed the ssDNA tail of the DNA substrate bound to the top of domains 1A and 2A, as seen in the Rep helicase structure. The duplex region of the DNA was shown to bind along the side of domain 2B. In each structure, the 1B and 2B domains adopted the same configuration as the closed Rep structure. In the presence of ADP (the product complex), the duplex DNA is only loosely bound, but in the ADPNP-bound form (the substrate complex), the 1A-2A cleft has closed and the DNA duplex adjacent to the 2B domain appears more ordered, suggesting a stronger interaction with the protein.
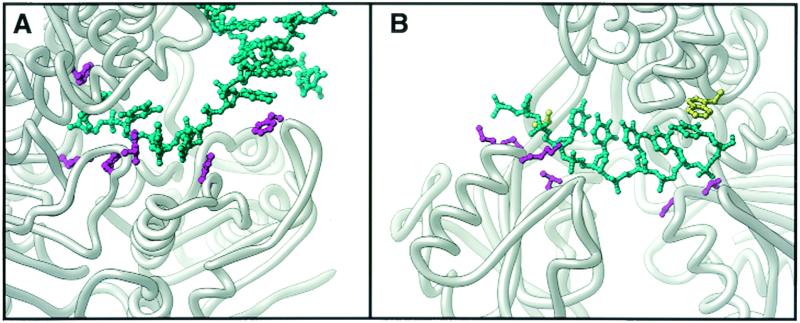
In both the substrate and product complexes, the ssDNA is bound to the top of the 1A and 2A domains. The protein-ssDNA interactions occur mainly through a series of hydrophobic contacts formed by aromatic side chains stacking against the DNA bases (Fig. 3A). Importantly, the frame of the bases relative to the pockets is shifted between the substrate and product complexes; that is, the ss tail is displaced 1 base toward the 5′ end of the DNA, corresponding to a 1-base step in the 3′-5′ direction. This movement appears to be caused by the flipping of a phenylalanine side chain into the initial binding pocket, with an accompanying displacement of the DNA base. This base flipping appears to be a consequence of the opening and closing of the cleft between domains 1A and 2A in response to ATP binding and hydrolysis. During this process, the relative affinity for ssDNA alternates between the 1A and 2A domains, allowing the ssDNA to slide over one or the other of the binding sites. The net result of these processes is that the DNA strand is shuffled along the top of the RecA domains.
FIG. 3.
Depiction of the manner of substrate binding in an SF1 (PcrA, panel A) and an SF2 (NS3 protein, panel B) helicase. The bound oligonucleotide is cyan, and the amino acid side chains involved in binding are purple. In the NS3 structure, the hydrophobic side chains that bookend the substrate are yellow.
This explains the DNA translocation action of the enzyme. It does not tell us anything about how the duplex itself is split apart. Examination of the structures shows that the duplex section of DNA could be simply sterically blocked from passing through the ssDNA-binding site so that translocation would simply shear the duplex apart as it encountered the protein. On closer inspection, though, it becomes apparent that this is only half of the story. The crystal structures suggest that the affinity of domain 2B for duplex DNA is also altered in response to ATP binding, which has also been shown to be the case biochemically (52). In the high-affinity (ADPNP-bound) state, the duplex is significantly distorted at the junction region, with a slight unwinding of the DNA resulting from this. Effectively, the binding of the DNA to the protein surface in the ADPNP-bound state provides the initial unwinding event, prior to the ss being translocated through the enzyme. These analyses very clearly demonstrate that the helicase activity results from translocation and active duplex destabilization, each mediated through different domains of the enzyme. They also suggest that the enzyme will translocate and unwind 1 bp of DNA per ATP hydrolyzed; the enzyme is said to have a step size of 1, a prediction that has recently been supported by pre-steady-state kinetic analyses (12). This, however, may not be true of all helicases, and the number of bases opened in response to one NTP hydrolysis could differ in other systems.
