Abstract
Alteromonas sp. strain O-7 secretes several proteins in response to chitin induction. We have found that one of these proteins, designated AprIV, is a novel chitin-binding protease involved in chitinolytic activity. The gene encoding AprIV (aprIV) was cloned in Escherichia coli. DNA sequencing analysis revealed that the open reading frame of aprIV encoded a protein of 547 amino acids with a calculated molecular mass of 57,104 Da. AprIV is a modular enzyme consisting of five domains: the signal sequence, the N-terminal proregion, the family A subtilase region, the polycystic kidney disease domain (PkdD), and the chitin-binding domain type 3 (ChtBD3). Expression plasmids coding for PkdD or both PkdD and ChtBD (PkdD-ChtBD) were constructed. The PkdD-ChtBD but not PkdD exhibited strong binding to α-chitin and β-chitin. Western and Northern analyses demonstrated that aprIV was induced in the presence of N-acetylglucosamine, N-acetylchitobiose, or chitin. Native AprIV was purified to homogeneity from Alteromonas sp. strain O-7 and characterized. The molecular mass of mature AprIV was estimated to be 44 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The optimum pH and temperature of AprIV were pH 11.5 and 35°C, respectively, and even at 10°C the enzyme showed 25% of the maximum activity. Pretreatment of native chitin with AprIV significantly promoted chitinase activity.
Chitin, an insoluble linear β-1,4-linked polymer of N-acetylglucosamine (GlcNAc), is an abundant organic compound in nature. This polysaccharide is found in the cell walls of fungi and the cuticles of insects and crustacea (11). Since carbon and nitrogen are generally limited in the marine environment, chitin is a particularly important nutrient source for marine organisms (7). Chitinolytic marine bacteria play a crucial role in the recycling of chitinous materials, such as zooplankton (42). To degrade chitin, chitinolytic bacteria produce two classes of enzymes: chitinases (EC 3.2.1.14) and β-N-acetylglucosaminidases (GlcNAcases; EC 3.2.1.30). Chitinases hydrolyze chitin to soluble oligosaccharides, mainly chitobiose, which are hydrolyzed to GlcNAc by GlcNAcases. The degradation products, mainly GlcNAc, are then taken up and used by cells as carbon and nitrogen source.
Alteromonas sp. strain O-7 was isolated at the Sagami Bay of Japan as an efficient producer of chitinolytic enzymes (29). We have been studying the chitinolytic system of the strain to clarify the roles of individual enzymes in chitin degradation, the relationship between structure and function, and the regulation of gene expression. We have already shown that the chitinolytic system of the strain consists of at least four chitinases (Chi85, ChiA, ChiB, and ChiC), three β-N-acetylglucosaminidases (GlcNAcase A, GlcNAcase B, and GlcNAcase C), chitinase-like enzyme (ChiD), and chitin-binding protein (Cbp1). These genes have been cloned and sequenced, and the role of each protein in chitin degradation has been clarified (31-33, 35, 37).
In crustacean cuticles, chitin is tightly associated with proteins, inorganic salts, such as calcium carbonate, and lipids, including pigments (24). Thus, both chitinases and proteases could be necessary for efficient degradation of crustacean cuticles in the natural environment. However, there is no report on proteases involved in chitin degradation by marine bacteria despite the extensive literature of microbial proteases. Recently, we found a novel serine protease that was expressed selectively by Alteromonas sp. strain O-7 in the presence of chitin. In this study, we report the cloning, sequence, and analysis of the gene coding for the chitin-binding protease (AprIV). Furthermore, the purification and characterization of the enzyme from the native strain are described.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Alteromonas sp. strain O-7 was cultured at 27°C in Bacto Marine Broth 2216 (Difco). E. coli JM109 and BL21(DE3) were grown at 37°C on Luria-Bertani medium supplemented with appropriate antibiotics for the selection of transformants and for the production of recombinant protein, respectively. Plasmids pUC18 and pUC19 were used as cloning vectors. Plasmid pGEX-6P-1 (Amersham Pharmacia Biotech) was used as the expression vector.
Nucleotide sequence determination.
Nucleotide sequencing was carried out by the dideoxy chain termination method by using the DYEnamic ET terminator cycle sequencing premix kit (Amersham Pharmacia Biotech) on a DNA sequencer (ABI Prism 310 Genetic Analyzer; Applied Biosystems). Sequence data were analyzed by using GENETYX-WIN program (Software Development Co., Ltd.).
N-terminal amino acid sequence of AprIV.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done by the method of Laemmli (15). Alteromonas sp. strain O-7 was cultured at 27°C in Bacto Marine Broth 2216 containing 0.2% powdered chitin from crab shell (Nacalai Tesque, Kyoto, Japan) until the optical density at 600 nm reached 1.5. The culture supernatant was collected by centrifugation (24,650 × g for 5 min at 4°C), and 0.1 volume of 20% trichloroacetic acid was added to the supernatant. After centrifugation (24,650 × g for 5 min at 4°C), the pellet was dissolved in SDS-PAGE sample buffer. Proteins were separated by SDS-PAGE and transferred to Sequi-Blot PVDF-Membrane (Bio-Rad Laboratories) with Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad Laboratories). The chitin-induced protein band corresponding to AprIV was directly sequenced by using an ABI Procise 491 HT protein sequencer (Applied Biosystems) that was connected to an on-line phenylthiohydantoin (PTH) derivative analyzer.
Cloning of the aprIV gene.
