Abstract
The Methanococcus jannaschii gene MJ0671 was cloned and overexpressed in Escherichia coli, and its gene product was tested for its ability to catalyze the pyridine nucleotide-dependent reduction of either 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate (compound 3) to 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5′-phosphate (compound 4) or 5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (compound 7) to 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (compound 5). Only compound 3 was found to serve as a substrate for the enzyme. NADPH and NADH functioned equally well as the reductants. This specificity for the reduction of compound 3 was also confirmed by using cell extracts of M. jannaschii and Methanosarcina thermophila. Thus, this step in riboflavin biosynthesis in these archaea is the same as that found in yeasts. The absence of the other genes in the biosynthesis of riboflavin in Archaea is discussed.
An unexpected finding emerging from the annotations of the genomic sequences of the methanoarchaea is the absence of many of the genes known to be involved in the biosynthesis of coenzymes (4, 13, 21). Notable among these are the missing genes encoding enzymes for the biosynthesis of riboflavin and flavin adenine dinucleotide (FAD). In the original annotations of the genomes of Methanococcus jannaschii and Methanobacterium thermoautotrophicum, only a few open reading frames in each organism were proposed as being involved in riboflavin biosynthesis. In M. jannaschii, MJ0055 was proposed to be a GTP cyclohydrolase II and MJ0671 was annotated as a riboflavin-specific deaminase (4), whereas in Methanobacterium thermoautotrophicum, the following assignments were made: MTH1499, GTP cyclohydrolase II; MTH235, riboflavin-specific deaminase; and MTH1390, riboflavin synthase β-subunit (21). Subsequent work by Koonin et al. proposed the following assignments for the genes in the M. jannaschii genome as possibly being involved in riboflavin biosynthesis: MJ0430 or MJ1102, ribD pyrimidine deaminase domain; MJ0671, ribD pyrimidine reductase domain; MJ0055, ribB 3,4-dihydroxy-2-butanone-4-phosphate synthetase; MJ0303, ribE 6,7-dimethyl-8-ribityllumazine synthase, and MJ0066 and/or MJ0973, FAD1 FAD synthetase (14). This later work more correctly defined the possible biosynthetic genes present in the genome of M. jannaschii but in no way established the functional identities of these genes. Furthermore, several expected genes in the pathway were not identified, such as GTP cyclohydrolase II, riboflavin synthase, and riboflavin kinase.
Despite the apparent absence of genes for these enzymes, early biosynthetic labeling studies supported the idea that the pathway to riboflavin in the Archaea was the same as that established in either yeasts or bacteria, where the riboflavin is derived from GTP and l-3,4-dihydroxy-2-butanone 4-phosphate (7). The question thus arises as to exactly what is the true nature of the archaeal pathway to riboflavin and FAD. Does biosynthesis proceed by a yeast pathway, a bacterial pathway, or an unknown pathway, and if so, what are the enzymes involved in the pathway? Are the enzymes not presently identified because they have such low sequence similarity that they cannot be readily detected, or have new enzymes evolved to catalyze the reactions that have been found to occur with the archaeal riboflavin synthase (6)? Information on the pathway is also relevant to coenzyme F420 biosynthesis, since compound 6 in the pathway (Fig. 1) is the branch point compound leading to both riboflavin and 7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO) (19), which is a precursor to coenzyme F420 (9).
