Abstract
Pink-pigmented facultatively methylotrophic bacteria (PPFMs), classified as Methylobacterium spp., are persistent colonizers of plant leaf surfaces. Reports of PPFM-plant dialogue led us to examine cytokinin production by PPFMs. Using immunoaffinity and high-performance liquid chromatography (HPLC) purification, we obtained 22 to 111 ng of trans-zeatin per liter from culture filtrates of four PPFM leaf isolates (from Arabidopsis, barley, maize, and soybean) and of a Methylobacterium extorquens type culture originally recovered as a soil isolate. We identified the zeatin isolated as the trans isomer by HPLC and by a radioimmunoassay in which monoclonal antibodies specific for trans-hydroxylated cytokinins were used. Smaller and variable amounts of trans-zeatin riboside were also recovered. trans-Zeatin was recovered from tRNA hydrolysates in addition to the culture filtrates, suggesting that secreted trans-zeatin resulted from tRNA turnover rather than from de novo synthesis. The product of the miaA gene is responsible for isopentenylation of a specific adenine in some tRNAs. To confirm that the secreted zeatin originated from tRNA, we mutated the miaA gene of M. extorquens by single exchange of an internal miaA fragment into the chromosomal gene. Mutant exconjugants, confirmed by PCR, did not contain zeatin in their tRNAs and did not secrete zeatin into the medium, findings which are consistent with the hypothesis that all zeatin is tRNA derived rather than synthesized de novo. In germination studies performed with heat-treated soybean seeds, cytokinin-null (miaA) mutants stimulated germination as well as wild-type bacteria. While cytokinin production may play a role in the plant-PPFM interaction, it is not responsible for stimulation of germination by PPFMs.
Although the best-studied plant-bacterium relationships involve pathogenesis or symbiosis, plants interact constantly with phylloplane bacteria and with the bacteria in the soil around their roots (rhizosphere). While it is not widely believed that epi- and endophytic bacteria have overt effects on plants, there is evidence (13) that plants interact with pink-pigmented facultatively methylotrophic bacteria (PPFMs), which are inhabitants of the phylloplane and rhizosphere. PPFMs are gram-negative members of the alpha subclass of the class Proteobacteria that belong to the genus Methylobacterium and are known to inhabit the leaf surfaces of a wide variety of plant species (11). One intriguing aspect of the plant-PPFM relationship is the possibility that PPFMs may provide cytokinins to the plant host or have cytokinin-like effects. Corpe and Basile (12) demonstrated that callus from Streptocarpus prolixus (gracilis) (Cape primrose) regenerated plantlets within 15 days when it was cultured cobiotically with PPFMs. PPFM-free controls exhibited no plant development after 30 days. However, Corpe and Basile did not indicate whether specific phytohormone regimens could mimic the PPFM effects or whether PPFMs produce phytohormones. We demonstrated previously that heat treatment of soybean seeds lowered the germination rate by about 70% (21) and that there was a concomitant 90% reduction in the PPFM titer (20). Imbibition in the presence of washed PPFMs and in the presence of PPFM spent medium restored the germination rates to values comparable to those of non-heat-treated controls (21). PPFM fresh medium did not have this effect, but medium supplemented with cytokinins (0.5 mg of benzyl adenine plus 0.5 mg of zeatin per liter) restored the germination rates to values similar to that observed after PPFM application.
Cytokinins can be produced by bacteria by at least two pathways. De novo synthesis involves the direct isopentenylation of AMP catalyzed by dimethylallyl transferase (DMAT), which was first characterized in Agrobacterium tumefaciens (17, 19, 27). The second pathway of bacterial cytokinin production involves turnover of modified tRNA and may also operate in higher plants. However, the contribution of tRNA turnover to the overall cytokinin pool in both bacteria and plants has been debated for a long time. The origin of cytokinins resulting from tRNA degradation involves a modified adenine immediately 3′ to anticodons recognizing codons beginning with uridine (Trp, Phe, and Tyr codons and some Cys, Leu, and Ser codons) (33, 42). This adenine is isopentenylated by isopentenyl tRNA transferase, the product of the miaA gene. In some bacteria this modified adenine is subsequently methylthiolated and/or hydroxylated. It is hypothesized that upon turnover of tRNA the modified adenine residue is released as a free cytokinin.
