Abstract
An ADP-ribosylating toxin named Aeromonas salmonicida exoenzyme T (AexT) in A. salmonicida subsp. salmonicida, the etiological agent of furunculosis in fish, was characterized. Gene aexT, encoding toxin AexT, was cloned and characterized by sequence analysis. AexT shows significant sequence similarity to the ExoS and ExoT exotoxins of Pseudomonas aeruginosa and to the YopE cytotoxin of different Yersinia species. The aexT gene was detected in all of the 12 A. salmonicida subsp. salmonicida strains tested but was absent from all other Aeromonas species. Recombinant AexT produced in Escherichia coli possesses enzymatic ADP-ribosyltransferase activity. Monospecific polyclonal antibodies directed against purified recombinant AexT detected the toxin produced by A. salmonicida subsp. salmonicida and cross-reacted with ExoS and ExoT of P. aeruginosa. AexT toxin could be detected in a wild type (wt) strain of A. salmonicida subsp. salmonicida freshly isolated from a fish with furunculosis; however, its expression required contact with RTG-2 rainbow trout gonad cells. Under these conditions, the AexT protein was found to be intracellular or tightly cell associated. No AexT was found when A. salmonicida subsp. salmonicida was incubated in cell culture medium in the absence of RTG-2 cells. Upon infection with wt A. salmonicida subsp. salmonicida, the fish gonad RTG-2 cells rapidly underwent significant morphological changes. These changes were demonstrated to constitute cell rounding, which accompanied induction of production of AexT and which led to cell lysis after extended incubation. An aexT mutant which was constructed from the wt strain with an insertionally inactivated aexT gene by allelic exchange had no toxic effect on RTG-2 cells and was devoid of AexT production. Hence AexT is directly involved in the toxicity of A. salmonicida subsp. salmonicida for RTG-2 fish cells.
Aeromonas salmonicida subsp. salmonicida is the etiological agent of furunculosis of Salmonidae. This fish disease causes most severe losses in production farms of salmon and trout and leads to the use of large amounts of antibiotics in closed and open waters for prevention and therapy of furunculosis. To develop efficient strategies to prevent outbreaks of A. salmonicida subsp. salmonicida, it is essential to know the main mechanisms of pathogenicity of this pathogen. Several potential virulence factors of A. salmonicida subsp. salmonicida have been described thus far. They include the surface array layer protein (7); hemolysins ASH1, ASH3, and ASH4 (12); H-lysin (29); salmolysin (19); serine protease AspA (32); and the glycerophospholipid:cholesterol acyltransferase (GCAT) complexed with lipopolysaccharide (18). Recent reports demonstrate the role of the S layer in adhesion (11) of A. salmonicida subsp. salmonicida. The other potential virulence factors of A. salmonicida subsp. salmonicida that are currently known do not seem to play a primary role in pathogenesis. GCAT and aspA gene deletion mutants showed that neither GCAT nor aspA is essential for acute A. salmonicida subsp. salmonicida-induced furunculosis (30). However, AspA is essential for pro-GCAT processing in broth cultures and might also be involved in activation of other secreted enzymes or toxins.
Several pathogenic bacteria use ADP ribosylation as a key mechanism to modify the properties of host cell proteins and thus to modulate their function and induce disease. Hence ADP ribosylation of eukaryotic regulatory proteins is the underlying pathogenic mechanism of a heterogeneous family of bacterial protein toxins. ADP-ribosylating toxins are broadly distributed among highly pathogenic bacteria and are the primary cause of various severe human diseases such as diphtheria, cholera, and pertussis. Among this family of toxins, the ADP-ribosyltransferase toxin called exoenzyme S (ExoS) of Pseudomonas aeruginosa is one of the most prominent representatives. It is secreted via a type III-dependent secretion mechanism (33, 34). Recently it was shown that ExoS is a bifunctional toxin (23) containing an N-terminal part, which resembles that of the Yersinia YopE toxin and which catalyzes rho-dependent actin depolymerization, and a C-terminal ADP-ribosylating domain. Unlike most bacterial toxins, ADP-ribosylating toxin ExoS does not have a rigid target protein specificity. Intracellular expression of the amino-terminal domain of ExoS elicits the disruption of actin, while expression of the carboxyl-terminal domain of ExoS produces factor-activating exoenzyme S (FAS)-dependent ADP-ribosyltransferase activity and is highly cytotoxic to eukaryotic cells (20).
Here we characterize an ADP-ribosylating protein derived from A. salmonicida subsp. salmonicida which has a significant sequence similarity to the ExoS and the related ExoT toxins of P. aeruginosa and we demonstrate its role in toxicity to fish cells.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmid cloning vectors.
Aeromonas sp. strains (Table 1) were routinely cultured on blood agar plates (Trypticase soy agar supplemented with 0.1% CaCl2 and 5% sheep blood) at 37°C except for A. salmonicida subsp. salmonicida, which was grown at 19°C. A. salmonicida subsp. salmonicida strain JF2267 was freshly isolated from an arctic char (Salvelinus alpinus) with typical furunculosis symptoms. The strain was identified as A. salmonicida subsp. salmonicida by a routine diagnostic agglutination test using rabbit anti-A. salmonicida subsp. salmonicida-specific antiserum and by sequence analysis of the rrs (16S rRNA) genes as described by Kuhnert et al. (15). For genetic modifications, A. salmonicida subsp. salmonicida was grown on Luria-Bertani (LB) agar plates (2) supplemented as necessary with 50 μg of kanamycin/ml or with 50 μg of chloramphenicol/ml. Liquid cultures of A. salmonicida subsp. salmonicida were made in TSB (2.75 g of Trypticase soy broth/100 ml, 1% glycerol, 0.1 M l-glutamic acid, pH 7.3). Ca2+-depleted medium was made by addition of 1 mM nitrilotriacetic acid (NTA; Titriplex I; pH 7.3) to TSB.
TABLE 1.