Alteromonas sp. strain O-7 total DNA was digested with various restriction enzymes and electrophoresed on a 1.0% agarose gel. The fragments in the range of 1.0 to 2.0 kb were excised from the gel and purified with GenElute gel purification kit (Sigma). These fragments were self-ligated and used as a template DNAs. The degenerate inverse primers [aprN, 5′-AT(A/G)CC(A/G)TA(A/T)GG(A/G/T)GT(A/G/T)GTTTC(A/T)GC-3′; aprC, 5′-CAAGC(A/T)AA(C/T)CAAGG(C/T)(A/T)(C/G)(A/T)GATGG-3′] were synthesized based on the N-terminal amino acid sequence of AprIV. PCR amplification was performed by KOD-Plus-DNA polymerase (Toyobo, Osaka, Japan) for 30 cycles consisting of 94°C for 15 s, 53°C for 30 s, and 68°C for 2 min. The 1.5-kb product was amplified by using the SphI digested template and phosphorylated by T4 polynucleotide kinase (Toyobo). The phosphorylated fragment was digested with SphI. The resulting 1.0-kb (pSpX10) and 0.5-kb (pXSp05) fragments were cloned into SmaI and SphI sites of pUC19. However, analyses of the entire nucleotide sequences of the inserted DNAs indicated that the 3′ downstream region of aprIV gene was missing. To clone the 3′downstream region of aprIV gene, Southern hybridization was performed with 1.0-kb (pSpX10) or 0.5-kb (pXSp05) fragment as a probe. The probes were labeled with alkaline phosphatase according to the manufacturer's instructions (AlkPhos DIRECT; Amersham Pharmacia Biotech). Both probes hybridized with the same 4.5-kb chromosomal fragment digested with EcoRV. The DNA fragments corresponding to 4.5-kb were excised from the gel and purified with a GenElute gel purification kit. These were ligated into the dephosphorylated SmaI site, and the recombinant plasmids were introduced into competent E. coli JM109. The library was screened by colony hybridization with the labeled probes as described previously (34).
Northern blot and primer extension analysis.
Northern blot and primer extension analyses were performed as described previously (36). The amount of transcript of aprIV was measured by Image Gauge version 3.45 (Fuji Film, Tokyo, Japan). Then, 10 μg of RNA was used to map the 5′ end of the aprIV transcript. Reverse transcription was initiated from the fluorescein isothiocyanate (FITC)-labeled primer complementary to the 5′ end of the aprIV coding region.
Construction of expression plasmids.
The expression plasmid, pGstPkdDChtBD, coding for PkdD and ChtBD (PkdD-ChtBD) of AprIV, was constructed as follows. Two oligonucleotide primers (pkdDF, 5′-AAGCGGCGGTTGAATTCCTAACAAATAATG-3′; chtBDR, 5′-GACTAATTTGTACTCGAGACATTGCGCGAC-3′) were synthesized, which were modified to contain EcoRI and XhoI recognition sites to facilitate cloning in frame into the glutathione S-transferase (GST) fusion protein expression vector pGEX-6P-1 (Amersham Pharmacia Biotech). PCR was performed with the plasmid pAP45 as a template for 30 cycles consisting of 94°C for 15 s, 57°C for 30 s, and 68°C for 1 min. The amplified DNA was digested by EcoRI and XhoI, and the resulting fragment (424 bp) was inserted into the corresponding sites of pGEX-6P-1. The expression plasmids, pGstPkdD and pGstChtBD, coding for PkdD and ChtBD, respectively, were constructed in the same manner as pGstPkdDChtBD, with four primers: pkdDF, pkdDR (5′-TAGTTGCACTCCACTCGAGGATACCATTAC-3′), chtBDF (5′-GGGAATTTGGTGAATTCGCAACAAGTTCAC-3′), and chtBDR. The nucleotide sequences of the PCR fragments were confirmed by DNA sequencing.
Purification of the recombinant proteins.
E. coli BL21(DE3) cells harboring pGstPkdDChtBD, pGstPkdD, and pGstChtBD were induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at the mid-exponential growth phase and further incubated for 1.5 h at 37°C. Cells were harvested by centrifugation and washed and resuspended with phosphate-buffered saline. The cells were disrupted by sonication, and the lysate was centrifuged at 10,000 × g for 10 min. The GST fusion proteins (GST-PkdD-ChtBD and GST-PkdD), except for GST-ChtBD, were purified from the supernatant by affinity chromatography with glutathione-Sepharose 4B (Amersham Pharmacia Biotech). The purified fusion proteins were treated with PreScission protease (Amersham Pharmacia Biotech) for 12 h at 4°C to obtain PkdD-ChtBD and PkdD. The N-terminal amino acid sequences of PkdD-ChtBD and PkdD were confirmed by protein sequencing.
Western blotting and immunodetection.
As described for the determination of N-terminal amino acid sequence, proteins from the culture supernatant were separated by SDS-PAGE and transferred to Sequi-Blot PVDF-Membrane. The membrane was incubated for 1 h at room temperature with anti-PkdD-ChtBD polyclonal mouse antiserum diluted to 1:2,000 in phosphate-buffered saline containing 2.0% skim milk (Difco). Bound antibody was detected as described previously (36). The amount of production of AprIV was measured by Image Gauge version 3.45.
Purification of AprIV.
Alteromonas sp. strain O-7 was cultured in Bacto Marine Broth 2216 containing 0.2% powdered chitin from crab shell (Nacalai Tesque) until the optical density at 600 nm reached 1.5. The supernatant was dialyzed against 50 mM Tris-HCl buffer (pH 8.0) and applied to a DEAE-Toyopearl column (3.0 by 15 cm; Tosoh, Tokyo, Japan). Active fractions in the flowthrough were collected, and ammonium sulfate was added to the fraction until the final concentration reached 1.3 M. The fraction was applied to a phenyl-Toyopearl 650 M column (1.0 by 20 cm; Tosoh) equilibrated with 50 mM Tris-HCl buffer (pH 8.0) containing 1.3 M ammonium sulfate. Two active fractions corresponding to native AprIV and proteolytic AprIV (AprIVΔC) were eluted at concentrations of ca. 0.6 and 0.1 M ammonium sulfate, respectively.
Enzyme assay.
Protease activity and properties of AprIV were examined as described previously (28). Chitinase activity was measured as described elsewhere (31).
Effects of AprIV and AprIVΔC on chitinase activity.