In plants and apparently in bacteria as well, the isopentenyl side chain of the adenine residue is cis hydroxylated, and thus cis-zeatin riboside and/or methylthiolzeatin is released upon tRNA degradation (7, 31, 32). This isomer is nearly biologically inactive as a plant hormone, in contrast to de novo -synthesized trans-cytokinins (22). Because of the difference in stereoisomerism between the two paths of cytokinin biosynthesis, it has been assumed that tRNA turnover makes a minor contribution, at best, to the plant cytokinin pool. However, trans-hydroxylated zeatin has been found in the tRNAs and culture filtrates of Bradyrhizobium japonicum (40) and the rhizosphere bacterium Azotobacter vinlandii (1, 43). The trans isomer in the tRNA of Agrobacterium that was described by Chapman and Morris (6) was later shown by workers in the same laboratory to be the cis isomer (32). tRNA-derived trans-zeatin levels are much lower than the cytokinin levels produced de novo by pathogenic bacteria. However, B. japonicum and A. vinlandii are not associated with the gross morphological changes seen during pathogenesis (legume nodulation in symbiotic nitrogen fixation relationships is not associated with cytokinin production by rhizobia [26, 37, 39, 46]). Therefore, low-level production of cytokinins may not be an isolated phenomenon.
The possibility of widespread cytokinin production by plant commensal bacteria and the growth- and development-promoting activities of PPFMs described above and elsewhere (12, 21) led us to examine cytokinin production by phylloplane PPFMs. In this study, we characterized production of trans-zeatin by four leaf-associated PPFMs and by a type culture of M. extorquens. Furthermore, we demonstrated that this production is derived from the release of trans-zeatin during tRNA turnover. This is the first detailed characterization of cytokinin production by a phylloplane commensal bacterium, and it is the first time that bacterial secretion of trans-zeatin has been definitively linked to a tRNA source.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used are listed in Table 1. Leaf PPFM isolates (from Arabidopsis, barley, maize, and soybean) were obtained previously by plating ground leaves from growth chamber-grown plants onto ammonium mineral salts (AMS) minimal medium containing methanol as the sole carbon source (AMS/methanol medium), as described by Holland and Polacco (20). 16S ribosomal DNA restriction endonuclease analysis placed the isolates in the genus Methylobacterium (16). The M. extorquens type strain (ATCC 43645) was obtained from the American Type Culture Collection; this strain is a soil isolate that was described by Urakami and Komagata (45) and Bousfield and Green (5). For analysis of cytokinin production, AMS/methanol medium (750 ml) containing 30 μg of cycloheximide per ml in 2-liter flasks was inoculated with 100 ml of a starter culture, and bacteria were grown to the stationary phase (1 to 4 days, depending on the bacterial strain) at 28 to 30°C and 200 to 230 rpm. Polypropylene flasks were used to prevent loss of cytokinins due to adhesion to the glass surfaces of culture flasks. Culture aliquots were plated on AMS/methanol medium to check for contamination immediately before harvest. Cultures were clarified by centrifugation for 5 min at 8,000 × g, and clarified supernatants were transferred to 1-liter polypropylene bottles and stored at −70°C until culture purity was confirmed. Cell pellets were stored in 450 mM NaCl-10 mM MgCl2-1 mM EDTA-5 mM β-mercaptoethanol-20 mM Tris-Cl (pH 7.5) prior to tRNA extraction. The culture conditions used for isolation of nucleic acids and for genetic manipulation were similar, but the culture volume was 100 ml in baffled 250-ml flasks.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− Δ(lacZYA-argF)U169 supE44 hsdR17 (rB− mB+) recA1 gyrA96 endA1 thi-1 relA1 deoR φ80d(lacΔZ)M15 lambda− | |
| HB101 | supE44 hsdS20(rB− mB−) recA13 ara14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | |
| Methylobacterium strains | ||
| M. extorquens ATCC 43645 | Type strain | ATCCa |
| M. extorquens exl-11 | miaA::pAYC61, Tcr | This study |
| Leaf isolates | ||
| Arabidopsis leaf isolate 1 | Methylobacterium sp. | 20 |
| Barley leaf isolate 1 | Methylobacterium sp. | 20 |
| Maize leaf isolate 1 | Methylobacterium sp. | 20 |
| Soybean leaf isolate 2 | Methylobacterium sp. | 20 |
| Plasmids | ||
| pBluescript SK+ | AprlacZ′ | Stratagene, Inc. |
| PGEM-T-easy | AprlacZ′ | Promega, Inc. |
| pAYC61 | Apr Tcrmob+ IncColE1 | 8 |
| pRL2 | PAYC61::600-bp miaA fragment | This study |
| pBBR1tp | TprlacZ′ mob+ | 14 |
| pRK2013 | ColE1-Tra(RK2)+ Kmr | 15 |
| pUC4-KIXX | Kmr | Stratagene, Inc. |
ATCC, American Type Culture Collection, Manassas, Va.