No other helicase has been studied structurally in the same detail as PcrA, but the determination of other helicase structures has allowed further elucidation of the molecular mechanisms involved. Four crystal structures of representative SF2 helicases have now been determined, and they reveal some expected similarities but also some intriguing differences. The first SF2 helicase structure determined (68) was that of the hepatitis C virus NS3 protein helicase domain (henceforth referred to as NS3). This monomeric 3′-5′ helicase is expressed as a polyprotein with a serine protease, to which it appears to remain fused in vivo. Despite this, the helicase domain is active on its own and appears to be specific for RNA-RNA duplexes, as might be expected in an enzyme critical for the replication of the viral RNA genome (21). The NS3 crystal structure showed the familiar tandem arrangement of RecA-like domains, with the adenylate kinase type of connectivity (Fig. 2C). There was, however, no equivalent to the PcrA 1B and 2B domains; instead, the C terminus of the protein forms a separate domain that is unrelated in structure to those of other helicases. The relative disposition of this domain to the RecA-like domains was also different from that of PcrA. As with the SF1 structures, all seven helicase motifs were in the RecA-like domains and the nucleotide-binding pocket was in the same spatial location. A subsequent structure (23) of the same enzyme with a bound deoxyuridine octamer was also determined. Here, the NA was bound along the top of the domain 1A and 2A equivalents, but unlike the PcrA and Rep structure, the protein-DNA interactions were predominantly through the DNA phosphate backbone (Fig. 3B). Although there were hydrophobic interactions with two of the substrate bases, nothing similar to the series of hydrophobic pockets seen in PcrA was evident in the binding site. These observations suggest that a very different mode of NA binding is utilized in NS3, despite the similar fold of the motor domain as a whole. The authors suggested that opening and closing of the RecA-like domains occur in response to ATP binding or hydrolysis, and so, a modified version of the PcrA mechanism could be proposed with the difference being in the way the enzyme grips the NA substrate.
A second group of SF2 helicases the structures of which have recently been determined are the eukaryotic initiation factor 4a (eIF4A) RNA helicases. These work as a part of multiprotein complexes to unwind secondary structure in the 5′-untranslated regions of mRNA in order to allow binding to the 40S ribosomal subunit (20). The structures of S. cerevisiae eIF4A (6) and a structurally homologous putative RNA helicase from the archaeon Methanococcus jannaschii (54) show the tandem RecA-like domains connected by a flexible linker region (Fig. 2D). In these enzymes, there are no additional domains, simply the NTP-hydrolyzing motor. Biochemical studies have shown that these helicases are capable of binding dsRNA (43) and may also unwind from a blunt end (44). None of the structures are in complex with RNA, but it would be reasonable to assume that any duplex would bind in a similar way to the DNA oligomer seen in the NS3 structure.
Another interesting example of an SF2 enzyme is the UvrB protein, one component of the nucleotide excision repair pathway in prokaryotes (59). The first stages of this process require three proteins, UvrA, UvrB, and UvrC, to recognize and excise damaged bases from DNA. The UvrB protein interacts with all of the other proteins utilized in the repair process and, though not active as a helicase on its own, is capable of acting as a rather poor helicase in complex with UvrA (17). The proposed function of the enzyme is to, in complex with UvrA, track along DNA in an ATP-dependent manner until it encounters DNA damage. At this point, UvrB forms a stable, specific complex with the DNA and recruits UvrC in order to make the incisions in the phosphodiester backbone. It seems probable that UvrB causes some local denaturation of the DNA in order to initiate the process. The structures (28, 58) of UvrB show a four-domain protein in which two of the domains are formed from the insertions in the RecA-like domains (Fig. 2E). These additional domains are involved in interaction with other proteins in the repair pathway and also suggest a mechanism by which the initial preincision DNA complex is formed; this is examined in more detail below. An examination of potential DNA-binding sites on the protein showed a conserved, positively charged patch around helicase motif IV that was proposed to interact with the DNA backbone (58). A structural alignment with NS3 suggested that the DNA could bind in the same orientation as the NS3 oligomer and that it would be possible to accommodate a duplex in the DNA binding site, consistent with the biochemical data for the protein.