The cuticles of prawn shell (Penaeoida) were stripped off and dried at 50°C for 24 h. The cuticles were crushed into powder in a mixer and filtered with no. 48 mesh (Iidaseisakusho, Osaka, Japan). The crude powdered chitin (10 mg) was washed twice with 1 ml of 50 mM Tris-HCl buffer (pH 8.0) and centrifuged. The pellets were suspended with 100 μl of the same buffer containing 5 mU of purified AprIV or AprIVΔC and incubated at 27°C for 1 h. The reaction mixtures were then centrifuged, and the removed proteins from the chitin were assayed by the method of Bradford with bovine serum albumin as a standard (2). The chitin was suspended with 1 ml of the buffer containing 1 mM phenylmethylsulfonyl fluoride and kept at room temperature for 30 min. The suspension was centrifuged and then resuspended with 400 μl of the buffer containing 0.5 μg of purified chitinase (Chi85) from the strain. The reaction mixture was incubated at 50°C for 6 h. Samples were taken at 1-h intervals. The reaction was terminated by cooling the mixture on ice, followed by centrifugation at 24,650 × g for 5 min. Reducing sugar in the supernatant was measured by a modification of Schales's procedure (23). The deproteinized chitin was prepared according to the method of Hackman (10). The effects of AprIV and AprIVΔC on chitinase activity were also examined by using the deproteinized chitin as a substrate.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this study are in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB071535.
RESULTS
Cloning and nucleotide sequence of aprIV.
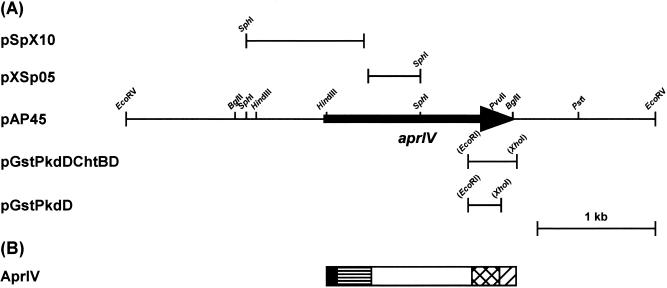
Alteromonas sp. strain O-7 was cultured in the presence of chitin, and extracellular proteins were separated by SDS-PAGE. The N-terminal amino acid sequence (AETTPYGINMVQANQVSDGY) of a 44-kDa chitin-induced protein was determined. The BLAST search program revealed that the protein, designated AprIV, is homologous to enzymes belonging to subtilase family of microbial serine proteases (26). Thus, to isolate the gene encoding AprIV, degenerate inverse primers (aprN and aprC) were synthesized based on the N-terminal amino acid sequence. The amplified DNA was ca. 1.5 kb, and the nucleotide sequences of the SphI digests (pSpX10 and pXSp05 [Fig. 1]) were analyzed. However, the PCR product appeared to lack the 3′ downstream region of the aprIV gene. To obtain the full-length gene, colony hybridization was performed by using a 1.0-kb DNA fragment of pSpX10 or a 0.5-kp DNA fragment of pXSp05 as a probe. Among 1,200 transformants, one positive clone (pAP45) which hybridized to both probes was isolated by colony hybridization. The clone was analyzed by restriction endonuclease digestion and found to contain a 4.5-kb insert with the full-length aprIV gene (Fig. 1).
FIG. 1.
Restriction map of pAP45 and domain structure of AprIV. (A) Restriction map of pAP45. Arrow indicates the ORF and the direction of transcription. aprIV, the gene encoding serine protease. (B) Diagram of domain structure of AprIV: signal peptide, ▪; N-terminal proregion, ▤; protease region, □; PkdD, ▩; ChtBD, ▨.
The nucleotide sequence of the SphI-PstI fragment (2.8 kb) from pAP45 was determined. The open reading frame (ORF) has an ATG start codon at position 657, which is preceded by a possible ribosome-binding site (AGGTG) at a distance of four nucleotides. An inverted repeat, which was composed of an 8-bp stem and a loop of five bases, was located 17 bp downstream of the aprIV gene terminal codon (TAA). The aprIV gene encodes a protein of 547 amino acids with a calculated molecular mass of 57,104 Da. The first 29 amino acids of the ORF encode a typical signal peptide sequence composed of an N-terminal positively charged amino acid followed by a hydrophobic region and a signal peptidase cleavage site, which fits the “−3, −1” rule of von Heijne (38). The N-terminal amino acid sequence of AprIV coincided precisely with the sequence starting from alanine 127 of the deduced polypeptide. These results suggest that the N-terminal sequence of AprIV (residues Asp30-Leu126) is a proregion, as is commonly found in the subtilase family (22).
BLAST search analysis of the deduced amino acid sequence of AprIV revealed that the gene encoded a modular enzyme consisting of five domains: a signal sequence (29 amino acids), an N-terminal proregion (97 amino acids), a protease region (289 amino acids), PkdD (75 amino acids), and ChtBD (57 amino acids). The protease region (residues Ala127-Ala404) of AprIV is classified into family A subtilases (26). Most family A subtilases are produced by the genus Bacillus. In gram-negative bacteria, it was reported that SapSh from psychrotrophic Shewanella sp. strain Ac10 (13) and VapT from alkaliphilic Vibrio metschnikovii strain RH530 (14) belong to this family and that, unlike Bacillus proteases, approximately 150 amino acid residues are inserted between the active site residues His and Ser of these enzymes. However, this insertion sequence was missing in AprIV. PkdD, which was first identified in human polycystin-1 (6), is also found in bacterial collagenases (16), proteases (19), cellulases (1), and chitinases (21). This domain has a β-sandwich fold, and the sequence, WDFGDG, is highly conserved in the domain (4). Residues 416 to 490 of AprIV showed similar sequence with PkdD, with the conserved sequence at residues 445 to 450. The PkdD of AprIV showed sequence similarity with those of lysyl endopeptidase (AP1) from Achromobacter lyticus (56.4% identity) (20), xanthomonapepsin from Xanthomonas campestris (48.7% identity) (19), collagenase (ColH) from Clostridium histolyticum (38.5% identity) (16), and human polycystin (34.6% identity) (5). As shown in Fig. 2, the C-terminal domain (residues from 500 to 543 amino acids) showed sequence homology with type 3 ChtBD (ChtBD3), as classified by the NCBI conserved domain database (CDD, http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). The ChtBD of AprIV had similarities with those of chitinase from Aeromonas punctata (42.9% identity) (27), chitodextrinase from Vibrio furnissii (35.7% identity) (12), cellulase from Erwinia chrysanthemi (23.8%) (9), and chitinase from Serratia marcescens (23.8% identity) (41).