Escherichia coli cultures were grown in Luria broth, unless indicated otherwise (30). Antibiotics were added at the following concentrations: ampicillin, 75 μg/ml; kanamycin, 45 μg/ml; and tetracycline, 15 μg/ml. All antibiotics were obtained from Sigma Chemical Co. (St. Louis, Mo.).
E. coli transformation was performed either by electroporation or by heat shock as described previously (3). The electroporation device used was an Electroporator II (Invitrogen, Carlsbad, Calif.). Plasmids were transferred from E. coli DH5α into Methylobacterium by three-way mating as described by Chistoserdov et al. (8) by using mobilization helper plasmid pRK2013 in E. coli HB101. Bacterial cells were collected by filtration on 0.45-μm-pore-size nitrocellulose filters (Gelman Metricel; diameter, 25 mm; Fisher Scientific Co., St. Louis, Mo.), which were transferred onto nutrient agar plates and incubated for 24 h at 30°C. Cells were resuspended in 5 ml of AMS/methanol medium, and aliquots were plated on appropriate selective media.
To label cytokinins in vivo, 100-ml portions of AMS/methanol medium in silanized 250-ml flasks (six replicates) were each inoculated with 1 ml of a starter culture of the Arabidopsis leaf PPFM isolate. Three of the replicates were supplemented with 50 μCi of [2,8-3H]adenine (specific activity, 20 to 40 Ci/mmol; NEN Life Sciences, Boston, Mass.) at the time of inoculation. These three replicates were harvested during logarithmic growth (22 h), and the supernatants were clarified and used for cytokinin recovery. The remaining three isolates were supplemented with 50 μCi of [3H]adenine during logarithmic growth (22 h) and harvested in the stationary phase (44 h). Growth was monitored spectrophotometrically (optical density at 600 nm), and cell numbers were confirmed by plating in a pilot experiment to obtain a standard growth curve for the strain used.
tRNA isolation, hydrolysis, and dephosphorylation.
The method used to isolate bacterial tRNA was modified from the method of Kelmers et al. (23). Cell pellets were first partially lysed with a French press at 1,200 lb/in2, and following phenol extraction, any remaining DNA was hydrolyzed by treatment with 200 U of DNase (Promega, Inc., Madison, Wis.) for 3 h at 37°C. The remaining nucleic acids were purified by ethanol precipitation, resuspended in low-salt buffer (250 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 5 mM β-mercaptoethanol, 20 mM Tris-Cl [pH 7.5]), and applied to DEAE-cellulose. Purified tRNA was eluted with high-salt buffer (650 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 5 mM β-mercaptoethanol, 20 mM Tris-Cl [pH 7.5]) and was quantified by determining A260. All subsequent steps were performed in polypropylene tubes to avoid loss of cytokinin. tRNA samples were hydrolyzed overnight in 0.3 N KOH at 37°C. The pH was adjusted to 9 with 6 N HCl, MgCl2 was added to a concentration of 20 mM, and the nucleotides were dephosphorylated with 20 U of calf intestinal phosphatase (Boehringer Mannheim, Hamburg, Germany; or Promega, Inc.) at 37°C. Due to the formation of magnesium phosphate precipitate, an additional 20 U of enzyme and 10 mM MgCl2 were added after 6 h, and incubation was continued overnight. The samples were then diluted 10-fold with immunoaffinity column buffer (40 mM ammonium acetate, pH 7.0) to obtain a volume of 10 ml, and cytokinins were isolated, fractionated, and identified as described below.
Immunoaffinity purification of cytokinins.