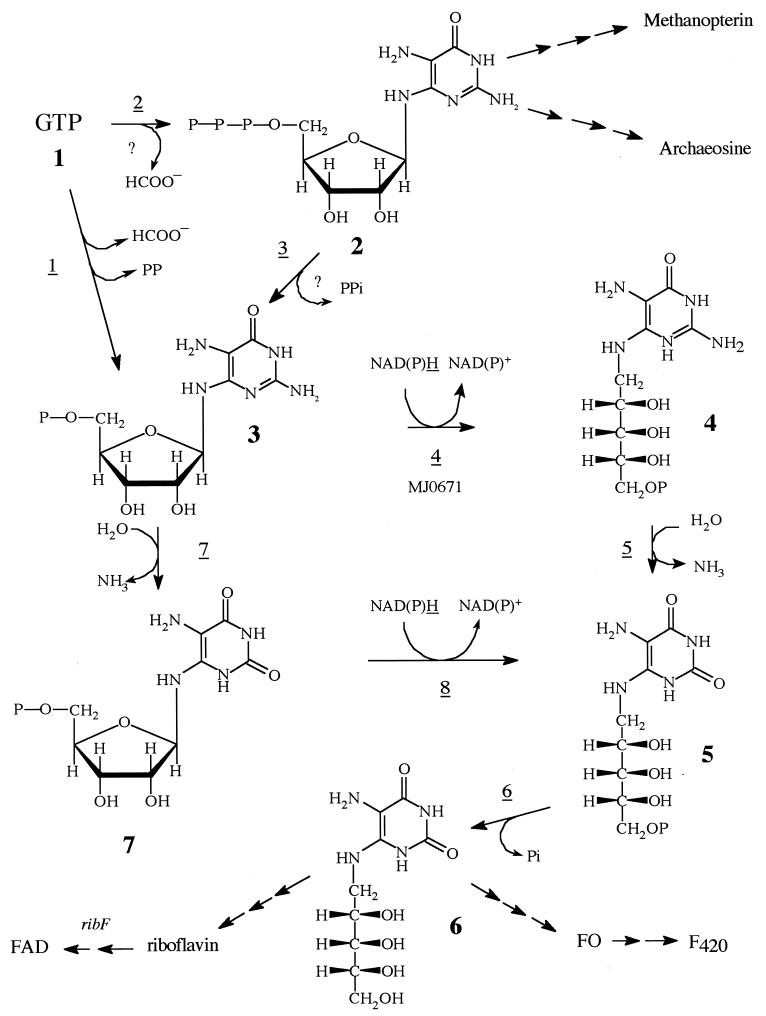
FIG. 1.
The fungal, bacterial, and archaeal pathways to compound 6. The fungal pathway would consist of reactions 1, 4, 5, and 6 and would use the following enzymes: reaction 1, GTP cyclohydrolase II; reaction 4, pyrimidine reductase; reaction 5, pyrimidine deaminase; and reaction 6, unknown phosphatase. The bacterial pathway would consist of reactions 1, 7, 8, and 6 and would use the following enzymes with the same names as those found in the fungal pathway but different substrate specificities as indicated in the figure. The archaeal pathway would consist of reactions 2, 3, 4, 5, and 6 and would use the following enzymes: reaction 2, proposed GTP cyclohydrolase III; reaction 3, nucleoside triphosphate pyrophosphatase; reaction 4, the MJ0671 pyrimidine reductase; reaction 5, an unknown pyrimidine deaminase; and reaction 6, an unknown phosphatase.
In this paper we demonstrate that M. jannaschii gene MJ0671 encodes a pyrimidine nucleotide reductase that catalyzes the conversion of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate (compound 3) into 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5′-phosphate (compound 4). The enzyme did not catalyze the pyrimidine nucleotide-dependent reduction of 5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (compound 7) to 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (compound 5). Thus, this early step in riboflavin biosynthesis in M. jannaschii is most similar to those found in yeasts and fungi (2). The characterization of this reaction defines an intermediate that must be operational in the early steps of riboflavin biosynthesis in the Archaea.
MATERIALS AND METHODS
Chemicals.
6,7-Dimethyl-8-ribityllumazine was prepared as previously described (1) and chemically converted into the 5′-phosphate derivative as described by Eberhardt et al. (6). 2-Amino-4-chloro-6-hydroxy-5-nitropyrimidine was prepared as described by Davoll and Evans (5). Ribitylamine was prepared by catalytic hydrogenation of ribose oxime (18). A chromatographically purified Escherichia coli alkaline phosphatase suspension in (NH4)2SO4 was obtained from the Sigma-Aldrich Chemical Co.
2,5-Diamino-6-ribitylamino-4(3H)-pyrimidinone (compound 8) and 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5′-phosphate (compound 4) were prepared by catalytic hydrogenation of the respective 5-nitro isomers as previously described (17). The 2-amino-6-hydroxy-5-nitroribitylaminopyrimidine (5) and the 5-nitro-6-ribitylamino-2,4(1H,3H) pyrimidinedione (17) were prepared by previously published procedures.
5-Amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (compound 6) was prepared by the catalytic hydrogenation of 4-ribitylamino-5-nitroso-2,6-dihydropyrimidine as described by Plaut and Harvey (18). 5-Amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (compound 5) was prepared by phosphorylation of 5-nitro-6-ribitylamino-2,4(1H,3H) pyrimidinedione followed by reduction (17).