Aeromonas strains used
| Species | Strain | No. of aexT- positive strainsa/ no. of strains tested |
|---|---|---|
| A. salmonicida subsp. salmonicida | ATCC 33658T | 1/1 |
| A. salmonicida subsp. salmonicida | JF2267b | 1/1 |
| A. salmonicida subsp. salmonicida | JF2580c | |
| A. salmonicida subsp. salmonicida | Field isolates | 10/10 |
| A. bestiarum | CDC9533-76 | 0/1 |
| A. bestiarum | Field isolates | 0/2 |
| A. caviae | ATCC 15468T | 0/1 |
| A. caviae | Field isolates | 0/3 |
| A. encheleia | DSM 11577T | 0/1 |
| A. eucrenophila | NCMB74T | 0/1 |
| A. eucrenophila | Field isolate | 0/1 |
| A. hydrophila | ATCC 7966T | 0/1 |
| A. hydrophila | Field isolates | 0/15 |
| A. jandaei | ATCC 49568T | 0/1 |
| A. media | ATCC 33907T | 0/1 |
| A. schubertii | ATCC 43700T | 0/1 |
| A. schubertii | Field isolate | 0/1 |
| A. sobria | CIP 74-33T | 0/1 |
| A. trota | ATCC 49657T | 0/1 |
| A. trota | Field isolate | 0/1 |
| A. veronii | ATCC 35624T | 0/1 |
| A. veronii | Field isolates | 0/4 |
Determined by Southern blotting using DIG-labeled RAEXT as the probe.
Isolated freshly from an arctic char with typical symptoms of furunculosis. Identification was done phenotypically and by 16S rRNA gene sequencing.
aexT mutant derived from JF2267 by allelic exchange with an aexT::Kmr construct.
Escherichia coli strains XL1-blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn 10 (Tetr)]c) (5) and BL21(DE3) (F′ dcm ompT hsdS [rB− mB−] gal λ[DE3]) (27) were used for cloning and expression of cloned genes respectively. Plasmids pBluescript II SK(−) and pBC-KS(−) (Stratagene, La Jolla, Calif.) were used as cloning vectors. Plasmid pETHIS-1 is a T7 promoter-based expression vector and allows addition of polyhistidine tails at the N-terminal end or at both the N- and C-terminal ends of proteins (26). The source of the aminoglycoside 3′-phosphotransferase gene aph(3′)-Ia conferring resistance to kanamycin (Kmr) was plasmid pSSVI186-IN (31). E. coli strains were grown at 37°C in LB broth supplemented when necessary with ampicillin (50 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (25 μg/ml) for selection and maintenance of recombinant plasmids. For blue-white differentiation of recombinant clones with pBluescript II SK(−), 125 μM X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) were added.
P. aeruginosa strain ATCC 27853 was grown for 8 h on LB plates at 20°C. To induce ExoS and ExoT secretion, cells were then incubated for 18 h at 20°C in 20 ml of TSB supplemented with 10 mM NTA (Titriplex I; pH 7.3) for chelation of Ca2+ ions.
PCR, cloning, preparation of gene probes, and genetic methods.
PCR was carried out with a DNA thermal cycler (GeneAmp 9600; PE Biosystems, Norwalk, Conn.) in 50-μl reaction mixtures containing 10 mM Tris-HCl, pH 8.3, 1.5 mM MgCl2, 50 mM KCl, 350 μM (each) deoxynucleoside triphosphate, 0.25 μM forward and reverse primers, 0.5 U of Taq polymerase, and 5 ng of template DNA. The DNAs were amplified for 35 cycles with 30 s of denaturation at 94°C, 30 s of annealing at corresponding temperatures (Table 2), and 1 min of extension at 72°C. For fragments above 1 kb, the extension time was extended by 1 min per kb. When DNA fragments were produced by PCR for subsequent cloning and expression, the Expand-Long-Template PCR kit (Roche Molecular Biochemicals, Rotkreuz, Switzerland) containing polymerase with proofreading capacity was used instead of Taq polymerase. In addition, an extension step of 7 min at 72°C was added at the end of the last cycle in order to ensure full-length synthesis. For the production of digoxigenin (DIG)-labeled probes, PCR mixtures were supplemented with 40 μM DIG-11-dUTP (Roche Molecular Biochemicals).
TABLE 2.
Oligonucleotide primers
| Primer | Sequencea | Position | Annealing temp (°C) |
|---|---|---|---|
| EXOS-L | cgcgaattcACTGGCTGGGCAAACTG | 1128-1144b | 52 |
| EXOS-R | cgcgaattCCCGCTGACATCGATTC | 2034-2019b | 52 |
| RASEXOS-L | GGCGCTTGGGCTCTACAC | 1537-1554c | 60 |
| RASEXOS-R | GAGCCCGCGCATCTTCAG | 2089-2072c | 60 |
| BASEXOSH8L | cgcgaattCGGCGAAACATCACAAGA | 645-662c | 59 |
| BASEXOSH8R | ggactagTCCCGCCAGCATAAAAAAC | 2165-2147c | 59 |
| AEXTDWN1 | CCTGCACTGAGCACCCTCT | 2191-2173c | 56 |
| KNTN903RINV1 | GAGTTTTTCTAATCAGAATTGGT | 56 |
A DNA fragment (called REXOS) corresponding to the catalytic portion of the P. aeruginosa exoS gene (14, 16) was amplified with primer pair REXOS-L and REXOS-R (Table 2), both containing EcoRI restriction site linkers. When genomic DNA (100 μg) of P. aeruginosa ATCC 27853 was used as template for PCR, 10% dimethyl sulfoxide was added. PCR fragments were purified with the QIAquick PCR purification kit (Qiagen, Basel, Switzerland). Plasmid pBluescript II SK(−) and purified PCR fragments were digested with EcoRI and ligated for 2 h at room temperature before transformation of E. coli K-12 strain XL1-blue. Plasmid constructs were sequenced to exclude artifacts.
To obtain pure, plasmid contaminant-free probes, the cloned exoS-derived fragment (REXOS) was excised with EcoRI and purified twice over agarose gels with the Jet-Sorb kit (Genomed GmbH, Bad Oeynhausen, Germany). This fragment was then used as the template for PCR with primers REXOS-L and REXOS-R (Table 2) for production of DIG-labeled probe REXOS.
A DNA fragment (called RAEXT) corresponding to the putative catalytic portion of the A. salmonicida subsp. salmonicida aexT gene was amplified with primer pair RASEXOS-L and RASEXOS-R (Table 2) and labeled with DIG. Genomic DNA derived from A. salmonicida subsp. salmonicida ATCC 33658T served as the template.
All cloning procedures and genetic methods such as Southern blot analysis were performed essentially in accordance with standard protocols (2). DNA was extracted by the method of Pitcher et al. (22) and manipulated by conventional methods (2). The CaCl2 method was used for preparation of competent cells (25). Sequencing reactions were performed with a Taq Dye Deoxy Terminator cycle sequencing kit (PE Biosystems), and reaction products were analyzed on an ABI Prism 310 genetic analyzer (PE Biosystems).