Another SF2 helicase structure, that of RecG, has recently been determined (50). The function of this enzyme is to rescue stalled replication forks by initially regressing them to a point at which one of a number of damage bypass pathways may operate (31). The structure of the enzyme (Fig. 2F) has been determined in complex with its preferred substrate, a three-way DNA fork. The C terminus of the protein forms the RecA-like motor domains, while the N terminus forms a large fold divided into three subdomains linked to the C terminus by a long α-helix. The DNA fork is bound to this domain in a manner that shows how the enzyme accomplishes duplex separation (see later sections). The leading arm of the fork is the duplex along which the enzyme is proposed to translocate. Unfortunately, the crystal structure does not reveal how this duplex binds to the RecA-like domains because the DNA substrate utilized was a little too short to contact them. However, it is clear from the disposition of the leading arm that one strand of the duplex could lie across the DNA-binding cleft in exactly the same way as the ss seen in the NS3 structure.
All of the structural data for the SF2 helicases suggest a common mode of NA binding, that is, via the phosphodiester backbone, in a non-sequence-specific manner. In principle, this would allow the helicase to bind both ss and ds substrates, a specificity that may be modulated by the disposition of additional domains on the enzyme. This mode of binding would also allow the helicase to bypass base lesions, a characteristic that will be discussed further. In contrast, the two SF1 helicase structures determined show that they appear to bind to the substrate via hydrophobic interactions with the bases and thus translocate along ss NA.
This may well be a general mechanistic distinction between the SF1 and SF2 helicases and suggests that the SF2 helicases are capable of translocation along ss or ds NA while the requirement for SF1 helicases to contact the bases limits them to ss substrates.
There are some biochemical data that support this hypothesis. Studies have shown that for the SF1 helicases such as Rep, UvrD, and PcrA, the rate of ATP hydrolysis is dramatically stimulated by ssDNA, with the kcat for ATP hydrolysis by Rep increasing by 1,000-fold on addition of ssDNA (65, 66) or 400-fold in the case of PcrA (4). By comparison, dsDNA appears to be a poor effector of PcrA ATPase activity (11). With the SF2 helicases, a different effect is seen. The ATPase activities of RecG (26), UvrB (35), and NS3 (36, 38) helicases have all been demonstrated to be stimulated by ds, as well as ss, NA. Generally, the level of stimulation by ds NA is slightly less than or similar to that of ss NA, although RecG shows greater stimulation by dsDNA. As mentioned above, there is no reason why the SF2 helicases could not operate on ss substrates. It should be borne in mind, however, that the structural data for SF1 helicase-DNA complexes are limited to two highly related examples from a diverse family, so it is possible that other modes of binding occur within the SF1 group, thus allowing ds translocation. The structural determination of more enzyme-NA complexes should help to clarify this issue.
Why should two different mechanisms have evolved? The answer may be related to the cellular function of the enzymes; for helicases involved in some repair processes, it may be advantageous to read the bases, for example, to detect lesions such as a pyrimidine dimer or abasic site. An interesting example of this is the SF1 RecBCD enzyme, which rapidly translocates DNA until a specific sequence (the chi sequence) is reached (51). This sequence specificity demonstrates the ability of the enzyme to read the bases as it moves, scanning for a particular feature. In other situations, such as chromosomal replication, speed and processivity are of greater concern, so the only interaction with the DNA is via the phosphate backbone. This mode of contact would also allow the helicase to bypass damage so that it could be subsequently repaired by a different pathway. It is noteworthy that such an ss-ds distinction is seen in the hexameric helicase family, certain members of which, such as RuvB, are capable of translocating a duplex (53), while others, such as DnaB (22), T7gp4 (49, 71), and RepA (34) appear to move along ssDNA. Structural data for these enzymes suggest that both classes contact the DNA backbone, in contrast to the monomeric helicases, but this has yet to be definitively proven.
HELICASE ACTION—ADAPTATION TO A THEME
The conservation of the core motor domains in all of the monomeric helicases studied to date strongly suggests that the additional, structurally diverse domains are the determinants of the specificity of the enzyme and uniquely target any given helicase to a particular cellular process. A corollary is that it is the extra domains themselves that generally confer helicase activity per se upon the enzyme. Without these domains, the enzyme core is simply capable of translocating along the NA without necessarily unwinding it. It is an extra added domain(s) that recognizes a particular NA structure and provides or enhances the strand separation functionality. This domain(s) need not come from the same polypeptide chain. For example, the eIF4A protein, which is essentially only the RecA-like motor with no additional domains, is a poor helicase on its own. However, the helicase activity of eIF4A is considerably enhanced by the presence of eIF4B (20) or eIF4H (41, 42), which may bind to eIF4A, thus adding the missing domains, aiding duplex separation.