Generation of recombinant enzymes.
The following genes have been cloned and their protein products expressed in E. coli: M. jannaschii MJ0671, E. coli ribA (GTP cyclohydrolase II), and E. coli bifunctional ribD (riboflavin-specific deaminase-reductase). Relevant sequences were amplified by PCR using genomic DNA from M. jannaschii (David E. Graham, Urbana, Ill.) or E. coli as the template. The following synthetic oligonucleotide primers were used: MJ0671, 5′ GGTGGTCCATGGTGATGGTAATGG 3′ and 5′ GATCGGATCCTTATTTCTTTACTTTAAATTCC 3′; ribA, 5′ GGTGGTCATATGCAGCTTAAAGGTGGTGTG 3′ and 5′ GATCGGATCCTTATTTGTTCAGCAAATGG 3′; and ribD, 5′ GGTCATATGCAGGACGAG 3′ and 5′ GTCGGATCCTTATGCACCCACTAAATGC 3′. The E. coli ribA (GTP cyclohydrolase II) and the bifunctional ribD groups of primers were constructed to insert the NdeI and BamHI restriction sites at their 5′ and 3′ ends. For MJ0671, the insert restriction sites are NcoI and BamHI. The PCR was performed with 1 μg of genomic DNA as template with 20 μmol of each primer, 3.75 U of AmpliTaq DNA polymerase, and 10 μl of 10× PCR buffer (Perkin-Elmer) in a final volume of 100 μl. Each cycle was set for 1 min of denaturation at 95°C, 2 min of annealing at 55°C for MJ0671, and 2 min of annealing at 60°C for E. coli ribA and ribD. Extension of 3 min at 72°C and 35 reaction cycles were carried out in a DNA thermal cycler. After purification of the PCR products via absorption and desorption to a QIAquick spin column (Qiagen), the PCR products were digested with NdeI and BamHI (Life Technologies) and then cloned into NdeI-BamHI digested pET17b and pET19b plasmid vectors (Novagen) to obtain the reconstructed plasmids pET17b-ribA and pET19b-ribD. The MJ0671 PCR product was digested with NcoI and BamHI and then cloned into NcoI- and BamHI-digested pET-19b plasmid vector to get the reconstructed plasmid pET19b-MJ0671. The recombinant plasmids were transformed to E. coli BL21(DE3) cells that were grown in Luria-Bertani broth supplemented with 100 μg of ampicillin per ml at 37°C to an absorbance of 0.9 to 1.0 at 600 nm. Protein production was induced with 28 mM lactose. After induction for 4 h at 37°C, the cells were harvested by centrifugation (4000 × g, 5 min) and frozen at −20°C until used. The presence of heterologously produced protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide) of the SDS-soluble cellular proteins. In the case of RibA and RibD overexpressions, 80% and 30%, respectively, of the total proteins were the desired proteins, whether recovered from either the whole cells or the soluble cell extracts (see below). For the MJ0671 overexpression, SDS analysis of the whole cells showed 70% of the total protein to be the desired protein. Analysis of the cell extract showed that the major portion of this protein was not soluble after sonication. Heating the cell extract as described below left essentially only the desired protein in solution. The measured molecular masses of these proteins as determined by SDS-PAGE for RibA, RibD, and MJ0671 were 21.6, 40.7, and 25.1 kDa, respectively. These values are in agreement with the predicted masses of RibA, RibD, and MJ0671 of 21.8, 40.3, and 25.0 kDa, respectively, as based on their gene sequences. The Bio-Rad low molecular weight protein standards, consisting of phosphorylase b, bovine serum albumin, ovalbumin, carbonic anhydrase, soybean trypsin inhibitor, and lysozyme, were used as markers.
Preparation of methanoarchaeal cell extracts.
Cell extracts of Methanosarcina thermophila strain TM-1 were prepared as previously described (16). Cell extracts of Methanobacterium jannaschii (Biswarup Mukhopadhyay, Urbana, Ill.) were prepared by sonication of cells (1.0 g wet weight) suspended in 10 ml of N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) buffer (50 mM TES, 10 mM MgCl2, 20 mM mercaptoethanol [pH 7.0]) followed by centrifugation (10 min, 16,000 × g). The M. jannaschii extracts were stored frozen at −20°C under argon until used. These extracts typically contained 10 to 20 mg of soluble protein/ml. For further purification, the crude extracts (100 μl) were passed through a Sephadex G-25 column (0.5 by 7 cm) and the proteins were eluted with the TES buffer. These purified extracts were stored frozen at −20°C under argon and used for the experiments described here.