Transformation of A. salmonicida subsp. salmonicida with plasmid DNA was accomplished by electroporation using the Gene Pulser (Bio-Rad Laboratories, Hercules, Calif.; settings: U (voltage) = 1.25 V, Rpar (parallel resistance) = 400 Ω, C (condenser capacity) = 25 μFD) and the 0.1-cm Gene Pulser cuvette (Bio-Rad). A. salmonicida subsp. salmonicida grown on solid LB agar medium was resuspended in LB broth, washed three times with 15% glycerol (sterile), and finally suspended in 15% glycerol at 1010 cells/ml. For each electroporation 125 μl of bacterial suspension and 5 μl of plasmid DNA (500 μg/ml) were used. The pulse length was 8 ms.
Construction of phage λ gene library from A. salmonicida subsp. salmonicida.
Genomic DNA (0.1 μg) from A. salmonicida subsp. salmonicida ATCC 33658T was partially digested with restriction enzyme Sau3A to generate fragments in the 3- to 4-kb range, which were then ligated to λ ZapExpress digested with BamHI (Stratagene). The ligated DNA was packed into prophage λ with the Gigapack III packaging extract (Stratagene). E. coli XL1-blue MRF′ (Stratagene) was used as a host. Phage plaques were lifted onto nylon filters and screened with DIG-labeled probes. Positive plaques were isolated and stored overnight at 4°C in 0.5 ml of SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris [pH 7.5], 0.01% gelatin) containing 20 μl of chloroform. The in vivo excision of plasmids from selected phagemid plaques was done according to the instructions with the λ ZapExpress kit (Stratagene).
Sequence data analyses.
Sequence alignment and editing were done with Sequencher software (Gene Codes Corporation, Ann Arbor, Mich.). Sequence comparisons were performed with BLAST (1), and sequences were aligned with the Wisconsin Package (Genetics Computer Group, Inc. [GCG], Madison, Wis.). The theoretical isoelectric pH (pI) and molecular masses of proteins were calculated with the GCG software.
Expression of recombinant AexT.
To characterize the AexT protein and to produce polyclonal, monospecific antibodies directed against AexT, we expressed a polyhistidine-tailed AexT peptide named AexT-His in recombinant E. coli strain BL21(DE3). The entire coding region inclusive of the stop codon of the aexT gene was amplified by PCR using primers BASEXOSH8L and BASEXOSH8R (Table 2) and genomic DNA of A. salmonicida subsp. salmonicida as the template. The purified PCR product was digested with restriction enzymes EcoRI and SpeI and cloned into EcoRI- and SpeI-digested vector pETHIS-1 to obtain plasmid pJFFASAexT-His, encoding N-terminally polyhistidine-tailed AexT (AexT-His) under the control of the T7 promoter. For the expression of the aexT-His gene, plasmid pJFFASAexT-His was transformed into E. coli strain BL21(DE3).
E. coli BL21(DE3) cells harboring plasmid pJFFASAexT-His were inoculated in 50 ml of LB broth with ampicillin at 37°C to an optical density at 600 nm of 0.3 and induced by addition of 0.2 mM IPTG (final concentration). Cells were then grown for an additional 3 h. After this time, the cells were sedimented by centrifugation at 5,000 × g for 10 min, resuspended in 5 ml of buffer, pH 7.9, containing 10 mM Tris-HCl, 1 M urea, 250 mM NaCl, 2.5 mM imidazole, 3 M guanidinium HCl, 0.2 mM phenylmethylsulfonyl fluoride, and sonicated with a microtip for 20 min at 50% output interval in a Sonifier 250 (Branson Ultrasonics, Danbury, Conn.). This sonicated fraction was directly loaded onto a prewashed 1.25-ml-bed-volume Ni-NTA column (Qiagen) and was washed twice with 5 ml of binding buffer (2 M urea, 20 mM Tris, 500 mM NaCl, 5 mM imidazole, 60 mM guanidinium HCl, pH 7.9). Elution of polyhistidine-tailed protein AexT-His was performed with 40 ml of a gradient elution buffer of 5 to 500 mM imidazole in 2 M urea-20 mM Tris-500 mM NaCl-60 mM guanidinium HCl, pH 7.9. The gradient had a flow rate of 0.25 ml/min, and fractions of 1 ml were collected with a HiLoad system (Pharmacia LKB). The fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (17). Those containing purified fusion protein AexT-His were pooled and dialyzed overnight against 5 liters of 0.85% NaCl-10 mM Tris-HCl, pH 7.5.
Immunization of rabbits with purified proteins. Monospecific polyclonal antibodies against AexT were obtained by immunization of a rabbit with the purified AexT-His protein as described for other polyhistidine-tailed proteins (4). Purified and dialyzed recombinant protein solution (100 μg/ml) was mixed 1:1 with complete Freund's adjuvant (Difco Laboratories, Detroit, Mich.), and 2 ml of the emulsion was then injected subcutaneously into a rabbit. The rabbit was given a booster immunization with the same amount of protein emulsified with Freund's incomplete adjuvant 21 days later. On day 45 after the first immunization, the rabbit was bled, and serum was prepared and stored at −20°C.
Immunoblot analyses.
Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (Bio-Rad Laboratories). For immunoblotting, Western blots were blocked with 1% milk buffer for 30 min and then incubated with the rabbit antiserum (1:1,500) in milk buffer overnight at 4°C. After a thorough wash with water, phosphatase-labeled conjugate (goat anti-rabbit immunoglobulin G heavy plus light chains [catalog no. 075-1506; Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.]) diluted 1:2,000 in milk buffer was added, and the reaction was visualized 90 min later by incubation with BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium.
Construction of an AexT-deficient A. salmonicida subsp. salmonicida JF2267 mutant.