FIG. 2.
Comparison of the amino acid sequence of ChtBD of AprIV with those of other proteins. ApChiA, Aeromonas punctata chitinase A; EndoI, Vibrio furnissii chitodextrinase; Cbp1, ChiA, ChiC, and ChiD, chitin-binding protein, chitinase A, chitinase C, and chitinase-like enzyme from Alteromonas sp. strain O-7, respectively; Cel5, Erwinia chrysanthemi cellulase; SmChiB, Serratia marcescens chitinase B. The stWWst motif, where “s” and “t” represent small residues and turn-like residues, respectively, is shown by a shadowed box. Residues that are identical are indicated by bold letters.
Northern blot analysis.
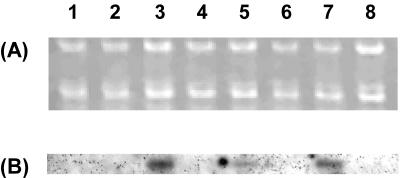
To confirm whether the expression of aprIV was induced by chitin and related compounds, Northern blot analysis was performed. Alteromonas sp. strain O-7 was cultured in Bacto Marine Broth containing either GlcNAc, N-acetylchitobiose, powdered chitin, or skim milk. The RNA transcript of aprIV was induced by 8.6-, 11.5-, and 14.3-fold in the presence of 1.0% N-acetylchitobiose, 0.5% powdered chitin, and 1.0% GlcNAc, respectively. However, GlcNAc and N-acetylchitobiose did not influence the expression of aprIV at the concentration of 0.5%. On the other hand, skim milk, which is often used to induce microbial proteases, did not induce the transcript (Fig. 3). These results indicate that chitin or GlcNAc induces AprIV production in Alteromonas sp. strain O-7.
FIG. 3.
Northern blot analysis of the aprIV transcript. Lanes: 1, control; 2 and 3, 0.5 and 1.0% GlcNAc, respectively; 4 and 5, 0.5 and 1.0% N-acetylchitobiose, respectively; 6, 0.5% glycol chitin; 7, 0.5% powdered chitin; 8, 1.0% skim milk. (A) Formaldehyde-agarose gel electrophoresis of the total RNA (1.2 μg) from the strain; (B) Northern blot analysis. The HindIII-SphI fragment (0.8 kb) from pAP45 was used as a probe.
Functions of PkdD and ChtBD.
Judging from the deduced amino acid sequence of AprIV, mature AprIV is a putative chitin-binding protease consisting of three discrete domains. To examine the function of PkdD and ChtBD, expression plasmids, coding for PkdD-ChtBD, PkdD or ChtBD were constructed by using the GST fusion protein expression vector pGEX-6P-1 as described in Materials and Methods (Fig. 1). The GST-ChtBD protein accumulated in the cells as an inclusion body. Then, the protein pellet was solubilized in 6 M guanidine hydrochloride and dialyzed against a series of buffers with stepwise decreases in the urea concentration (50 mM Tris-HCl buffer [pH 7.0] containing 6, 4, 2, and 0 M urea). However, the GST-ChtBD aggregated again during dialysis. Therefore, we could not purify the protein. On the other hand, GST-PkdD-ChtBD and GST-PkdD were purified from the supernatants of the cells by affinity chromatography with glutathione-Sepharose 4B, and these fusion proteins were treated by PreScission protease to obtain PkdD-ChtBD and PkdD. Each of the purified proteins (5 μg) was mixed with 5 mg of α-chitin, β-chitin, chitosan, cellulose, or Avicel. The binding assay mixtures were incubated on ice for 30 min, and then the pellets were directly dissolved with SDS-PAGE sample buffer. The amount of binding protein was measured by Image Gauge (data not shown). The PkdD-ChtBD of AprIV exhibited strong binding activity to α-chitin (80%) and β-chitin (70%) but weak affinity for chitosan (8%), cellulose (18%), and Avicel (16%). PkdD showed no binding to any polysaccharides tested. These results indicate that ChtBD of AprIV contributes to the binding to α-chitin and β-chitin.
Purification of AprIV and AprIVΔC.
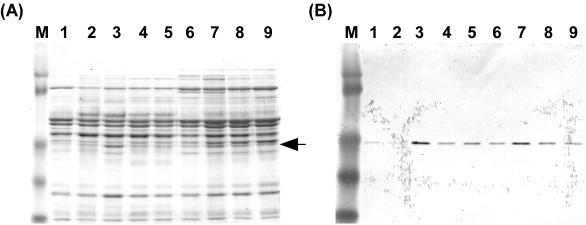
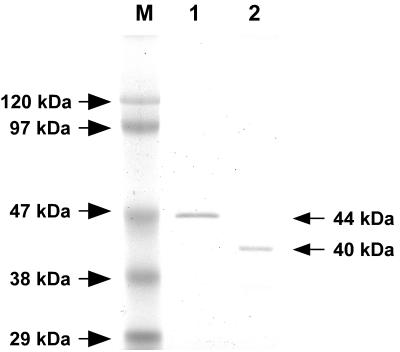
Before we began the purification of AprIV, we investigated the optimum conditions for AprIV production. Alteromonas sp. strain O-7 was cultured in Bacto Marine Broth containing GlcNAc, N-acetylchitobiose, glycol chitin, or powdered chitin, and then the production levels of AprIV in culture supernatants were analyzed by Western blotting with anti-PkdD-ChtBD polyclonal mouse antiserum (Fig. 4). AprIV production increased by 3.4-, 5.8-, and 6.5-fold in the presence of 1.0% N-acetylchitobiose, 0.2% powdered chitin, and 1.0% GlcNAc, respectively, compared to that of no addition. Thus, the strain was cultured in the medium containing 1.0% GlcNAc, and AprIV was purified by successive column chromatographies on DEAE-Toyopearl 650 M and phenyl-Toyopearl 650 M. The enzyme was purified ca. 4.4-fold with a yield of 1.8%. The yield of the enzyme was very low because the strain secreted several proteases into the medium. Two active fractions were obtained from phenyl-Toyopearl 650 M column chromatography. The molecular masses of these proteases estimated by SDS-PAGE were 44 and 40 kDa (Fig. 5), and the N-terminal amino acid sequences (AETTPYGINM) were identical. These results indicate that the 40-kDa AprIV (AprIVΔC) is produced by partial proteolysis of mature AprIV.