Preparation of recombinant enzymes.
Cell extracts with the overproduced proteins were prepared by sonication of the E. coli cells (∼300 mg wet weight) suspended in 3 ml of TES extraction buffer (50 mM TES, 10 mM MgCl2, 20 mM mercaptoethanol [pH 7.0]) followed by centrifugation (10 min, 16,000 × g). SDS-PAGE gel analysis of the resulting insoluble pellets and the solubilized proteins was used to establish the presence of the desired overexpressed proteins.
Heat purification of the solubilized M. jannaschii-derived proteins was performed in a solution containing 0.15 M potassium phosphate, 25 mM TES, 5 mM MgCl2, and 10 mM mercaptoethanol (pH 7.0). Crude extracts containing the desired proteins were heated at 80°C for 15 min and centrifuged (10 min, 16,000 × g) to remove the denatured E. coli proteins. The resulting protein solutions were used for the work reported here. E. coli extracts not containing the overexpressed genes were treated in the same manner and used for control experiments. The crude extracts containing the overproduced E. coli enzymes RibA and RibD were desalted in a Sephadex G-25 column and kept at 4°C until use.
Production of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate (compound 3) using the E. coli recombinant GTP cyclohydrolase II.
To 100 μl of the TES extraction buffer was added 10 μl of 0.1 M GTP, 10 μl of 0.1 M dithiothreitol (DTT), and 10 μl of the Sephadex-purified RibA-containing E. coli extract (7.5 mg of protein/ml). After incubation for 30 min at 37°C, 5 μl of 0.5 M EDTA and 2 volumes of 0.3 M potassium phosphate buffer (pH 7.0) were added. The separation of compound 3 from the proteins was obtained by centrifugation of the solution through an Amicon Centricon-10 concentrator (1 h at 4°C). Portions of the resulting filtrate were used as substrates in the subsequent incubations without any further purification. Quantitation of the amount of compound 3 in any given solution was accomplished by adding 100 μl of 0.2 M Tris buffer (pH 8.0) containing 25 μl of 0.058 M 2,3-butanedione- in MeOH and heating for 30 min at 100°C (12). If required, the precipitated proteins were removed by centrifugation, and the resulting 6,7-dimethylpterin was quantitated by high-performance liquid chromatography (HPLC). All steps were carried out under conditions designed to minimize the exposure of the samples to light.
Production of 5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (compound 7) using the E. coli enzymes RibA and RibD.
A 50-μl portion of the Sephadex-purified RibA (7.5 mg of protein/ml) preparation and a 50-μl portion of the Sephadex-purified RibD (1.5 mg of protein/ml) preparation were combined and incubated with 2 mM GTP, 2 mM DTT, 10 mM MgCl2, and 20 mM β-mercaptoethanol in 50 mM TES (pH 7.0) for 30 min at 37°C. The proteins were removed with Amicon Centricon-10 concentrators, and the filtrate containing compound 7 was used as substrate in the subsequent incubation. For HPLC analysis of the products, the solutions were derivatized by reaction with 2,3-butanedione as described above.
Enzymatic analysis of the pyrimidine nucleotide reductase.
Cell extracts of Methanosarcina thermophila (100 μl), M. jannaschii (100 μl), or E. coli containing the overproduced MJ0671 gene product (20 μl) were added to 100 μl of a 200 μM solution of compound 3, and the resulting solution was brought to a concentration of 2.5 mM in NADH or 2.5 mM NADPH and 2.5 mM DTT by the addition of 0.1 M solutions of these compounds. The control reactions were performed without reductant or with E. coli extracts not containing the overproduced reductase. The solutions were incubated for different periods at 50°C, derivatized with 2,3-butanedione, and analyzed by HPLC as described above for compound 3.
Characterization of enzymatic reaction products.