An AexT-deficient A. salmonicida subsp. salmonicida JF2267 mutant was constructed by allelic gene replacement with an insertionally inactivated aexT gene. Plasmid pJFFBAS211, derived from the phage bank and containing the entire aexT gene and sequences downstream of aexT, was used as a source for aexT. From this plasmid, aexT was subcloned as a 1.5-kb HindIII-SpeI fragment onto vector pBC-KS, a plasmid derived from the gene for ColE1, which confers chloramphenicol resistance. Inactivation of the aexT gene was obtained by insertion of the aph(3′)-Ia kanamycin resistance gene on a 1.3-kb BglII-BamHI fragment from plasmid pSSVI186-IN into the BamHI site located on the middle of the aexT gene (Fig. 1). The resulting plasmid, pJFFaexT::Km4, was then used for electroporation of A. salmonicida subsp. salmonicida JF2267. Selection for allelic gene replacement of aexT by the aexT::Kmr mutation was obtained by selection of kanamycin-resistant chloramphenicol-sensitive A. salmonicida subsp. salmonicida on LB agar plates containing kanamycin. Kanamycin-resistant colonies were selected and passaged twice on LB agar plates containing kanamycin. Four single colonies, which were shown to be sensitive to chloramphenicol, were retained, and the correct insertion of the aexT::Kmr allele and the absence of the wild-type (wt) aexT gene was verified by PCR using primer pairs RASEXOS-L and RASEXOS-R and AEXTDWN and KNTN903RINV1 (Fig. 1). Note that primer AEXTDWN matched a DNA sequence downstream the aexT gene (Fig. 1) which was not carried by plasmid pJFFaexT::Km4, used for the construction. Primer KNTN903RINV1 matched the aph(3′)-Ia gene of pJFFaexT::Km4. Strain JF2580 was shown to contain the aexT::Kmr allele at the locus of aexT and was retained as the aexT mutant. Strain JF2580 contains no functional aexT gene, as evidenced by PCR.
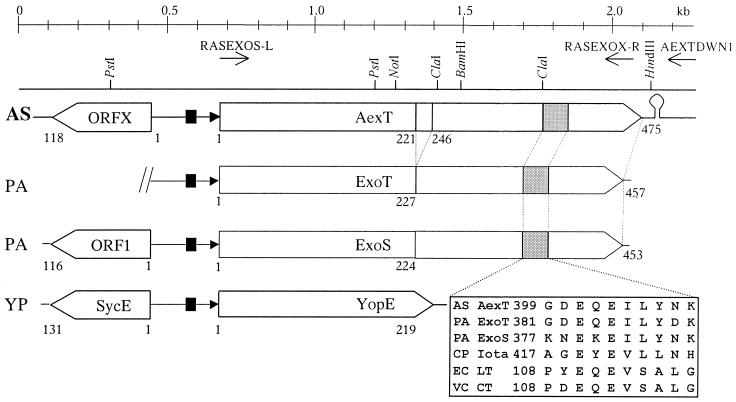
FIG. 1.
Genetic map of the gene encoding AexT and ORFX of A. salmonicida subsp. salmonicida in alignment with the corresponding genes of P. aeruginosa and Y. pestis. Maps were constructed from the current sequence data and from EMBL/GenBank accession no. AF288366 for A. salmonicida subsp. salmonicida AexT, L27629 for P. aeruginosa ExoS, L46800 for P. aeruginosa ExoT, and AF053946 for Y. pestis YopE. The last is also representative of the analogous genes of Y. pseudotuberculosis and Y. enterocolitica (8). Top line, scale in kilobase pairs; line below, physical map of the locus showing a few sites for restriction enzymes (arrows, locations of the oligonucleotide primers that were used for verification of the insertion of the Kmr cassette in the aexT mutant. Boxes with arrowheads, ORFs. Numbers indicate corresponding amino acid positions. The putative biglutamic acid active sites (grey boxes) are shown in detail. Black boxes, transcription activator (ExsA) binding site; black triangles, consensus sequences for the transcription promoter. Abbreviations: AS, A. salmonicida subsp. salmonicida; PA, P. aeruginosa; YP, Y. pestis; CP, Clostridium perfringens; EC, E. coli; VC, V. cholerae; Iota, iota toxin; LT, heat-labile toxin; CT, cholera toxin.
Infection of fish cell cultures with A. salmonicida subsp. salmonicida.
Rainbow trout (Oncorhynchus mykiss) gonad cells (RTG-2; ATCC CCL-55) were grown in 75-cm2 tissue culture flasks (Techno Plastic Products AG, Trasadingen, Switzerland) at 22°C in minimum essential medium (GibcoBRL Life Technologies, Basel, Switzerland) supplemented with 2 mM l-glutamine (GibcoBRL Life Technologies), 1× nonessential amino acids (GibcoBRL Life Technologies), 3 g of sodium bicarbonate/liter, and 10% fetal bovine serum. Three days before infection, the cells were trypsinized and subsequently seeded into 24-well culture plates (Techno Plastic Products AG) at 6 × 105 cells per 2-cm2 well in 1 ml of supplemented medium. Monolayered RTG-2 cells were then infected at multiplicities of infection of 2:1 and 20:1 (ratio of bacteria to fish cells) with the different A. salmonicida subsp. salmonicida cultures resuspended in phosphate-buffered saline (PBS), pH 7.4. In four control wells, 100 μl of PBS, pH 7.4, was added. As a further control, 1.2 × 106 bacteria of A. salmonicida subsp. salmonicida JF2267 were added to 1 ml of supplemented cell culture medium. After 2, 8, and 24 h of infection at 19°C, the fish cells were inspected and photographed under a green-filtered phase-contrast microscope (Axiovert 100; Zeiss, Jena, Germany). Detachment of the cells from the flask was achieved by vigorous shaking and scraping off of the cells from the wells. The suspended cells were centrifuged for 10 min at 5,000 × g. The pellet was then resuspended in 50 μl of SDS sample loading buffer (2) for SDS-PAGE and immunoblot analysis. Supernatant was mixed with 1/2 volume of SDS sample loading buffer. Equivalent amounts of pellets and supernatants were applied to SDS-PAGE gels. As a control, A. salmonicida subsp. salmonicida JF2267 and JF2580 were incubated in cell culture medium without cells for the same periods.
ADP-ribosyltransferase assays.
ADP-ribosyltransferase assay mixtures contained 100 μM [14C]NAD (specific activity: 6 Ci/mol) and 0.2 M sodium acetate, pH 6, in a total of 20 μl. As a source of FAS, 4 μl (approximately 200,000 cells) of noninfected RTG-2 fish cells were used in the reaction mixture. The reaction was started by adding 4 μl of a solution containing approximately 0.5 μg of recombinant AexT-His protein or 4-μl aliquots of supernatants from either P. aeruginosa ATCC 27853 or A. salmonicida subsp. salmonicida cultures. An aliquot of pure growth medium was used for background determination. The reaction was performed at 20°C for 1 h and stopped by the addition of 500 μl of 10% trichloroacetic acid (TCA). The mixtures were blotted onto filters (GS; 0.22-μm pore size; Millipore, Bedford, Mass.) with a vacuum pump and washed five times with 0.75 ml of 10% TCA. The filters were air dried, and scintillation liquid (Emulsifier Scintillator Plus; Packard Instrument Company, Meriden, Conn.) was added. Scintillation was detected as counts per minute on a liquid scintillation counter (Wallac 1410; Pharmacia, Dübendorf, Switzerland). Experiments were performed in triplicate, and scintillation was counted three times per experiment. Background counts were subtracted, and results with their standard deviations are given in counts per minute. Due to the high background of ADP-ribosyltransferase activity of the fish cells themselves, the activity of AexT in fish cells infected with A. salmonicida subsp. salmonicida could not be measured.