Cytokinins in culture supernatants (850 ml) were first concentrated by binding to C18 silica (Bondesil; Varian Inc., Harbor City, Calif.) at pH 7.0 and then eluted with 15 to 20 ml of methanol. Eluates were dried in vacuo, resuspended in 400 μl of dimethyl sulfoxide, and diluted with 40 mM ammonium acetate (pH 7.0) to obtain 40-ml preparations. Samples were passed through DEAE-cellulose (DE-52; bed volume, 10 ml; Whatman) connected in tandem to immunoaffinity columns, each containing 0.5 to 1.0 ml of microcrystalline cellulose (Whatman) to which purified clone 16 (anti-trans-zeatin and anti-trans-ribosylzeatin) monoclonal antibody (44) had been conjugated (29). The columns were each equilibrated with 120 ml of immunoaffinity buffer and, after sample application, were washed with an additional 20 ml of this buffer passed through both columns and 10 ml passed through only the immunoaffinity column. Cytokinins were eluted from the immunoaffinity columns with 15 ml of methanol and dried in vacuo . Isopentenyladenine (iP) and isopentenyladenosine (iPA) not isolated by low-affinity binding to clone 16 monoclonal antibody were isolated similarly by using the clone 12 broad-range cytokinin monoclonal antibody described by Trione et al. (44). Similarly, cis-zeatin was isolated by using a monoclonal antibody for this isomer obtained from Gary Banowetz (USDA Agricultural Research Service, Oregon State University, Corvallis). Since this monoclonal antibody also binds the trans isomer at a 10-fold-lower affinity than it binds the cis isomer (data not shown), the trans isomers were first removed from supernatants and tRNA hydrolysates by passing the preparations over the clone 16 anti-trans-zeatin monoclonal antibody immunoaffinity material and then over the anti-cis-zeatin monoclonal antibody immunoaffinity material. To isolate any remaining iP or iPA, the flowthrough was passed over the broad-range clone 12 antibody.
For the 100-ml supernatant obtained from a [3H]adenine-fed culture, the C18 concentration step was omitted. Unlabeled adenine (1 μg) was added to the supernatant, which was passed directly over the linked DEAE-cellulose immunoaffinity columns. The efficiency of cytokinin recovery by this method was estimated by adding [3H]zeatin riboside trialcohol ([3H]ZRTA) to uninoculated control medium and measuring the recovery of label after high-performance liquid chromatography (HPLC) fractionation. tRNA hydrolysates were passed directly over immunoaffinity columns without concentration on DEAE precolumns.
HPLC fractionation and RIA of cytokinins.
The immunoaffinity-purified cytokinins were fractionated by HPLC performed with a Beckman Ultrasphere octadecylsilica column (5 μm; 4.6 by 250 mm) equilibrated in triethylammonium acetate buffer (40 mM acetic acid adjusted to pH 3.4 with triethylamine [29]). They were eluted at a flow rate of 1.0 ml/min with a linear acetonitrile gradient (the initial gradient was either 5 to 15% or 10 to 15% for 15 min and was followed by a 15 to 35% gradient for 15 min and a 35 to 100% gradient for 1 min). Under these conditions, cytokinin standards (Sigma) eluted from the column in the following order: trans-zeatin, dihydrozeatin, cis-zeatin, trans-zeatin riboside, dihydrozeatin riboside, cis-zeatin riboside, kinetin, iP, and iPA. The minimum separation between these cytokinins was 0.5 min. [3H]ZRTA eluted between dihydrozeatin and cis-zeatin. The fraction volumes were 0.5 or 1.0 ml, depending on the chromatographic conditions. Fractions were dried to completion in vacuo after neutralization with 20 μl of triethylamine. Cytokinins in individual fractions were quantified by a radioimmunoassay (RIA) as described by MacDonald and Morris (29) by using the clone 16 anti-trans-zeatin riboside monoclonal antibody (final dilution, 1:10,000). As described above, this monoclonal antibody has negligible affinity for cis-zeatin (44) and low affinity for iP and iPA. HPLC fractions from [3H]ZRTA recovery and [3H]adenine precursor studies were not subjected to RIA but were counted directly by the liquid scintillation technique in 1.5 ml of Scintisafe 30 (Fisher Scientific Co.). Most analyses were performed in triplicate (separate cultures and analyses); the only exceptions were the barley leaf isolate analyses, which were performed in duplicate.
DNA manipulation.
The plasmids used are listed in Table 1. Small-scale plasmid preparations were obtained as described by Li and Schweizer (25). Larger-scale preparations were obtained by using a modification of the polyethylene glycol precipitation method (9). Plasmids used for sequencing were prepared with a Gibco Concert Rapid Plasmid Miniprep (GibcoBRL, Inc., Rockville, Md.) used in accordance with the manufacturer's instructions.