HPLC analysis of the 2,3-butanedione-derivatized compounds was performed with an Axxi-Chrom octyldecyl silane column (5 μm, 4.6 mm [internal diameter] by 25 cm) by using a NaOAc-methanol gradient at a flow rate of 0.5 ml/min. For the first 5 min, the eluent consisted of 25 mM NaOAc (pH 6.0), 0.02% NaN3, and 5% methanol, and then the methanol concentration was raised linearly to 80% over the next 40 min. The elution of 6,7-dimethylpterin was monitored fluorometrically with an excitation wavelength of 365 nm and an emission wavelength of 445 nm. The 8-ribityldimethylpteridine and 8-ribityldimethyllumazine derivatives were monitored with an excitation wavelength of 408 nm and an emission wavelength of 485 nm. The retention times of the 2,3-butanedione-derivatives derived from compounds 3, 4, and 5 were 27 min, 9 min, and 8 min, respectively. The retention times of the 2,3-butanedione derivatives of the dephosphorylated compound 4 (compound 8) and compound 6 were 20 min and 18 min, respectively. The amounts of the individual compounds were determined by their areas under the HPLC peaks compared to those of known samples.
To separate 6,7-dimethylpterin and 6,7-dimethylumazine, the HPLC analysis was performed isocratically in a solution containing 25 mM potassium phosphate buffer (pH 2.5) and 15% methanol at a flow rate of 0.5 ml/min. The 6,7-dimethyllumazine was observed at an excitation wavelength of 340 nm and an emission wavelength of 480 nm. The retention times of 6,7-dimethylpterin and 6,7-dimethyllumazine under these conditions were 30 and 36 min, respectively.
Thin-layer chromatography analysis of the 2,3-butanedione derivatives was performed with silica gel 60 F254 plates (E. Merck, Darmstadt, Germany). The solvent system was acetonitrile/water/formic acid at a ratio of 80:20:10 (vol/vol/vol). The 2,3-butanedione derivatives had the following Rf values: compound 7, 0.68; compound 3, 0.56; compound 6, 0.37; compound 8, 0.35; compound 4, 0.10; and compound 5, 0.10. Upon exposure to UV light, the pterin-like derivatives show a blue fluorescence, while the lumazine-like derivatives show a greenish fluorescence.
Alkaline phosphatase treatment of samples.
To verify that the newly produced compound 4 was phosphorylated, the 2,3-butanedione derivative of compound 4, from an incubation with extracts containing MJ0671, was purified by preparative HPLC using the analytical Axxi-Chrom octyldecyl silane column. After careful evaporation of the liquid at 40°C, with a stream of nitrogen, the sample was dissolved in 100 μl of 0.1 M glycine buffer (pH 10.4) containing 1 mM MgCl2 and 1 mM ZnCl2. Alkaline phosphatase (0.17 U) was added after incubation for 1 h at 37°C, and the sample was assayed by HPLC by using the same reversed-phase column to establish the nature of the products.
RESULTS
Incubation of cell extracts with compound 3 and compound 4.
Incubation of cell extracts of M. jannaschii or Methanosarcina thermophila with compound 3 and NADH or NADPH, followed by conversion of the products into the dimethylpterin and/or dimethyllumazine derivatives, produced one major new fluorescent compound that was generated only in the presence of compound 3. This product corresponded to that derived from compound 4. Both of the cell extracts produced compound 4 with either NADPH or NADH as the reductant (Table 1). In the case of M. jannaschii, 2.8 nmol of compound 3/h/mg of protein was reduced with NADPH and 1.6 nmol of compound 3/h/mg of protein was reduced with NADH. For Methanosarcina thermophila, 2.2 nmol of compound 3/h/mg of protein was reduced with NADPH and 2.4 nmol of compound 3/h/mg of protein was reduced with NADH. In each case, 12 to 31% of compound 3 was converted into compound 4. Compound 4 was not observed in incubations conducted in the absence of reduced pyridine nucleotide coenzyme or in control incubations with buffer only. No product was observed in similar incubations with compound 7. None of the archaeal extracts produced compound 7 or used compound 7 as a substrate.
TABLE 1.
Formation of compound 4 from compound 3 by using cell extracts of M. thermophila and M. jannaschii and the enzyme derived from the M. jannaschii gene MJ0671
| Samplea | Compound 4b (% conversion)c
|
|
|---|---|---|
| NADPH | NADH | |
| M. jannaschii | 2.8 (22%) | 1.6 (12%) |
| Methanosarcina thermophila | 2.2 (18%) | 2.4 (31%) |
| E. coli extract containing MJ0671 | 111 (15%) | 77 (8%) |
| E. coli extract not containing MJ0671 | <0.002 (0.08%) | <0.002 (0.08%) |
Incubations with the archaeal cell extracts were performed for 1 h at 50°C, whereas the incubations with the E. coli extracts were performed for 15 min at 50°C.