Nucleotide sequence accession numbers.
The sequence of the aexT gene and its neighboring ORFX-containing gene was submitted to the GenBank/EMBL database and was given accession no. AF288366. The sequence of the rrs (16S rRNA) of A. salmonicida subsp. salmonicida strain JF2267 has accession no. AF200329.
RESULTS
Cloning and characterization of the aexT gene.
Analyses of different Aeromonas sp. with broad-range ADP-ribosylating toxin probes revealed a signal for a potential ADP-ribosyltransferase gene in A. salmonicida subsp. salmonicida. Specifically, this signal was obtained with probe REXOS, which is derived from the catalytic domain of ExoS. This probe was then used to screen a λ phage gene library of A. salmonicida subsp. salmonicida ATCC 33658T. Three positive overlapping clones were found and were joined together to form a continuous DNA fragment of 2,260 bp. The derived DNA sequence of this fragment revealed a complete open reading frame (ORF) of 1,428 bp showing high similarity with ADP-ribosylating toxin ExoT of P. aeruginosa. In analogy to ExoT, the protein encoded by this ORF was called Aeromonas exoenzyme T (AexT), and its corresponding gene was called aexT. The cloned fragment contains an additional ORF, named ORFX, which shows similarity to the sycE gene of Yersinia sp. and to ORF1, which precedes exoS of P. aeruginosa (Fig. 1). ORFX is preceded by a ribosomal binding site (RBS) and followed by a putative rho-independent transcription termination site. The sequenced DNA fragment encoding AexT and ORFX revealed a high G+C content of 60%, which is above the average G+C content of A. salmonicida subsp. salmonicida, 55% (3). The ORF of aexT contains an ATG initiation codon and a TGA stop codon. The 87 bp preceding ATG show 71% identical nucleotide positions to the sequence preceding exoS and exoT in P. aeruginosa. The putative RBS, AGAAG, is positioned 10 bp upstream of the ATG. The putative promoter sequence −10 box (TAGACT) and the canonical −35 box (CCGATA) of aexT are located at the same positions as those for exoS and exoT. Upstream of the promoter −10 and −35 box sequences, there is a consensus binding site (TACAAAAA) similar to the one found upstream of exoS and exoT, which in P. aeruginosa is known to be bound by transcriptional regulator ExsA (9, 13). An inverted repeat is located 25 bp downstream of the stop codon of the aexT gene, representing the putative transcription termination sites similar to those of exoS and exoT (Fig. 1). PCR amplification and DNA sequence analysis were used to find the same genes and regulatory elements with identical nucleotide sequences in strain JF2267, a virulent strain of A. salmonicida subsp. salmonicida, which was freshly isolated from an arctic char (S. alpinus) with typical furunculosis symptoms (Table 1). Strain JF2267, in contrast to strain ATCC 33658T, was shown to be virulent and was used for further studies. Southern blot analyses of genomic DNA of various Aeromonas spp. (Table 1) with a DIG-labeled probe for aexT (RAEXT) revealed a single copy of aexT in all A. salmonicida subsp. salmonicida strains tested (Table 1). None of the other Aeromonas strains analyzed showed hybridization signals with the aexT probe.
Analyses of the AexT sequence.
The amino acid sequence for AexT was deduced from the nucleotide sequences using the universal genetic code. AexT has a calculated pI of 5.13 and a molecular mass of 50.1 kDa. BLAST searches (1) revealed the similarity of AexT to ExoT and ExoS over the whole length. In addition, similarity to the YopE cytotoxin of Yersinia pseudotuberculosis (EMBL/GenBank accession no. P08008), Yersinia pestis (accession no. P31493), and Yersinia enterocolitica (accession no. M34280) was found within the N-terminal 210 amino acids (aa) of AexT (Fig. 1). Comparisons of the amino acid sequence of AexT with those of ExoT and ExoS revealed AexT to be 62.8% identical to ExoT (57.9% identical to ExoS) and 67.5% similar to ExoT (62.8% similar to ExoS). A segment of 25 aa in the middle of AexT is not found in the other ADP-ribosylating toxins. Gap comparisons of ExoT with ExoS showed them to be identical in 75.1% of the positions and similar in 77.7%. The N-terminal domain amino acids of AexT were 33.5% identical and 37.4% similar to those of cytotoxin YopE of Y. pseudotuberculosis and 26.8% identical and 32.8% similar to those of YopE of Y. pestis (Fig. 1). The biglutamic acid active site (GDEQEILYNK) found for various ADP-ribosylating toxins (23) is also conserved within the C-terminal domain of AexT (Fig. 1).
Toxicity of A. salmonicida subsp. salmonicida strains and expression of AexT.