FIG. 4.
Western blot analysis of AprIV. (A) Coomassie blue staining. Lanes: M, prestained molecular weight marker; 1, control; 2 and 3, 0.5 and 1.0% GlcNAc, respectively; 4 and 5, 0.5 and 1.0% N-acetylchitobiose, respectively; 6, 0.5% glycol chitin; 7, 8, and 9, 0.2, 0.5, and 1.0% powdered chitin, respectively. The arrow indicates AprIV protein. (B) Western blot analysis.
FIG. 5.
SDS-PAGE of AprIV and AprIVΔC. Lanes: M, marker proteins; 1, AprIV; 2, AprIVΔC.
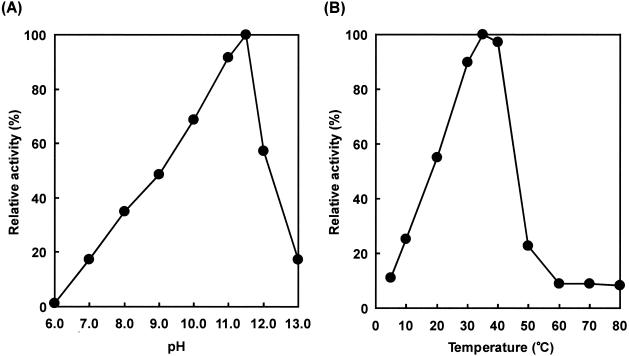
Characterization of AprIV.
The enzymatic characterization of the purified AprIV was investigated. The optimum pH and temperature were pH 11.5 and 35°C, respectively, when casein was used as a substrate (Fig. 6). The optimum temperature of AprIV was much lower than that of other family A subtilases, and even at 10°C the enzyme showed 25% of the maximum activity, indicating that AprIV is a cold-adapted enzyme like SapSh from psychrotrophic Shewanella sp. strain Ac10 (13). The activity of AprIV was inhibited by 1 mM PMSF and 1 mM EDTA and was stabilized by the addition of 1 mM Ca2+. Thus, AprIV is a chelator-sensitive and cold-adapted alkaline serine protease. AprIVΔC showed almost the same enzymatic properties as mature AprIV except for binding to chitin (data not shown).
FIG. 6.
Effects of pH and temperature on protease activity of AprIV. (A) The following buffer systems were used: 50 mM acetate buffer (pH 6.0), 50 mM Tris-HCl buffer (pH 7.0 to 9.0), and 50 mM glycine-NaOH buffer (pH 10.0 to 13.0). The reaction mixtures were incubated at 35°C for 30 min. The amount enzyme was 13.6 U. (B) The reaction was carried out at the various temperatures for 30 min at pH 11.5 (50 mM glycine-NaOH buffer). The amount of enzyme was 13.6 U.
Effects of AprIV and AprIVΔC on chitinase activity.
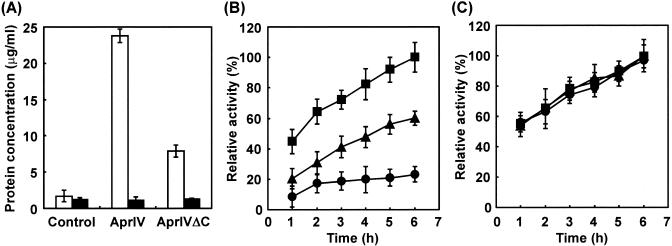
We investigated the effects of AprIV and AprIVΔC on chitin degradation. Since commercially available chitin was treated with NaOH to remove proteins, deproteinized chitin was not used as a substrate of AprIV (data not shown). Thus, we prepared native chitin from the exoskeleton of prawn. AprIV significantly removed proteins from native chitin in comparison with AprIVΔC (Fig. 7A). When chitin pretreated with or without AprIV was used as substrate, Chi85, the major chitinase of Alteromonas sp. strain O-7, hydrolyzed AprIV-treated chitin more effectively than intact chitin (Fig. 7B). On the other hand, when AprIVΔC-treated chitin was used as substrate, the stimulation of chitinase activity partly reduced. These results indicate that ChtBD of AprIV plays an important role in targeting the enzyme to the substrate by increasing its local concentration on chitin-linked proteins. Furthermore, when deproteinized chitin from the exoskeleton of prawn was used as substrate, AprIV showed no effect on chitinase activity (Fig. 7C).
FIG. 7.
Particitation of AprIV and AprIVΔC in chitin degradation. (A) Protease activities of AprIV and AprIVΔC were measured with the substrates native chitin (open bar) and deproteinized chitin (solid bar). Control experiments were carried out without enzyme. (B and C) The effects of AprIV and AprIVΔC on chitinase activity were examined by using native chitin (B) or deproteinized chitin (C). Symbols: •, Chi85; ▴, Chi85 plus AprIVΔC; ▪, Chi85 plus AprIV. Aliquots were taken at the indicated times and centrifuged, and chitinase activity was measured. The value after incubation for 6 h with AprIV and Chi85 was taken as 100%. Data from three independent experiments are shown; standard deviations are indicated by vertical lines.