To prepare PPFM genomic DNA, 100 ml of a stationary-phase culture was pelleted by centrifugation and resuspended in 1 ml of TE (1 mM EDTA, 10 mM Tris-Cl; pH 8.0); this was followed by addition of 750 μl of n-butanol (H2O saturated) and gentle mixing for 5 min. The cells were repelleted, washed in TE, and then resuspended in 1 ml of TE to which 80 μl of a 100-mg/ml lysozyme solution (Calbiochem, Inc., San Diego, Calif.) and 20 U of RNAseOne (Promega, Inc.) were added. After incubation at 37°C for 1 to 2 h, 120 μl of 20% (wt/vol) sodium dodecyl sulfate (SDS) and 100 μl of a 20-mg/ml proteinase K (Sigma) solution were added, and the preparation was incubated for another 30 to 60 min at 50°C. The preparation was extracted once with chloroform, ammonium acetate (pH 7.5) was added to a concentration of 100 mM, and the DNA was precipitated with isopropanol (0.67 volume). The resulting DNA was spooled out of the preparation, rinsed in 70% ethanol, resuspended in 200 μl of TE, extracted once with phenol-chloroform, and reprecipitated with ethanol (2 volumes) and 0.3 mM sodium acetate (pH 5.5). The yield was approximately 150 μg of PPFM DNA per 100 ml of culture. The DNA (1 μg/μl) was stored in TE at −20°C.
Restriction digestions were performed by following the instructions of the manufacturers (Promega, Inc.; Boehringer Mannheim; and New England Biochemical, Beverly, Mass.). Fragments were analyzed by horizontal agarose gel electrophoresis. Restriction fragments were recovered from agarose by using either a 3′-5′ Glasselect gel recovery kit (5′-Eppendorf, Boulder, Colo.) or a GibcoBRL Concert Matrix gel extraction system. Ligation was performed by using either a rapid ligation kit (Boehringer Mannheim) or Promega 2× rapid ligation buffer with 10 U of T4 DNA ligase (Promega, Inc.). All ligation reaction mixtures were incubated in an ice-water mixture for 16 to 24 h.
Southern blotting was performed by alkaline transfer onto positively charged nylon membranes (Hybond-N [Amersham-Pharmacia, Inc., Piscataway, N.J.] or MSI MagnaCharge [Osmonics, Inc., Minnetonka, Minn.]). DNA was transferred without gel pretreatment in a transfer buffer containing 0.4 N NaOH for 4 to12 h. Each membrane was washed until it was neutralized in 2× SSC (1× SSC is 150 mM NaCl plus 15 mM sodium citrate), and the DNA was fixed by air drying at room temperature. Hybridization was performed at 65°C by using Sigma PerfectHybe hybridization solution according to the manufacturer's instructions. Two successive 65°C 1-h washes were done at high stringency (1× SSC, 0.1% SDS) and then at very high stringency (0.1× SSC, 0.1% SDS). All probes were labeled with [α-32P]dCTP (NEN Life Sciences) by the random prime method by using a Gibco RadPrime labeling kit according to the manufacturer's instructions.
Isolation of the M. extorquens miaA gene.
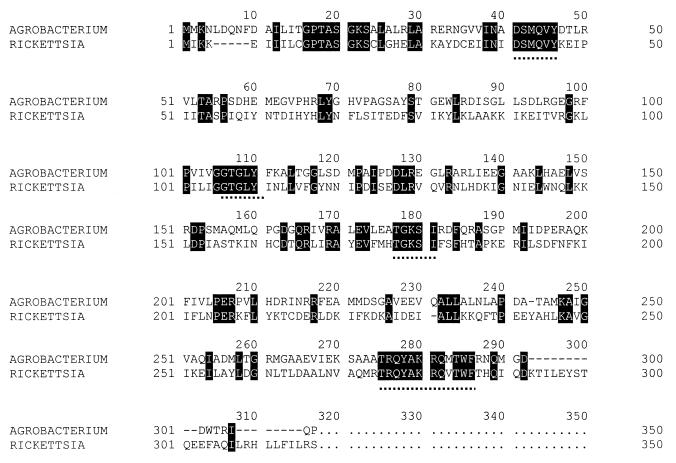
For initial amplification of an internal fragment of the M. extorquens miaA gene, amino acid sequences encoded by the miaA genes of Agrobacterium tumefaciens and Rickettsia prowazekii, the two organisms most closely related to Methylobacterium with known miaA genes, were aligned (Fig. 1). Partially degenerate sense and antisense primers for four conserved regions of the DNAs were constructed. A total of four sense and four antisense primers were constructed. The codon biases used for construction were based on M. extorquens AM1 codon usage determined by analysis of five known M. extorquens AM1 genes, mxaB, mxaE, mxaF, mxaH, and pykA. All other primers used in this study were synthesized for known DNA sequences and are described below. Primers were synthesized by Gibco Life Sciences, Inc.
FIG. 1.