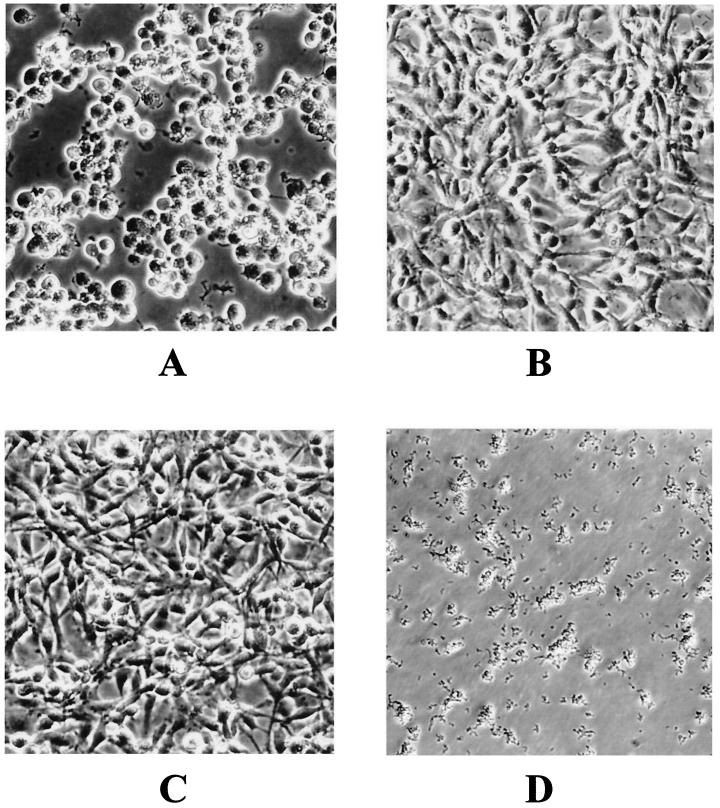
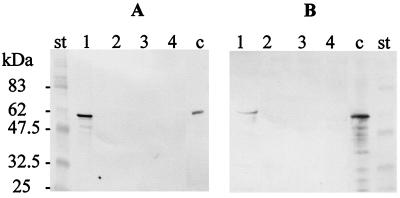
The toxicity of A. salmonicida subsp. salmonicida strain JF2267 and that of its aexT mutant derivative JF2580 were assessed by infecting cultured RTG-2 rainbow trout gonad cells. Infection with A. salmonicida subsp. salmonicida strain JF2267 caused a toxic effect resulting in characteristic cell rounding and detachment of cells within 2 h (Fig. 2). In contrast, RTG-2 cells infected with the aexT mutant showed no morphological changes at all, like the control cells incubated with PBS (Fig. 2). The same results were obtained after 8 h of incubation. After 24 h of incubation, the cells incubated with strain JF2267 had lysed, while those incubated with aexT mutant JF2580 or with PBS still remained unchanged. The results were the same with a multiplicity of infection of 2:1 or 20:1 (ratio of bacteria to fish cells). To assess the production of AexT by A. salmonicida subsp. salmonicida JF2267 and JF2580 after 2 h of incubation with RTG-2 cells (multiplicity of infection, 2:1) or after 2 h of incubation in cell culture medium alone, pellets as well as supernatants of the cells infected with the bacteria or the bacteria in cell culture medium alone were analyzed on immunoblots with monospecific polyclonal anti-AexT antibodies. The immunoblot analysis showed a strong reaction of a band at 58 kDa, corresponding to the native AexT protein, for the pellet of the RTG-2 cells infected with A. salmonicida subsp. salmonicida JF2267. No reaction was found in pellets of bacteria incubated in cell culture medium alone or in samples containing the pellets of RTG-2 cells infected with aexT mutant JF2580 or of RTG-2 cells alone (Fig. 3A). When the supernatants of the samples were analyzed, a weak reaction with anti-AexT antibodies was found in the sample of RTG-2 cells infected with JF2267 but not in the sample of RTG-2 cells incubated with aexT mutant JF2580 or in RTG-2 cells alone or in strain JF2267 in culture medium alone (Fig. 3B). These results indicate that A. salmonicida subsp. salmonicida JF2267 produced AexT after interaction with RTG-2 fish cells but that no detectable amounts of AexT were found after incubation with cell culture medium alone. Most of the AexT that was produced by A. salmonicida subsp. salmonicida JF2267 after contact with RTG-2 cells was found in pelleted cellular and bacterial material, showing that AexT was located inside the cells or was associated with the cells. The aexT mutant did not produce any AexT under any of the conditions tested.
FIG. 2.
Toxicity of AexT-producing A. salmonicida subsp. salmonicida to RTG-2 fish cells. The cells were photographed 2 h after inoculation. (A) RTG-2 cells inoculated with AexT-producing (wt) A. salmonicida subsp. salmonicida strain JF2267. (B) RTG-2 cells inoculated with isogenic aexT mutant JF2580. (C) RTG-2 cells inoculated with 100 μl of PBS buffer. (D) Strain JF2267 in culture medium without fish cells.
FIG. 3.
Biosynthesis of AexT by A. salmonicida subsp. salmonicida. The immunoblots were reacted with anti-AexT antibodies and contained RTG-2 cells inoculated with JF2267 (lanes 1), RTG-2 cells inoculated with mutant JF2580 (lanes 2), RTG-2 cells with PBS (lanes 3), and A. salmonicida subsp. salmonicida JF2267 in culture medium (lanes 4). Lanes c, purified recombinant AexT-His as a control; lanes st, molecular mass standard. (A) Pellets containing cells and bacteria. (B) Supernatants.
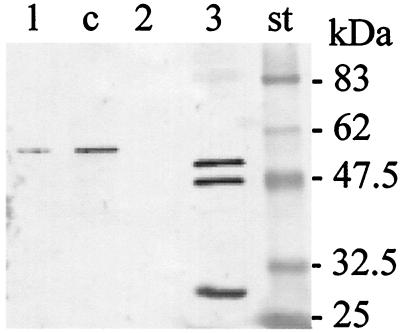
When A. salmonicida subsp. salmonicida JF2267 was grown in standard liquid TSB, no AexT could be revealed by immunoblot analysis. To analyze whether depletion or repletion of TSB medium of iron or calcium ions could affect production of AexT, A. salmonicida subsp. salmonicida strain JF2267 was cultured under various conditions consisting of TSB supplemented with either 10 mM CaCl2 or 0.01 to 1 mM EDTA or 1 mM EDTA plus 1 mM phenylmethylsulfonyl fluoride or 10 or 100 mM NTA or 0.1 mM FeIIICl3 or 0.1 mM FeIICl2 or 0.1 mM EDDA (ethylenediamine-di[o-hydroxyphenylacetic acid]) or 20 mM sodium oxalate. Some AexT protein could be detected in concentrated culture supernatant when the medium was supplemented with 10 mM NTA to complex free Ca2+ (Fig. 4). No AexT was found in strain JF2267 in the other supplemented growth media or in aexT mutant JF2580 under any of these conditions. When supernatant from a culture of P. aeruginosa ATCC 27853T grown in Ca2+-depleted medium was analyzed on immunoblots with anti-AexT antibodies, distinct reactions could be detected with proteins of 49 and 53 kDa, representing the ExoS and ExoT proteins, respectively, as expected from the amino acid sequence similarities to AexT (Fig. 4). In spite of the presence of an intact copy of the aexT gene, A. salmonicida subsp. salmonicida strain ATCC 33658T did not show expression of AexT under any of the above-mentioned conditions. Furthermore RTG-2 cells were inert to infection with ATCC 33658T.
FIG. 4.
Expression of AexT by A. salmonicida subsp. salmonicida in low-Ca2+ medium and serological cross-reactions with ExoS and ExoT. Bacterial cultures were grown in Ca2+-depleted TSB medium. Lane 1, A. salmonicida subsp. salmonicida wt strain JF2267; lane 2, A. salmonicida subsp. salmonicida aexT mutant JF2580; lane 3, P. aeruginosa strain ATCC 27853. Culture supernatants were concentrated 20-fold and analyzed on immunoblots with anti-AexT antibodies. Lane c, purified recombinant AexT-His as a control; lane st, molecular mass standard. The identity of the band at 30 kDa reacting with P. aeruginosa ATCC 27853 (lane 3) is not determined.