This division of labor in helicases may be more clearly seen when one examines the helicase-DNA complex structures determined to date. In PcrA, for example, it has been observed both structurally (61) and biochemically (52) that the 2A and 2B domains significantly distort and hence aid unwinding of the duplex DNA. Since PcrA translocates ssDNA, the simple steric hindrance of the 2A/2B domains against the duplex should be sufficient to strip the DNA strands away from each other. However, it has been shown that the binding of domain 2B to the duplex is essential for helicase activity, showing that the distortion induced in the DNA is a critical element of the reaction as described previously.
The mechanism by which the SF2 helicases destabilize and separate duplexes is less clear, mainly due to a lack of structures of the enzymes in complex with their preferred substrates. The NS3-ssDNA complex (23) provides information on the possible translocation mechanism but is less informative about the way in which duplex NA might be melted. In this enzyme, the additional domains take the form of a pair of long antiparallel strands inserted into the domain 2A equivalent and an α-helical C-terminal domain. Examination of the structure shows that two hydrophobic residues, a valine and a tryptophan, are contributed by the extended loop and C-terminal domain, respectively, which bookend the bases at either end of the oligonucleotide substrate. The interaction between these residues and the bases may force duplex separation as the substrate is translocated or stabilize the newly formed ss.
The excision repair enzyme UvrB demonstrates an alternative use for the additional domains. As previously described, the UvrA2B complex is thought to translocate DNA in an ATP-dependent manner until damaged dsDNA is encountered. At this point, a tight complex is formed and the DNA is partially unwound in preparation for incision and subsequent steps of the repair pathway. The crystal structures (28, 58) of the enzyme give some clues as to how this may occur. One of the insertions in the motor domains takes the form of an extended β-hairpin, which pokes straight out into the proposed DNA-binding site. The authors suggest that the hairpin enters the DNA duplex between the two strands in the damaged duplex, thus locking it to the enzyme and forming the observed tight complex. This hairpin insertion also stabilizes the partially unwound state of the duplex, although it is not entirely clear how this initial unwinding occurs. The other additional domains of the enzyme appear to be involved in recruiting other proteins of the repair pathway. The requirement of UvrA for helicase activity may be to prevent reannealing of the duplex DNA as it passes over the pin, although there are no structural data that support this. It seems that UvrB retains some elements of a helicase, such as translocation and initial duplex destabilization, but has sacrificed processive helicase activity for a more specific action that is modulated by other proteins.
A rather different approach to duplex unwinding is observed in RecG. The C-terminal half of the protein forms the familiar RecA-like domains (50). Connected to these by a long helix is a large N-terminal domain. This takes the form of a clamp type of structure similar to that seen in the Holliday junction resolving protein RuvA (2, 40). The edges of this clamp form pins that sterically block duplex DNA from passing through the central channel of the domain. In effect, the motor domains seem to pull the DNA through the clamp, stripping apart the duplex in the process. The exposed bases at the ssDNA-dsDNA interface are stacked against aromatic amino acid side chains, thus stabilizing the unwound junction. Since RecG operates at a three-way fork, it is capable of unwinding two duplexes simultaneously (one on each arm of the fork) and feeding the unwound strands together to form a new, fourth duplex. This four-way junction is the equivalent of the Holliday junction intermediate observed in homologous recombination and is probably processed in prokaryotes by the RuvABC system (reviewed in reference 64). The similarity to the RuvABC recombination system is instructive, since the RecG monomer combines the motor elements of RuvB in its C-terminal domains with a RuvA type of functionality in the N-terminal domain. Unlike the RuvABC system, the DNA is not topologically linked to the protein (as is the case with the toroidal RuvB protein), which suggests a lower processivity for the system. This is entirely consistent with the physiological role of RecG, which only requires it to act over a short distance. The RuvABC complex, by contrast, may need to migrate a Holliday junction several kilobases. Whereas the RuvABC and UvrABC systems split the components required for biological activity among several proteins, RecG unites a bipartite functionality within a single enzyme.