The values are nanomoles of compound 4 formed per hour per milligram of protein with the indicated reductant.
The values for the percentages of conversion of compound 3 to compound 4 during the incubation are based on the ratios of the derivatives formed from compound 3 and compound 4 present at the end of the incubations.
Compound 4 was not further metabolized to compound 5 by any cell extract. In each solution to which compound 3 was added, 6,7-dimethylpterin was also detected. This resulted from the reaction of unreacted compound 3 with butanedione. The evidence supporting the assignment of the observed peak as the desired dimethylpteridine derivative of compound 4 is as follows. The product compound had the same HPLC retention time as the known compounds both before and after removal of the phosphate with the E. coli alkaline phosphatase. Both the phosphorylated and dephosphorylated compounds had the same fluorescent excitation maximum (λmax = 412 nm) and emission maximum (λmax = 483 nm) as those of the known compounds; these values were also the same as the published values (3). The dephosphorylated compound 4 (compound 8) was not used as the substrate for a deamination reaction by the archaeal extracts.
Incubations of E. coli cell extract containing the MJ0671-encoded enzyme produced compound 4 only when the incubation was conducted in the presence of compound 3 and NADH or NADPH. In the absence of compound 3 or a reduced pyridine nucleotide, no formation of compound 4 was observed. The evidence supporting the assignment of the observed peak as the desired dimethylpterin derivative of compound 4 was the same as that described above. Incubations with E. coli cell extracts with compound 3 without the overexpressed gene produced no detectable amount of products. No evidence was obtained for the production of 6,7-dimethyllumazine arising from compound 7, which would have been the product after the deamination of compound 3 following the bacterial pathway. Furthermore, compound 7 was not reduced to compound 5 as expected for the bacterial route.
DISCUSSION
Because of the absence of sequences homologous to GTP cyclohydrolase II, the first enzyme in the biosynthesis of riboflavin, or GTP cyclohydrolase I, the first enzyme in the biosynthesis of pterins, it was proposed that a new enzyme, GTP cyclohydrolase III, catalyzed the first step in the biosynthesis of riboflavin, F420, and methanopterin in the methanoarchaea (12). GTP cyclohydrolase II, although listed as an annotated gene in both the M. jannaschii and Methanobacterium thermoautotrophicum genomes (4, 21), in fact corresponds only to the 3,4-dihydroxy-2-butanone-4-phosphate synthetase portion of this bifunctional enzyme (10). GTP cyclohydrolase III was proposed to catalyze the release of the C-8 carbon atom of the imidazole ring of GTP with the formation of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-triphosphate (compound 2). Compound 2 would then serve as a precursor for the formation of the pterin ring in methanopterin or, after the loss of pyrophosphate, would produce 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate (compound 3), an established intermediate in the biosynthesis of riboflavin (8). Evidence for this pathway was supported by the experimental demonstration that cell extracts of Methanobacterium thermoautotrophicum and Methanosarcina thermophila incubated with GTP produced the expected intermediates (12). We have not been able to confirm this pathway in M. jannaschii, which indicates that yet another pathway for the ring opening of the imidazole ring of GTP may be present in this archaeon. It is expected, however, that the pathway from GTP to riboflavin and FO must lead through compound 3, since this intermediate is further metabolized, as shown in Fig. 1.
At present there are two known routes whereby compound 3 can be converted into 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (compound 5), which after dephosphorylation produces 5-amino-6-ribitylamino-2,4(1H, 3H)-pyrimidinedione (compound 6). Compound 6 is a very important biosynthetic intermediate, since it not only gives rise to riboflavin but also, via a separate pathway, to the 7,8-didemethyl-8-hydroxy-5-deazariboflavin moiety of F420 (Fig. 1) (15).