Biochemical activity of recombinant AexT.
To characterize AexT biochemically, we determined the ADP-ribosyltransferase activity of a purified recombinant AexT-His protein that was obtained from E. coli strain BL21(DE3) transformed with plasmid pJFFASAexT-His. Purified and renatured AexT-His revealed ADP-ribosylating activity resulting in a signal of 123 ± 11 cpm for 0.5 μg of recombinant protein in our standard assay. In comparison, 4 μl of culture supernatant of P. aeruginosa strain ATCC 27853, grown under Ca2+-depleted conditions and used as a positive control standard, produced 4,286 ± 125 cpm under the same experimental conditions. The ADP-ribosylating activity in supernatants of A. salmonicida subsp. salmonicida JF2267 cultures grown in Ca2+-depleted medium could not be measured with accuracy in this test. The counts per minute were at the limit of sensitivity of the method of measurement, although the AexT protein could be detected by immunoblotting (Fig. 4). aexT mutant strain JF2580 and strain ATCC 33658T showed no ADP-ribosyltransferase activity, as expected from immunoblot analyses. Moreover ADP-ribosyltransferase could not be determined in infected fish cells due to high background activity.
DISCUSSION
In this study we present genetic, biochemical, and biological evidence for a new toxin, AexT, of A. salmonicida subsp. salmonicida, which belongs to the family of ADP-ribosylating toxins. The aexT gene was cloned by screening a gene library of A. salmonicida subsp. salmonicida with broad-range ADP-ribosylating toxin gene probes. The aexT gene was found to be specific for A. salmonicida subsp. salmonicida, where it was found in a single copy in virulent strain JF2267, in strain ATCC 33658T, and in a further 10 field isolates. The aexT gene, in contrast, was not found in 18 other Aeromonas species. ADP-ribosylating toxins are generally very potent bacterial toxins, known to be primarily responsible for the high pathogenicity of P. aeruginosa, Vibrio cholerae, Corynebacterium diphtheriae, and Bordetella pertussis. The amino acid sequence of AexT of A. salmonicida subsp. salmonicida closely resembles those of ExoS and ExoT of P. aeruginosa, and antibodies directed against AexT cross-reacted with ExoS and ExoT. Additionally, AexT shows N-terminal similarity with YopE of Yersinia spp. ExoT, ExoS, and YopE are well-characterized cytotoxins, which catalyze rho-dependent actin depolymerization and which induce rounding of eukaryotic cells grown in culture (20, 24). AexT was shown to have ADP-ribosyltransferase activity, as measured for recombinant purified AexT-His. The ADP-ribosyltransferase activity of AexT in culture supernatants of virulent A. salmonicida subsp. salmonicida strain JF2267 was at the limits of detection and could not be measured accurately. This might be due to the low specific ADP-ribosyltransferase activity of AexT compared to that of ExoT in P. aeruginosa and to the small amount of AexT expressed under culture conditions as revealed by immunoblot analysis.
Infection of RTG-2 fish cell cultures with A. salmonicida strain JF2267 induced strong morphological changes of the cells (Fig. 2), which were accompanied by the production of AexT, which was found to be intracellular or tightly cell associated, as revealed on immunoblots with anti-AexT antibodies. These morphological changes of the cells were similar to those reported for cells infected with ExoS-producing P. aeruginosa (10) and YopE-producing Yersinia species (21). Prolonged incubation of RTG-2 cells with A. salmonicida subsp. salmonicida JF2267 led to cell lysis. In contrast, isogenic aexT mutant JF2580, which was derived from JF2267 by allelic exchange with an in vitro-mutated aexT::Kmr gene, had no effect at all on RTG-2 fish cells, even after prolonged incubation or infection with a 10-fold-greater amount of bacteria (multiplicity of infection, 20:1). Furthermore, no expression of AexT was detected when JF2580 was used for the infection of fish cells. Hence AexT is a main factor responsible for the toxic effect of A. salmonicida subsp. salmonicida strain JF2267 on RTG-2 fish cells.
The expression of aexT was shown to be induced by contact with fish cells or, to a minor extent, by low Ca2+ concentrations in the medium. Incubation of A. salmonicida subsp. salmonicida JF2267 in cell culture medium alone did not result in any production of AexT toxin. This indicated that AexT production in A. salmonicida subsp. salmonicida occurred specifically during infection of the host. Furthermore we have shown that, upon infection of RTG-2 cells with virulent A. salmonicida subsp. salmonicida JF2267, most of the AexT toxin was found in the cellular material and only minor amounts were found in the culture medium. This indicates that AexT is directly translocated to the fish cells, supposedly via a specific secretion mechanism, or tightly associates with the surfaces of the fish cells. The dependence of A. salmonicida subsp. salmonicida strain JF2267 on fish cells or on Ca2+-restricted conditions for the expression and secretion of the AexT protein toxin suggests that regulation of expression of the aexT gene and secretion of AexT might be coupled to a type III secretion system. This assumption is further strengthened by ORFX, present upstream of aexT, which shows high similarity to gene sycE (specific Yop chaperone E) of Y. pestis. SycE serves as a secretion signal and is a part of the type III secretion pathway for secretion of YopE (6). Furthermore aexT was shown to be preceded by a consensus sequence for the binding of a transcriptional activator, known in P. aeruginosa as ExsA, which is involved in type III-dependent gene expression (9).
Interestingly, AexT was not produced by A. salmonicida subsp. salmonicida strain ATCC 33658T, as shown by immunoblot analysis with anti-AexT antibodies, and did not affect the morphology of fish cells, in spite of the presence of the aexT gene and the sequences upstream of aexT. As the aexT gene and sequences upstream are the same in AexT-producing strain JF2267 and in strain ATCC 33658T, we deduce that the alteration responsible for the loss of AexT production in strain ATCC 33658T resides outside the aexT operon. In this respect, it has to be noted that A. salmonicida subsp. salmonicida strains ATCC 33658T and JF2267 have the same hemolytic activity, as estimated on blood agar plates, implying that the toxic effect for RTG-2 cells is not due to the A. salmonicida subsp. salmonicida hemolysins. The loss of expression of aexT, as observed in A. salmonicida subsp. salmonicida strain ATCC 33658T, probably caused by passages under in vitro cultivation, might be a frequent event in A. salmonicida subsp. salmonicida strains and could explain the currently observed variations in virulence as well as differences in efficacy of protection of whole-cell antigen vaccines (28).