RELATED SYSTEMS
Assuming that the seven helicase motifs are the characteristic conserved core of an NA-translocating protein, it is instructive to examine how such a motor might be utilized in a variety of other cellular processes. In eukaryotes, a widespread group of enzymes containing these motifs are the ATP-dependent chromatin-remodeling factors, including the SWI2/SNF2 and ISWI families (see, for example, references 60 and 62). These enzymes function to destabilize the interaction between DNA and histone octamers during transcription. They all contain a conserved ATPase core domain (16), including the helicase motifs, with differing N- and C-terminal domains. It has been demonstrated that these proteins do not function in the strand displacement assays diagnostic of helicases in vitro (8), The exact mechanism by which these proteins facilitate chromatin remodeling is not clear. However, it has been shown that the remodeling complexes, in an ATP-dependent manner, are capable of generating superhelical torsion in DNA that may be utilized in the remodeling process (18). The mechanism by which this torsion is generated has yet to be unambiguously determined, but it seems likely that one domain of the remodeling factor tightly grips the DNA to form a local closed section, while torsional stress is applied to the duplex within the same section. This torsional stress may be generated by a rotational wrench action around the long axis of the duplex or by linear tracking along the helical backbone, which will produce the same effect. It is easy to see that the linear tracking activity could be produced by the helicase core motor in exactly the same way as is seen in the SF2 helicase family. The additional domains in the chromatin-remodeling proteins, instead of being used to destabilize the duplex, are likely to include the block required to generate torsion and to couple this torsion to the remodeling activity. Another likely function of the additional domains is to ensure that the enzyme complex is loaded onto the DNA in the correct orientation; reverse loading of the complex would introduce negative supercoiling in the DNA if positive supercoiling were required and vice versa.
Another common system in which DNA translocation is a central activity is that of the type I restriction endonucleases (recently reviewed in reference 32). These elaborate enzymes recognize a target sequence within a given stretch of DNA and then create a ds break anywhere from 40 bp to many kilobases away from the target. At all times, the enzyme remains bound to the target sequence; thus, the cleavage sites are reached by translocating the DNA relative to the enzyme, forming loops that may be directly observed by electron microscopy (72). Given the nature of the activity of the enzyme, it is not surprising that the so-called R subunit of all type I restriction endonucleases contains the seven SF2 helicase motifs (10, 15). This is consistent with the notion that the SF2 core acts as a ds translocase without opening the duplex (19). In this regard, it is noteworthy that experiments with psoralen-cross-linked DNA (13) showed that, at low levels, the interstrand cross-links did not impede translocation, suggesting that the enzyme tracked along the DNA backbone without opening the strands.
If the translocase model is extended to the hexameric helicases, more examples may be seen. The recently determined structure of the bacterial conjugation protein TrwB (14) showed remarkable similarity to the hexameric helicases, and the proposed function of the enzyme, to transfer a single DNA strand between bacteria during conjugation, can be seen to be the translocation activity of a helicase. In a similar fashion, the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system (70) also resembles the hexameric helicases, but here the function of the protein is to transport a pathogenicity factor, in this case a protein, to the host cell.
CONCLUSIONS
The examples outlined above demonstrate the economy of Nature in reusing a series of common building blocks to achieve a variety of ends. In the helicase systems, a widely occurring fold with ATP-hydrolyzing functionality has been employed to build a motor capable of translocating NA polymers. By building on this core, the NA specificity of the enzyme may be defined. Additionally, these domains may either provide or enhance the enzyme's ability to open the NA duplex. The nontranslocase parts of the enzymatic machine may be composed of additional domains attached to the core or may be a separate polypeptide chain(s). As well as recognizing particular substrates, these domains may also function to affect other biochemical characteristics of the system, such as rate or processivity, or to recruit other proteins to form a larger complex. It seems likely that the classification of helicases into SF1 or SF2 on the basis of conserved motifs may reflect a preference for ss or ds substrates generally bound to the enzyme via the bases of phosphodiester backbone, respectively. It may be that these two modes of binding have evolved to allow the enzyme to read the bases or bypass damaged NA, respectively. Further elucidation of the structures of these enzymes in complexes with their physiological substrates would help to clarify this theory.
Finally, it should be clear that the presence of helicase motifs does not always imply strand separation functionality. Any situation in which there is active translocation of a polynucleotide substrate in response to nucleotide hydrolysis may involve the participation of this diverse family of enzymes.