Primer extension analysis.
Primer extension analysis was carried out to determine the transcriptional start point and to locate the promoter of aprIV. The analysis determined the transcriptional start point to be the C that was 27 bp upstream from the initiation codon (Fig. 8). TTGACA (positions −35 to −30) and TTTAGT (positions −12 to −7) with a 17-bp space similar to the consensus sequence typical of prokaryotic promoters seem to be the promoter of aprIV.
FIG. 8.
Determination of the transcription start site of aprIV. (A) Primer extension and nucleotide sequencing were performed with the same FITC-labeled primer. The nucleotide sequence around the transcription start site is shown in lanes A, C, G, and T. The transcriptional start site is shown by an arrow (lane P). (B) Nucleotide sequence of the 5′ upstream regions of aprIV. The deduced amino acid sequence of AprIV is shown below the nucleotide sequence. The transcriptional start site is indicated by “+1.” The position of FITC-labeled primer is shown below the nucleotide sequence by an arrow. The putative −35 and −10 regions are shown by boxes. The conserved sequences are shown by shadowed boxes. The inverted repeat sequences are indicated by convergent arrows.
DISCUSSION
Alteromonas sp. strain O-7 secretes four serine proteases and three metalloproteases (17). Of these, the genes for three serine proteases (AprI, AprII, and AprIII) and two metalloproteases (MprI and MprII) have been cloned and sequenced, and the relationship between structure and function of these enzymes has been investigated (17, 30, 34). Furthermore, we found that the aprII gene was regulated by Fur protein, which plays an essential role in the iron acquisition system (36). In the present study, we clarified that the fourth serine protease, designated AprIV, is a novel chitin-binding protease involved in chitinolytic activity. The aprIV gene encoded a modular novel chitin-binding protease (AprIV) composed of a signal peptide, an N-terminal proregion, a family A subtilase region, PkdD, and type 3 ChtBD. Recently, ChtBDs have been classified into types 1, 2, and 3 in the NCBI conserved domain database. Types 1, 2, and 3 consist of ChtBDs from fungi and plants, viruses and insects, and bacteria, respectively. The ChtBD of AprIV is 57 amino acid residues long and exhibits homology with ChtBDs classified into type 3. Sequence alignment of ChtBD of AprIV with those of type 3 revealed a consensus sequence (AKWWTQ) that is well conserved in bacterial ChtBDs. Brun et al. demonstrated that the consensus sequence of cellulase was essential for the cellulose-binding domain-cellulose interaction based on the three-dimensional structure (3). Two Trp residues in the consensus sequence of AprIV might be involved in the stacking against the pyranose rings of GlcNAc residue in chitin. A number of serine proteases have been reported but, to date, there are only two examples of a chitin-binding protease. Streptomyces griseus protease C is a chymotrypsin-like serine protease (25), and the carboxy-terminal domain shares extensive homology with ChtBDs of Bacillus circulans chitinases A1 and D (39, 40). However, the carbohydrate-binding capabilities and its activity in the degradation of chitin-linked proteins have not been examined experimentally. Furthermore, ChtBD of Streptomyces griseus protease C does not have the highly conserved sequence of type 3 ChtBD. A modular chitin-binding protease (Sp22D) from the malaria vector, Anopheles gambiae, consists of two ChtBDs, mucin-like repeats, a tandem array of low-density lipoprotein receptor-like domains, a scavenger receptor cystein rich domain, and a trypsin-like domain (8). It was proposed that Sp22D might serve as sentinel to detect exposed chitin and then trigger appropriate physiological or developmental responses based on domain organization (5). AprIV clearly differs from these two chitin-binding proteases in both structure and function, that is, the protease region of AprIV is classified in the subtilisin family, and ChtBD of AprIV possesses the highly conserved sequence found in ChtBD type3 (Fig. 2). Furthermore, we demonstrated that ChtBD of AprIV selectively adsorbed chitin and that chitinase activity was significantly promoted by the pretreatment of native chitin with AprIV. Removal of proteins in the surface cuticles by AprIV appears to facilitate the approach between chitinase and chitin molecules.
Native AprIV was purified to homogeneity by conventional chromatographic techniques. Its optimum temperature was 35°C, much lower than that of family A subtilases except for protease (SapSh) from psychrophilic Shewanella sp. strain Ac10, whose optimum temperature was 45°C (13). In general, the psychrophilic enzymes exhibit high thermal lability. After incubation of AprIV at 30°C for 30 min, the enzyme lost 67% of its activity.
The results of Northern and Western analyses demonstrated that the synthesis of AprIV is induced in the presence of chitin, N-acetylchitobiose, or GlcNAc. However, the induction rate determined by Northern blotting was different from that determined by Western blotting, although the order of the inducer specificity was identical. This seems to be due to the binding to chitin and/or proteolytic degradation of AprIV. Chitinases of the strain are also induced in the presence of chitin, N-acetylchitobiose, or GlcNAc. These results suggest that the expression of AprIV and chitinases involved in chitin degradation system of Alteromonas sp. strain O-7 might be coordinately controlled by the same regulatory protein. The nucleotide sequence upstream of the aprIV gene was then aligned with those of the chitinase genes. The alignment revealed the conserved 8-bp sequence, 5′-ACAACATG-3′ (shown in Fig. 8), found in these genes, except in ChiD, where only a 5-bp portion was identical to the conserved sequence. These sequences were located at −16, −23, −11, and −23 bp upstream of the ATG initiation codons of aprIV, chiA, chiC, and chiD genes, respectively. In addition, an inverted repeat motif, which overlapped with the conserved sequence of these genes, was found. The inverted repeat of chiA consists of TACA-Nx-TGTA, and the motif is similar to that of aprIV and chiC. However, one side of the inverted repeat is missing in chiD. Although microbial chitinolytic enzymes and the related proteins are usually substrate inducible, the regulatory gene remains to be identified. Ni and Westpheling demonstrated that chitinase genes of Streptomyces share a similar pair of direct repeat sequences in the promoter region, which have been suggested to be involved in chitin induction (18). The promoter region that includes the direct repeat sequences was shown to interact with a sequence-specific DNA-binding factor by electromobility shift assay. It is under investigation whether the inverted motif of aprIV, chiA, chic, and chiD is the cis-acting sequence which regulates the expression of genes in chitinolytic system of Alteromonas sp. strain O-7.