Amino acid alignment for isopentenyl tRNA transferase (MiaA) from A. tumefaciens (18) and R. prowazekii (2). Partially degenerate primers based on the amino acid sequences located above the dotted lines were constructed based on known codon usage in M. extorquens AM1.
PCRs were carried out in solutions containing 10 mM Tris-Cl, 50 mM KCl, 2.0 mM MgCl2, 0.1% (wt/vol) Triton X-100, either 5% (vol/vol) dimethyl sulfoxide, 10% (vol/vol) formamide, or 1× TaqMaster PCR enhancer (Eppendorf, Inc., Hamburg, Germany), 10 U of Taq polymerase (Sigma; Fisher Biotech, Inc.; GibcoBRL, Inc.; or Eppendorf, Inc.), and 19 pmol of each primer. The reaction conditions used were as follows: 96°C for 5 min, followed by 35 cycles of 96°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min, and a final extension step consisting of 72°C for 10 min. The products were analyzed by agarose gel electrophoresis, and PCR fragments were recovered from the gel and ligated into plasmid pGEMT-easy (Promega, Inc.) for replication in E.coli.
To prepare an M. extorquens genomic library, total DNA was digested completely with PstI and cloned into pBluescript SK+ (Stratagene, Inc., La Jolla, Calif.). The library produced 3,000 CFU/ng of vector, 85% of which contained inserts, as indicated by the lack of blue pigment produced by hydrolysis of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside). IPTG (isopropyl-1-thio-β-d-galactoside) was added to induce lacZ activity (38). Library colony lifts were prepared on nylon filters as described by Sambrook et al. (38) and were probed as described above for Southern blotting. Approximately 13,000 independent clones were screened. Colonies that produced a hybridization signal with the miaA PCR fragment were checked for the expected insert size derived from a Southern blot against a complete PstI digest of M. extorquens total DNA.
Generation of a miaA knockout mutant of M. extorquens.
Since the 745-bp miaA fragment originally amplified from M. extorquens was located 80 bp from the 3′ end of the gene, a 600-bp internal fragment was generated by PCR by using primers with 5′ EcoRI sites (sense primer sequence, GAATTCGTCTACGCCGACCT; antisense primer sequence, GAATTCGTCCCGTCGAGAT). The resulting truncated fragment was ligated into pGEMT-easy and then reexcised with EcoRI and cloned into the EcoRI site of the multiple cloning region of suicide plasmid pAYC61 to generate plasmid pRL2. pRL2 was mobilized into M. extorquens by three-way mating, and tetracycline-resistant exconjugates were isolated. Integration of the plasmid into the miaA gene by a single homologous crossover was confirmed by PCR analysis with primers complementary to the vector and the internal fragment and by sequence analysis of the PCR products. Mutants were maintained in the presence of tetracycline to select against insert excision. tRNA and supernatant samples from two independent mutant cultures were examined for the presence of cytokinins as described above.
DNA sequencing and analysis.
DNA was sequenced at the University of Missouri DNA Core Facility by using ABI BigDye terminator chemistry and an ABI 377 automated sequencer (Applied Biosystems, Inc., Foster City, Calif.). Unresolved sequences were clarified by the dideoxy sequencing method with a Sequenase sequencing kit (U.S. Biochemical Corp., Cleveland, Ohio); 35S-labeled dATP (NEN Life Sciences, Inc.) was used for labeling, and the terminal deoxynucleotidyl transferase tailing method was used to extend prematurely truncated products (25). The sequence was resolved by electrophoresis in a 6% (wt/vol) urea-polyacrylamide gel as described by Sambrook et al. (38).
Soybean seed germination studies.
The effect of disruption of cytokinin production in the miaA PPFMs on the ability of these organisms to increase germination of heat-treated soybean seeds was determined by using methods described by M. A. Holland (personal communication). Replicates (50 seeds each) were first heated at 45°C for 48 h, and then imbibition was allowed to occur in one of the following preparations for 5 h at room temperature with gentle shaking: fresh AMS medium, fresh AMS medium containing 15 μg of tetracycline per ml, M. extorquens liquid culture, miaA mutant liquid culture, M. extorquens spent medium, or miaA mutant spent medium. All cultures were grown to the stationary phase before they were used, and spent media were clarified by centrifugation before they were used. After imbibition, the seeds were drained and allowed to germinate for 5 days at room temperature in the dark on single layers of germination paper in covered glass petri dishes, after which the numbers of germinated seeds (seeds clearly producing radicles) were determined. At least two replicates were used for most treatments. The exceptions were the experiments performed with clarified spent medium; in these experiments single replicates consisting of 50 seeds each were used.