REFERENCES
- 1.Ali, J. A., and T. M. Lohman. 1997. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275:377-380. [DOI] [PubMed] [Google Scholar]
- 2.Ariyoshi, M., T. Nishino, H. Iwasaki, H. Shinagawa, and K. Morikawa. 2000. Crystal structure of the Holliday junction DNA in complex with a single RuvA tetramer. Proc. Natl. Acad. Sci. USA 97:8257-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird, L. E., H. S. Subramanya, and D. B. Wigley. 1998. Helicases: a unifying structural theme? Curr. Opin. Struct. Biol. 8:14-18. [DOI] [PubMed] [Google Scholar]
- 4.Bird, L. E., J. A. Brannigan, H. S. Subramanya, and D. B. Wigley. 1998. Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling model. Nucleic Acids Res. 26:2686-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carson, M. 1991. Ribbons 2.0. J. Appl. Crystallogr. 24:958-961. [Google Scholar]
- 6.Caruthers, J. M., E. R. Johnson, and D. B. McKay. 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. USA 97:13080-13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y. Z., W. Zhuang, and E. W. Prohofsky. 1992. Energy flow considerations and thermal fluctuational opening of DNA base pairs at a replicating fork: unwinding consistent with observed replication rates. J. Biomol. Struct. Dyn. 10:415-427. [DOI] [PubMed] [Google Scholar]
- 8.Côté, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 9.Daune, M. 1999. Molecular biophysics: structure in motion. Part I, p. 38-100. Oxford University Press, New York, N.Y.
- 10.Davies, G. P., I. Martin, S. S. Sturrock, A. Cronshaw, N. E. Murray, and D. T. F. Dryden. 1999. On the structure and operation of type I restriction enzymes. J. Mol. Biol. 290:565-579. [DOI] [PubMed] [Google Scholar]
- 11.Dillingham, M. S. 1999. Ph.D. thesis. University of Oxford. United Kingdom.
- 12.Dillingham, M. S., D. B. Wigley, and M. R. Webb. 2000. Unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry 39:205-212. [DOI] [PubMed] [Google Scholar]
- 13.Eskin, B. 1973. Ph.D. thesis. University of California, Berkeley.
- 14.Gomis-Ruth, F. X., G. Moncalian, R. Perez-Luque, A. Gonzalez, E. Cabezon, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., and E. V. Koonin. 1991. Endonuclease (R) sub-units of type-I and type-III restriction modification enzymes contain a helicase-like domain. FEBS Lett. 291:277-281. [DOI] [PubMed] [Google Scholar]
- 16.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 17.Gordienko, I., and W. D. Rupp. 1997. The limited strand-separating activity of the UvrAB protein complex and its role in the recognition of DNA damage. EMBO J. 16:889-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Havas, K., A. Flaus, M. Phelan, R. Kingston, P. A. Wade, D. M. Lilley, and T. Owen-Hughes. 2000. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103:1133-1142. [DOI] [PubMed] [Google Scholar]
- 19.Janscak, P., and T. A. Bickle. 2000. DNA supercoiling during ATP-dependent DNA translocation by the type I restriction enzyme EcoAI. J. Mol. Biol. 295:1089-1099. [DOI] [PubMed] [Google Scholar]
- 20.Jaramillo, M., T. E. Dever, W. C. Merrick, and N. Sonenberg. 1991. RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol. Cell. Biol. 12:5992-5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadare, G., and A. L. Haenni. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan, D. L. 2000. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol. 301:285-299. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J. L., K. A. Morgenstern, J. P. Griffith, M. D. Dwyer, J. A. Thomson, M. A. Murcko, C. Lin, and P. R. Caron. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89-100. [DOI] [PubMed] [Google Scholar]
- 24.Korolev, S., J. Hsieh, G. H. Gauss, T. M. Lohman, and G. Waksman. 1997. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90:635-647. [DOI] [PubMed] [Google Scholar]
- 25.Korolev, S., N. Yao, T. M. Lohman, P. C. Weber, and G. Waksman. 1998. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 7:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd, R. G., and G. J. Sharples. 1993. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 12:17-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohman, T. M., and K. P. Bjornson. 1996. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65:169-215. [DOI] [PubMed] [Google Scholar]