In summary, the present data show that AexT is an ADP-ribosylating toxin of A. salmonicida subsp. salmonicida which has a toxic effect on fish cells upon infection. The aexT gene is induced upon interaction with a culture of fish cells, and the AexT protein causes cell damage.
Acknowledgments
We thank Lea Lagcher for cultivation of the fish cells and Sarah Burr for editorial help.
This work was supported by the Priority Program Biotechnology of the Swiss National Science Foundation (grant 5002-045027).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1999. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Belland, R. J., and T. J. Trust. 1988. DNA:DNA reassociation analysis of Aeromonas salmonicida. J. Gen. Microbiol. 134:307-315. [DOI] [PubMed] [Google Scholar]
- 4.Braun, M., P. Kuhnert, J. Nicolet, A. P. Burnens, and J. Frey. 1999. Cloning and characterization of two bistructural S-layer-RTX proteins from Campylobacter rectus. J. Bacteriol. 181:2501-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-378. [Google Scholar]
- 6.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757-765. [DOI] [PubMed] [Google Scholar]
- 7.Chu, S., S. Cavaignac, J. Feutrier, B. M. Phipps, M. Kostrzynska, W. W. Kay, and T. J. Trust. 1991. Structure of the tetragonal surface virulence array protein and gene of Aeromonas salmonicida. J. Biol. Chem. 266:15258-15265. [PubMed] [Google Scholar]
- 8.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M.-P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank, D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26:621-629. [DOI] [PubMed] [Google Scholar]
- 10.Ganesan, A. K., D. W. Frank, R. P. Misra, G. Schmidt, and J. T. Barbieri. 1998. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J. Biol. Chem. 273:7332-7337. [DOI] [PubMed] [Google Scholar]
- 11.Garduno, R. A., A. R. Moore, G. Olivier, A. L. Lizama, E. Garduno, and W. W. Kay. 2000. Host cell invasion and intracellular residence by Aeromonas salmonicida: role of the S-layer. Can. J. Microbiol. 46:660-668. [DOI] [PubMed] [Google Scholar]
- 12.Hirono, I., and T. Aoki. 1993. Cloning and characterization of three hemolysin genes from Aeromonas salmonicida. Microb. Pathog. 15:269-282. [DOI] [PubMed] [Google Scholar]
- 13.Hovey, A. K., and D. W. Frank. 1995. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 177:4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knight, D. A., B. Finck, V. S. M. Kulich, and J. T. Barbieri. 1995. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 63:3182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhnert, P., S. Capaul, J. Nicolet, and J. Frey. 1996. Phylogenetic positions of Clostridium chauvoei and Clostridium septicum based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 46:1174-1176. [DOI] [PubMed] [Google Scholar]
- 16.Kulich, S. M., T. L. Yahr, L. M. Mende Mueller, J. T. Barbieri, and D. W. Frank. 1994. Cloning the structural gene for the 49-kDa form of exoenzyme S (exoS) from Pseudomonas aeruginosa strain 388. J. Biol. Chem. 269:10431-10437. [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lee, K. K., and A. E. Ellis. 1990. Glycerophospholipid:cholesterol acyltransferase complexed with lipopolysaccharide (LPS) is a major lethal exotoxin and cytolysin of Aeromonas salmonicida: LPS stabilizes and enhances toxicity of the enzyme. J. Bacteriol. 172:5382-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nomura, S., M. Fujino, M. Yamakawa, and E. Kawahara. 1988. Purification and characterization of salmolysin, an extracellular hemolytic toxin from Aeromonas salmonicida. J. Bacteriol. 170:3694-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pederson, K. J., A. J. Vallis, K. Aktories, D. W. Frank, and J. T. Barbieri. 1999. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol. Microbiol. 32:393-401. [DOI] [PubMed] [Google Scholar]
- 21.Pettersson, J., A. Holmstrom, J. Hill, S. Leary, E. Frithz-Lindsten, A. von Euler-Matell, E. Carlsson, R. Titball, A. Forsberg, and H. Wolf-Watz. 1999. The V-antigen of yersinia is surface exposed before target cell contact and involved in virulence protein translocation. Mol. Microbiol. 32:961-976. [DOI] [PubMed] [Google Scholar]
- 22.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 23.Radke, J., K. J. Pederson, and J. T. Barbieri. 1999. Pseudomonas aeruginosa exoenzyme S is a biglutamic acid ADP-ribosyltransferase. Infect. Immun. 67:1508-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosqvist, R., A. Forsberg, and W. H. Wolf. 1991. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect. Immun. 59:4562-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schaller, A., R. Kuhn, P. Kuhnert, J. Nicolet, T. J. Anderson, J. I. MacInnes, R. P. A. M. Segers, and J. Frey. 1999. Characterization of apxIVA, a new RTX determinant of Actinobacillus pleuropneumoniae. Microbiology 145:2105-2116. [DOI] [PubMed] [Google Scholar]
- 27.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 28.Thornton, J. C., R. A. Garduno, S. J. Carlos, and W. W. Kay. 1993. Novel antigens expressed by Aeromonas salmonicida grown in vivo. Infect. Immun. 61:4582-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Titball, R. W., and C. B. Munn. 1985. The purification and some properties of H-lysin from Aeromonas salmonicida. J. Gen. Microbiol. 131:1603-1609. [DOI] [PubMed] [Google Scholar]
- 30.Vipond, R., I. R. Bricknell, E. Durant, T. J. Bowden, A. E. Ellis, M. Smith, and S. MacIntyre. 1998. Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect. Immun. 66:1990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viret, J. F. 1993. Meganuclease I-SceI as a tool for the easy subcloning of large DNA fragments devoid of selection marker. BioTechniques 14:325-326. [PubMed] [Google Scholar]
- 32.Whitby, P. W., M. Landon, and G. Coleman. 1992. The cloning and nucleotide sequence of the serine protease gene (aspA) of Aeromonas salmonicida ssp. salmonicida. FEMS Microbiol. Lett. 78:65-71. [DOI] [PubMed] [Google Scholar]
- 33.Yahr, T. L., J. T. Barbieri, and D. W. Frank. 1996. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J. Bacteriol. 178:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yahr, T. L., J. Goranson, and D. W. Frank. 1996. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol. Microbiol. 22:991-1003. [DOI] [PubMed] [Google Scholar]