RESULTS
Cytokinin production by PPFMs.
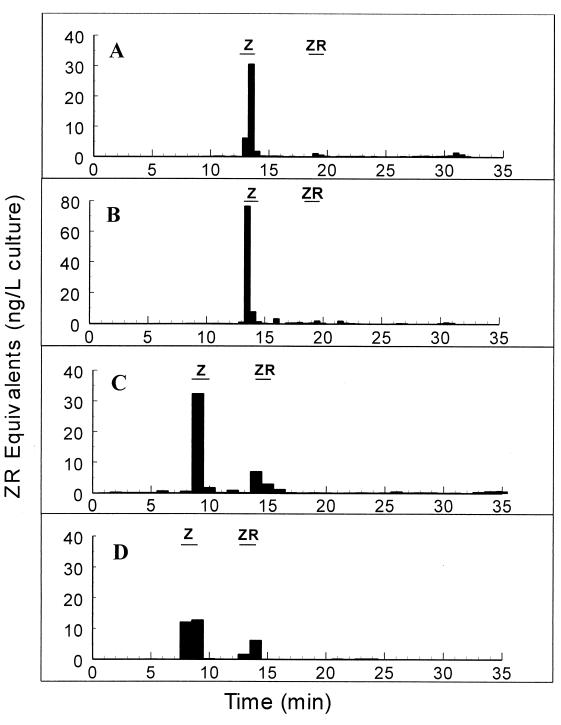
To determine whether leaf- and soil-dwelling Methylobacterium spp. produced cytokinins, immunoaffinity chromatography was performed with culture supernatants of PPFM leaf isolates obtained from Arabidopsis thaliana, barley, soybean, and maize, as well as with culture supernatants of the M. extorquens type culture. Putative cytokinins eluted from the columns were fractionated by HPLC and were quantified by RIA performed with cis- and trans-isomer-specific monoclonal antibodies to the hydroxylated cytokinins, such as zeatin and zeatin riboside (44). Representative RIA results and HPLC absorbance profiles (A254) for the trans isomers isolated from the Arabidopsis leaf PPFM culture supernatant are shown in Fig. 2A and B, respectively. RIA results for the remaining three leaf isolates and for M. extorquens are shown in Fig. 3. The A254 profiles for all isolates were similar.
FIG. 2.
RIA analysis (A) and HPLC absorbance profile (B) of cytokinins isolated by the immunoaffinity procedure from spent media from cultures of the Arabidopsis PPFM isolate. The retention times of trans-zeatin (Z) and trans-zeatin riboside (ZR) standards are indicated by bars. Control media showed no activity in any fraction. The data were not corrected for recovery.
FIG. 3.
Cytokinins secreted into liquid cultures by a soil isolate of M. extorquens (A) and by plant leaf PPFM isolates obtained from maize (B), soybean (C), and barley (D). The retention times of trans-zeatin (Z) and trans-zeatin riboside (ZR) standards are indicated by bars. The retention times varied due to changes in HPLC gradient conditions resulting from a change in equipment. (B and C) Averages based on triplicate determinations; (A and D) averages based on duplicate determinations. The values were not corrected for recovery.
The cytokinin trans-zeatin was present at detectable and replicable levels in all of the cultures tested, while detection of trans-zeatin riboside was variable and smaller quantities were present. No active fractions were detected in the uninoculated control media. Although the gradient conditions for different experiments were different, in every case the unknown cytokinin had the same retention time as the known cytokinin standard, and the identity of the cytokinin isolated was confirmed by gas chromatography-mass spectrometry by using known standards (28). The amount of total isolated trans-zeatin, corrected for the 49% level of recovery of the labeled standard in parallel experiments, is shown for each strain in Table 2. When the values were normalized for the level of tRNA recovery, 15 ± 2 and 19 ± 6 ng of cytokinin per mg of tRNA were recovered from the spent media of M. extorquens and the Arabidopsis leaf isolate, respectively. The M. extorquens culture supernatant was also analyzed to determine the presence of the cis isomer of zeatin. There were no absorbance peaks corresponding to the retention time of cis-zeatin, and fractions obtained at the expected cis-zeatin retention times exhibited no immunoactivity (28).
TABLE 2.