In the first route, compound 3 first undergoes an NADPH-dependent reduction to compound 4, followed by a deamination to compound 5. This pathway has been found to operate in yeasts and other eukaryotes (2). This differs from the bacterial pathway, where compound 3 first undergoes a deamination to compound 7, which is then reduced to compound 5 (2). Considering that cell extracts of M. jannaschii and Methanobacterium thermoautotrophicum were observed to convert compound 3 to compound 4, but not to compound 7, it is clear that the methanoarchaea use the eukaryotic pathway for this step in the biosynthesis of riboflavin. Furthermore, since the MJ0671-derived enzyme readily carries out this reaction, it is clear that this gene encodes the reductase in M jannaschii. The gene is similar to the Methanobacterium thermoautotrophicum MTH0235 and the Archaeoglobus fulgidus AF2007 genes, suggesting that it functions similarly in these archaea.
In most bacteria, the deaminase and the reductase are bifunctional enzymes, whereas they are separate enzymes in yeasts (20). Analogous to the proposed fungal route in riboflavin biosynthesis, the archaeal reductase is also separate from the deaminase. Nevertheless, the yeast deaminase is highly similar to the bacterial deaminases, while the archaeal enzyme catalyzing the deamination of compound 4 to compound 5 has not been identified. We have tested three recombinantly produced enzymes from M. jannaschii, which we considered as possible enzymes for catalyzing the deaminase reaction. The genes for these enzymes were selected with the consideration that they would catalyze hydrolysis reactions like that observed in the pyrimidine deaminase and would be members of the amidohydrolase family related to urease (11). The selected genes from M. jannaschii tested included MJ0430 and MJ1102, both of which were annotated as dCTP deaminases, and MJ0699, which was annotated as an N-ethylammeline chlorohydrolase. The enzyme encoded by MJ1102 was confirmed to be a dCTP deaminase. The enzyme encoded by MJ0430 did not function as a dCTP deaminase. None of these three enzymes were found to convert compound 4 to compound 5 (Graupner and White, unpublished results). Thus, either the reaction proceeds by another route, another enzyme has evolved to carry out the reaction, or the similarity of the enzyme to other known deaminases is so low that we cannot identify it by standard methods.
Before the work reported here was undertaken, only one gene in the methanoarchaea had been confirmed as being involved in riboflavin biosynthesis, and this was riboflavin synthase (6). The enzyme catalyzing the biosynthesis of riboflavin in Methanobacterium thermoautotrophicum was isolated, sequenced, and cloned by marker rescue with a ribC mutant of E. coli, and the gene was found to correspond to the Methanobacterium thermoautotrophicum gene MTH0134. The gene has a high similarity to the M. jannaschii gene MJ1184. It was stated in that work that the enzyme expressed by this gene “has no sequence similarity whatsoever to the enzymes from eubacteria and yeasts.” Thus, at least one enzyme in the riboflavin pathway has been shown to be so different in its sequence that it cannot be recognized by sequence comparisons as a riboflavin synthase. Of the other genes discussed in the introduction to the present study, we have confirmed that MJ0055 and MJ0303 serve their indicated functions, whereas neither MJ0066 nor MJ0973 serves as a FAD synthetase (Graupner and White, unpublished results).
We thus see a confusing picture of the relationships between the archaeal riboflavin biosynthetic enzymes and those found in other organisms. Of the eight genes identified as being involved in the biosynthesis of riboflavin in E. coli, only five have sequence similarities that are strong enough to be identified in the methanoarchaea by means of a BLAST search. These include the pyrimidine reductase and deaminase domains of ribD; the 3,4-dihydroxy-2-butanone-4-phosphate synthetase gene, ribB; the 6,7-dimethyl-8-ribityllumazine synthase gene, ribE; and the FAD synthetase gene, ribF. Of these genes, only the 3,4-dihydroxy-2-butanone-4-phosphate synthetase and the 6,7-dimethyl-8-ribityllumazine synthase genes have been confirmed. The absence of so many identifiable genes in the pathway may indicate that the entire pathway for FAD biosynthesis has evolved more than once. The question that must be addressed is this: has each of these unrecognizable enzymes evolved from a common source, or are they different enzymes catalyzing the same reactions that have evolved completely independently? This question may be answered only after the different riboflavin biosynthetic genes from many different organisms are sequenced and identified and the evolution of the diverse sequences can be mapped.
Acknowledgments
We thank D. R. Graham for help in preparation and editing of the manuscript.
This work was supported by National Science Foundation Grant MCB 9985712.