- 28.Machius, M., L. Henry, M. Planitkar, and J. Deisenhofer. 1999. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc. Natl. Acad. Sci. USA 96:11717-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marians, K. J. 2000. Crawling and wiggling on DNA: structural insights to the mechanism of DNA unwinding by helicases. Structure 8:R227-R235. [DOI] [PubMed]
- 30.Matson, S. W., D. W. Bean, and J. W. George. 1994. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays 16:13-22. [DOI] [PubMed] [Google Scholar]
- 31.McGlynn, P., and R. G. Lloyd. 2001. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA 98:8227-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray, N. E. 2000. Type I restriction systems: sophisticated molecular machines. Microbiol. Mol. Biol. Rev. 64:412-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuwald, A. F., L. Aravind, J. L. Spouge, and E. V. Koonin. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27-43. [PubMed] [Google Scholar]
- 34.Niedenzu, T., D. Roleke, G. Bains, E. Schersinger, and W. Saenger. 2001. Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol. 306:479-487. [DOI] [PubMed] [Google Scholar]
- 35.Oh, E. Y., and L. Grossman. 1987. Helicase properties of the Escherichia coli UvrAB protein complex. Proc. Natl. Acad. Sci. USA 84:3638-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paolini, C., R. De Francesco, and P. Gallinari. 2000. Enzymatic properties of hepatitis C virus NS3-associated helicase. J. Gen. Virol. 81:1335-1345. [DOI] [PubMed] [Google Scholar]
- 37.Patel, S. S., and K. M. Picha. 2000. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69:651-697. [DOI] [PubMed] [Google Scholar]
- 38.Preugschat, F., D. R. Averett, B. E. Clarke, and D. J. Porter. 1996. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J. Biol. Chem. 271:24449-24457. [DOI] [PubMed] [Google Scholar]
- 39.Putnam, C. D., S. B. Clancy, H. Tsuruta, S. Gonzalez, J. G. Wetmur, and J. A. Tainer. 2001. Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol. 311:297-310. [DOI] [PubMed]
- 40.Rafferty, J. B., S. E. Sedelnikova, D. Hargreaves, P. J. Artymuik, P. J. Baker, G. A. Sharples, A. A. Mahdi, R. G. Lloyd, and D. W. Rice. 1996. Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science 274:415-421. [DOI] [PubMed] [Google Scholar]
- 41.Richter, N. J., G. W. Rogers, J. O. Hensold, and W. C. Merrick. 1999. Further biochemical and kinetic characterisation of human eukaryotic initiation factor 4H. J. Biol. Chem. 274:35415-35424. [DOI] [PubMed] [Google Scholar]
- 42.Richter-Cook, N. J., T. E. Dever, J. O. Hensold, and W. C. Merrick. 1998. Purification and characterization of a new eukaryotic protein translation factor. Eukaryotic initiation factor 4H. J. Biol. Chem. 273:7579-7587. [DOI] [PubMed] [Google Scholar]
- 43.Rogers, G. W., N. J. Richter, and W. C. Merrick. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274:12236-12244. [DOI] [PubMed] [Google Scholar]
- 44.Rogers, G. W., W. F. Lima, and W. C. Merrick. 2001. Further characterization of the helicase activity of eIF-4A. Substrate specificity. J. Biol. Chem. 276:12598-12608. [DOI] [PubMed] [Google Scholar]
- 45.Roman, L. J., and S. C. Kowalczykowski. 1989. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry 28:2863-2873. [DOI] [PubMed] [Google Scholar]
- 46.Schmid, S. R., and P. Linder. 1992. D-E-A-D protein family of putative RNA helicases. Mol. Microbiol. 6:283-292. [DOI] [PubMed] [Google Scholar]
- 47.Shao, X., and N. V. Grishnin. 2000. Common fold in helix-hairpin-helix proteins. Nucleic Acids Res. 28:2643-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiratori, A., T. Shibata, M. Arisawa, F. Hanaoka, Y. Murakami, and T. Eki. 1999. Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast 15:219-253. [DOI] [PubMed] [Google Scholar]
- 49.Singleton, M. R., M. R. Sawaya, T. Ellenberger, and D. B. Wigley. 2000. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101:589-600. [DOI] [PubMed] [Google Scholar]