REFERENCES
- 1.Ahsan, M. M., T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1996. Cloning, DNA sequencing, and expression of the gene encoding Clostridium thermocellum cellulase CelJ, the largest catalytic component of the cellulosome. J. Bacteriol. 178:5732-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Brun, E., F. Moriaud, P. Gans, M. J. Blackledge, F. Barras, and D. Marion. 1997. Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi. Biochemistry 36:16074-16086. [DOI] [PubMed] [Google Scholar]
- 4.Bycroft, M., A. Bateman, J. Clarke, S. J. Hamill, R. Sandford, R. L. Thomas, and C. Chothia. 1999. The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease. EMBO J. 18:297-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danielli, A., T. G. Loukeris, M. Lagueux, H. M. Muller, A. Richman, and F. C. Kafatos. 2000. A modular chitin-binding protease associated with hemocytes and hemolymph in the mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 97:7136-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gluecksmann-Kuis, M. A., O. Tayber, E. A. Woolf, L. Bougueleret, N. Deng, G. D. Alperin, F. Iris, F. Hawkins, C. Munro, N. Lakey, G. Duyk, M. C. Schneider, L. Geng, F. Zhang, Z. Zhao, S. Torosian, S. T. Reeders, P. Bork, M. Pohlschmidt, C. Loehning, B. Kraus, U. Nowicka, A. L. S. Leung, and A.-M. Frischauf. 1995. Polycystic kidney disease: the complete structure of the PKD1 gene and its protein. Cell 81:289-298.7736581 [Google Scholar]
- 7.Gooday, G. W. 1990. The ecology of chitin degradation. Adv. Microb. Ecol. 11:387-430. [Google Scholar]
- 8.Gorman, M. J., O. V. Andreeva, and S. M. Paskewitz. 2000. Sp22D: a multidomain serine protease with a putative role in insect immunity. Gene 251:9-17. [DOI] [PubMed] [Google Scholar]
- 9.Guiseppi, A., B. Cami, J. L. Aymeric, G. Ball, and N. Creuzet. 1988. Homology between endoglucanase Z of Erwinia chrysanthemi and endoglucanases of Bacillus subtilis and alkalophilic Bacillus. Mol. Microbiol. 2:159-164. [DOI] [PubMed] [Google Scholar]
- 10.Hackman, R. H. 1954. Studies on chitin 1. Enzymic degradation of chitin and chitin esters. Aust. J. Biol. Sci. 7:168-178. [DOI] [PubMed] [Google Scholar]
- 11.Jannatipur, M., R. W. Soto-Gil, L. C. Childers, and J. W. Zynskind. 1987. Translocation of Vibrio harveyi N, N′-diacetylchitobiase to the outer membrane of Escherichia coli. J. Bacteriol. 169:3785-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keyhani, N. O., and S. Roseman. 1996. The chitin catabolic cascade in the marine bacterium Vibrio furnissii: molecular cloning, isolation, and characterization of a periplasmic chitodextrinase. J. Biol. Chem. 271:33414-33424. [DOI] [PubMed] [Google Scholar]
- 13.Kulakova, L., A. Galkin, T. Kurihara, T. Yoshimura, and N. Esaki. 1999. Cold-active serine alkaline protease from the psychrotrophic bacterium Shewanella strain Ac10: gene cloning and enzyme purification and characterization. Appl. Environ. Microbiol. 65:611-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon, Y. T., J. O. Kim, S. Y. Moon, Y. D. Yoo, and H. M. Rho. 1995. Cloning and characterization of the gene encoding an extracellular alkaline serine protease from Vibrio metschnikovii strain RH530. Gene 152:59-63. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Matsushita, O., C.-M. Jung, S. Katayama, J. Minami, Y. Takahashi, and A. Okabe. 1999. Gene duplication and multiplicity of collagenases in Clostridium histolyticum. J. Bacteriol. 181:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyamoto, K., H. Tsujibo, E. Nukui, H. Itoh, Y. Kaidzu, and Y. Inamori. 2002. Isolation and characterization of the genes encoding two metalloproteases (MprI and MprII) from a marine bacterium. Alteromonas sp. strain O-7. Biosci. Biotechnol. Biochem. 66:416-421. [DOI] [PubMed]
- 18.Ni, X., and J. Westpheling. 1997. Direct repeat sequences in the Streptomyces chitinase-63 promoter direct both glucose repression and chitin induction. Proc. Natl. Acad. Sci. USA 94:13116-13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oda, K., M. Ito, K. Uchida, Y. Shibano, K. Fukuhara, S. Takahashi. 1996. Cloning and expression of an isovaleryl pepstatin-insensitive carboxyl proteinase gene from Xanthomonas sp. T-22. J. Biochem. 120:564-572. [DOI] [PubMed] [Google Scholar]
- 20.Ohara, T., K. Makino, H. Shinagawa, A. Nakata, S. Norioka, and F. Sakiyama. 1989. The primary structure and structural characteristics of Achromobacter lyticus protease I, a lysine-specific serine protease. J. Biol. Chem. 264:3832-3839. [PubMed] [Google Scholar]
- 21.Perrakis, A., I. Tews, Z. Dauter, A. B. Oppenheim, I. Chet, K. S. Wilson, and C. E. Vorgias. 1994. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure 2:1169-1180. [DOI] [PubMed] [Google Scholar]
- 22.Rao, M. B., A. M. Tanksale, M. S. Ghatge, and V. T. Deshpande. 1998. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 62:597-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schales, O., and S. S. Schales. 1945. A simple method for the determination of glucose in blood. Arch. Biochem. 8:285-292. [PubMed] [Google Scholar]
- 24.Shimahara, K., Y. Takiguchi, K. Ohkouchi, K. Kitamura, and O. Okada. 1984. Chemical composition and some properties of crustacean chitin prepared by use of proteolytic activity of Pseudomonas maltophilia LC102, p. 239-255. In J. P. Zikakis (ed.), Chitin, chitosan and related enzymes. Academic Press, Orlando, Fla.