REFERENCES
- 1.Bacher, A. 1986. Heavy riboflavin synthase from Bacillus subtilis. Methods Enzymol. 122:192-199. [DOI] [PubMed] [Google Scholar]
- 2.Bacher, A., S. Eberhardt, W. Eisenreich, M. Fischer, S. Herz, B. Illarionov, K. Kis, and G. Richter. 2001. Biosynthesis of riboflavin, p. 1-49. In Vitamins and hormones, vol. 61. Academic Press, New York, N.Y. [DOI] [PubMed]
- 3.Bacher, A., S. Eberhardt, M. Fischer, S. Mortl, K. Kis, K. Kugelbrey, J. Scheuring, and K. Schott. 1997. Biosynthesis of riboflavin: lumazine synthase and riboflavin synthase. Methods Enzymol. 280:389-399. [DOI] [PubMed] [Google Scholar]
- 4.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon. Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 5.Davoll, J., and D. D. Evans. 1960. The synthesis of 9-glycitylpurines, 3-glycityl-[1,2,3]-trizolo[d]-pyrimidines, 9-glycitylpteridines, and 10-glycitylbenzo[g]pteridines, including riboflavin and riboflavin 2-imine. J. Chem. Soc. (London) 1960:5041-5049. [Google Scholar]
- 6.Eberhardt, S., S. Korn, F. Lottspeich, and A. Bacher. 1997. Biosynthesis of riboflavin: an unusual riboflavin synthase of Methanobacterium thermoautotrophicum. J. Bacteriol. 179:2938-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisenreich, W., B. Schwarzkopf, and A. Bacher. 1991. Biosynthesis of nucleotides, flavins, and deazaflavins in Methanobacterium thermoautotrophicum. J. Biol. Chem. 266:9622-9631. [PubMed] [Google Scholar]
- 8.Foor, F., and G. M. Brown. 1975. Purification and properties of the guanosine triphosphate cyclohydrolase II from Escherichia coli. J. Biol. Chem. 250:3545-3551. [PubMed] [Google Scholar]
- 9.Graupner, M., and R. H. White. 2001. Biosynthesis of the phosphodiester bond in coenzyme F420 in the methanoarchaea. Biochemistry 40:10859-10872. [DOI] [PubMed] [Google Scholar]
- 10.Herz, S., S. Eberhardt, and A. Bacher. 2000. Biosynthesis of riboflavin in plants. The ribA gene of Arabidopsis thaliana specifies a bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase. Phytochemistry 53:723-731. [DOI] [PubMed] [Google Scholar]
- 11.Holm, L., and C. Sander. 1997. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins 28:72-82. [PubMed] [Google Scholar]
- 12.Howell, D. M., and R. H. White. 1997. d-erythro-neopterin biosynthesis in the methanogenic archaea Methanococcus thermophila and Methanobacterium thermoautotrophicum ΔH. J. Bacteriol. 179:5165-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, J. C. Venter, et al. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 14.Koonin, E. V., A. R. Mushegian, M. Y. Galperin, and D. R. Walker. 1997. Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol. Microbiol. 25:619-637. [DOI] [PubMed] [Google Scholar]
- 15.Le Van, Q., B. Schwarzkopf, and A. Bacher. 1985. Biosynthesis of 7,8-didemethyl-8-hydroxy-5-deazariboflavin, the chromophoric moiety of coenzyme F420. J. Am. Chem. Soc. 107:8300-8301. [Google Scholar]
- 16.Nelson, M. J., and J. G. Ferry. 1984. Carbon monoxide-dependent methyl coenzyme M methylreductase in acetotrophic Methosarcina spp. J. Bacteriol. 160:526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen, P., and A. Bacher. 1988. Biosynthesis of riboflavin. A simple synthesis of the substrate and product of the pyrimidine deaminase and of structural analogs. Z. Naturforsch. Teil B 43:1358-1364.
- 18.Plaut, G. W. E., and R. A. Harvey. 1971. The enzymatic synthesis of riboflavin, p. 515-538. In Methods in enzymology, vol. 18B. Academic Press, New York, N.Y.
- 19.Reuke, B., S. Korn, W. Eisenreich, and A. Bacher. 1992. Biosynthetic precursors of deazaflavins. J. Bacteriol. 174:4042-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter, G., M. Fischer, C. Krieger, S. Eberhardt, H. Lüttgen, I. Gerstenschläger, and A. Bacher. 1997. Biosynthesis of riboflavin: characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis. J. Bacteriol. 179:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, J. N. Reeve, et al. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]