- 50.Singleton, M. R., S. Scaife, and D. B. Wigley. 2001. Structural analysis of DNA replication fork reversal by RecG. Cell 107:79-89. [DOI] [PubMed] [Google Scholar]
- 51.Smith, G. R., S. K. Amundsen, A. M. Chaudhury, K. C. Cheng, A. S. Ponticelli, C. M. Roberts, D. W. Schultz, and A. F. Taylor. 1984. Roles of RecBC enzyme and chi sites in homologous recombination. Cold Spring Harbor Symp. Quant. Biol. 49:485-495. [DOI] [PubMed] [Google Scholar]
- 52.Soultanas, P., M. S. Dillingham, P. Wiley, M. R. Webb, and D. B. Wigley. 2000. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 19:3799-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stasiak, A., I. R. Tsaneva, S. C. West, C. J. B. Benson, X. Yu, and E. H. Egelman. 1994. The Escherichia coli RuvB branch migration protein forms double rings around DNA. Proc. Natl. Acad. Sci. USA 91:7618-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Story, R. M., H. Li, and J. N. Abelson. 2001. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl. Acad. Sci. USA 98:1465-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Story, R. M., and T. A. Steitz. 1992. The structure of the E. coli RecA protein monomer and polymer. Nature 255:318-325. [DOI] [PubMed] [Google Scholar]
- 56.Subramanya, H. S., L. E. Bird, J. A. Brannigan, and D. B. Wigley. 1996. Crystal structure of a DExx box helicase. Nature 384:379-383. [DOI] [PubMed] [Google Scholar]
- 57.Taylor, A. F., and G. R. Smith. 1985. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J. Mol. Biol. 185:431-443. [DOI] [PubMed] [Google Scholar]
- 58.Theis, K., P. J. Chen, M. Skorvaga, B. Van Houten, and C. Kisker. 1999. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 18:6899-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Houten, B. 1990. Nucleotide excision repair in Escherichia coli. Microbiol. Rev. 54:18-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varga-Weisz, P. 2001. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene 20:3076-3085. [DOI] [PubMed] [Google Scholar]
- 61.Velankar, S. S., P. Soultanas, M. S. Dillingham, H. S. Subramanya, and D. B. Wigley. 1999. Crystal structures of complexes of PcrA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75-84. [DOI] [PubMed] [Google Scholar]
- 62.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gray. 1982. Distantly related sequences in the a- and b-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.West, S. C. 1996. The RuvABC proteins and Holliday junction processing in E. coli. J. Bacteriol. 178:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong, I., and T. M. Lohman. 1992. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science 256:350-355. [DOI] [PubMed] [Google Scholar]
- 66.Wong, I., M. Amaratunga, and T. M. Lohman. 1993. Heterodimer formation between Escherichia coli Rep and UvrD proteins. J. Biol. Chem. 268:20386-20391. [PubMed] [Google Scholar]
- 67.Yamada, K., N. Kunishima, K. Mayanagi, T. Ohnishi, T. Nishino, H. Iwasaki, H. Shinagawa, and K. Morikawa. 2001. Crystal structure of the Holliday junction migration protein RuvB from Thermus thermophilus HB8. Proc. Natl. Acad. Sci. USA 98:1442-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao, N. H., T. Hesson, M. Cable, Z. Hong, A. D. Kwong, H. V. Le, and P. C. Weber. 1997. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 4:463-467. [DOI] [PubMed] [Google Scholar]
- 69.Yarranton, G. T., and M. L. Gefter. 1979. Enzyme-catalyzed DNA unwinding: studies on Escherichia coli rep protein. Proc. Natl. Acad. Sci. USA 76:1658-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeo, H.-J., S. Savvides, A. B. Herr, E. Lanka, and G. Waksman. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6:1461-1472. [DOI] [PubMed] [Google Scholar]
- 71.Yu, X., M. M. Hingorani, S. S. Patel, and E. H. Egelman. 1996. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat. Struct. Biol. 3:740-743. [DOI] [PubMed] [Google Scholar]
- 72.Yuan, R., D. L. Hamilton, and J. Burckhardt. 1980. DNA translocation by the restriction enzyme from E. coli K. Cell 20:237-244. [DOI] [PubMed] [Google Scholar]