- 25.Sidhu, S. S., G. B. Kalmar, L. G. Willis, and T. J. Borgford. 1994. Streptomyces griseus protease C: a novel enzyme of the chymotrypsin superfamily. J. Biol. Chem. 269:20167-20171. [PubMed] [Google Scholar]
- 26.Siezen, R. J., and J. A. M. Leunissen. 1997. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6:501-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitrit, Y., C. E. Vorgias, I. Chet, and A. B. Oppenheim. 1995. Cloning and primary structure of the chiA gene from Aeromonas caviae. J. Bacteriol. 177:4187-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsujibo, H., K. Miyamoto, T. Hasegawa, and Y. Inamori. 1990. Purification and characterization of two types of alkaline serine proteases produced by an alkaliphilic actinomycete. J. Appl. Bacteriol. 69:520-529. [DOI] [PubMed] [Google Scholar]
- 29.Tsujibo, H., Y. Yoshida, C. Imada, Y. Okami, K. Miyamoto, and Y. Inamori. 1991. Isolation and characterization of a chitin-degrading marine bacterium belonging to the genus Alteromonas. Fish. Sci. 57:2127-2131. [Google Scholar]
- 30.Tsujibo, H., K. Miyamoto, K. Tanaka, M. Kawai, K. Tainaka, C. Imada, Y. Okami, and Y. Inamori. 1993. Cloning and sequence of an alkaline serine protease-encoding gene from the marine bacterium Alteromonas sp. strain O-7. Gene 136:247-251. [DOI] [PubMed] [Google Scholar]
- 31.Tsujibo, H., H. Orikoshi, H. Tanno, K. Fujimoto, K. Miyamoto, C. Imada, Y. Okami, and Y. Inamori. 1993. Cloning, sequence, and expression of a chitinase gene from a marine bacterium, Alteromonas sp. strain O-7. J. Bacteriol. 175:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsujibo, H., K. Fujimoto, H. Tanno, K. Miyamoto, C. Imada, Y. Okami, and Y. Inamori. 1994. Gene sequence, purification and characterization of N-acetyl-β-glucosaminidase from a marine bacterium, Alteromonas sp. strain O-7. Gene 146:111-115. [DOI] [PubMed] [Google Scholar]
- 33.Tsujibo, H., K. Fujimoto, H. Tanno, K. Miyamoto, Y. Kimura, C. Imada, Y. Okami, and Y. Inamori. 1995. Molecular cloning of the gene which encodes β-N-acetylglucosaminidase from a marine bacterium, Alteromonas sp. strain O-7. Appl. Environ. Microbiol. 61:804-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsujibo, H., K. Miyamoto, K. Tanaka, Y. Kaidzu, C. Imada, Y. Okami, and Y. Inamori. 1996. Cloning and sequence analysis of a protease-encoding gene from the marine bacterium Alteromonas sp. strain O-7. Biosci. Biotechnol. Biochem. 60:1284-1288. [DOI] [PubMed] [Google Scholar]
- 35.Tsujibo, H., H. Orikoshi, K. Shiotani, M. Hayashi, J. Umeda, K. Miyamoto, C. Imada, Y. Okami, and Y. Inamori. 1998. Characterization of chitinase C from a marine bacterium, Alteromonas sp. strain O-7, and its corresponding gene and domain structure. Appl. Environ. Microbiol. 64:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsujibo, H., K. Miyamoto, T. Okamoto, H. Orikoshi, and Y. Inamori. 2000. A serine protease-encoding gene (aprII) of Alteromonas sp. strain O-7 is regulated by the iron uptake regulator (Fur) protein. Appl. Environ. Microbiol. 66:3778-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsujibo, H., J. Miyamoto, N. Kondo, K. Miyamoto, N. Baba, and Y. Inamori. 2000. Molecular cloning of the gene encoding an outer-membrane-associated β-N-acetylglucosaminidase involved in chitin degradation system of Alteromonas sp. strain O-7. Biosci. Biotechnol. Biochem. 64:2512-2516. [DOI] [PubMed] [Google Scholar]
- 38.von Heijne, G. 1983. Patterns of amino acids near signal sequence cleavage sites. Eur. J. Biochem. 133:17-21. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe, T., K. Suzuki, W. Oyanagi, K. Ohnishi, and H. Tanaka. 1990. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J. Biol. Chem. 265:15659-15665. [PubMed] [Google Scholar]
- 40.Watanabe, T., W. Oyanagi, K. Suzuki, K. Ohnishi, and H. Tanaka. 1992. Structure of the gene encoding chitinase D of Bacillus circulans WL-12 and possible homology of the enzyme to other prokaryotic chitinases and class III plant chitinases. J. Bacteriol. 174:408-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe, T., K. Kimura, T. Sumiya, N. Nikaidou, K. Suzuki, M. Suzuki, M. Taiyoji, S. Ferrer, and M. Regue. 1997. Genetic analysis of the chitinase system of Serratia marcescens 2170. J. Bacteriol. 179:7111-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, C., A. M. Lee, B. L. Bassler, and S. Roseman. 1991. Chitin utilization by marine bacteria. A physiological function for bacterial adhesion to immobilized carbohydrates. J. Biol. Chem. 25:24260-24267. [PubMed] [Google Scholar]