Cytokinins recovered from supernatants of free-living PPFM cultures
| Isolate | Concn of cytokinins recovered (ng/liter of culture)a
|
||
|---|---|---|---|
| trans-Zeatin | trans-Zeatin riboside | Total | |
| Arabidopsis leaf | 76 ± 46 | 32 ± 24 | 108 ± 25 |
| M. extorquens | 53 ± 8 | 4 ± 2 | 57 ± 10 |
| Maize leaf | 114 ± 73 | 9 ± 8 | 123 ± 81 |
| Soybean leaf | 57 ± 49 | 18 ± 14 | 75 ± 60 |
| Barley leaf | 38 | 17 | 55 |
Values were corrected for 49% recovery of the tritated standard in parallel experiments. Most values are averages ± standard deviations based on triplicate determinations; the values for the barley leaf isolate were determined in duplicate and varied from the mean by no more than 25%.
Incorporation of [3H]adenine into cytokinins by PPFMs.
To confirm that trans-zeatin and trans-zeatin riboside are produced by PPFMs, we cultured the Arabidopsis PPFM in the presence of [3H]adenine. Labeled cytokinins were isolated by the trans-zeatin-specific immunoaffinity procedure, fractionated by HPLC, and identified on the basis of retention times that corresponded to the retention times of known standards. Two trans-zeatin labeling protocols were employed. Label was introduced at the time of inoculation, and cultures were harvested during logarithmic growth; label was also introduced during logarithmic growth, and cultures were harvested in the stationary phase (Fig. 4). Significant amounts of labeled trans-zeatin riboside, however, were recovered only during the latter half of the growth period. During both time periods, a small amount of label was incorporated into a fraction whose retention time corresponded to the retention time of iPA.
FIG. 4.
Incorporation of tritiated adenine into trans-zeatin (Z), trans-zeatin riboside (ZR), and iPA by an Arabidopsis leaf PPFM isolate. The cross-hatched bars indicate the results when tritiated label was added at the time of inoculation and cultures were harvested during logarithmic growth (22 h). The solid bars indicate the results when label was added during logarithmic growth and cultures were harvested during stationary growth (44 h). The values are averages based on three replicate determinations.
Similar results were obtained when the soybean leaf isolate was provided with [3H]adenosine (data not shown). When the values were corrected for the number of cells (CFU) present at the time of harvest, there was no difference in the level of incorporation of label into total cytokinin between the two culture phases (early incorporation, 130 ± 36 cpm/108 CFU; late incorporation, 121 ± 32 cpm/108 CFU). The only significant difference involved incorporation into trans-zeatin riboside; during the first half of batch culture there was little significant incorporation, but the level of incorporation increased to approximately 11 cpm/108 CFU during the stationary phase.
Isolation of trans-zeatin from the tRNA of PPFMs.
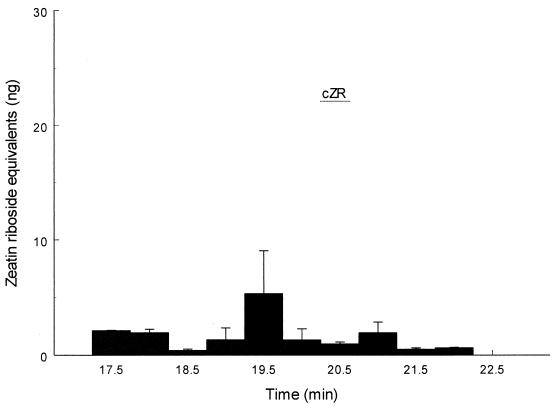
tRNA was isolated from the Arabidopsis PPFM isolate and from M. extorquens by a modification of the procedure outlined by Kelmers et al. (23), hydrolyzed and dephosphorylated (18), and then subjected to HPLC and RIA analysis as described above. Monoclonal antibodies to both stereoisomers of zeatin riboside were used in the analysis. Significant amounts of the trans isomer were detected by the RIA; approximately 6.2 ± 0.4 ng/mg of tRNA was isolated from M. extorquens (Fig. 5), and 17 ± 7 ng/mg of tRNA was recovered from Arabidopsis PPFM isolates (data not shown). These values are much lower than the 200 ng/mg reported for B. japonicum (40), but the recovery procedures differed; using our procedures, we recovered less than one-half of the previously reported levels of trans-zeatin riboside from B. japonicum tRNA (data not shown). No significant signal corresponding to the cis isomer was obtained, as shown by the lack of active fractions in an RIA for the cis isomer (Fig. 6). We also confirmed the presence of the trans isomer and the absence of the cis isomer for B. japonicum tRNA (data not shown).
FIG. 5.
RIA of trans-zeatin riboside (t-ZR) recovered from Arabidopsis PPFM tRNA. The values are averages based on two separate determinations. Twenty percent of the total tRNA hydrolysate from an 850-ml culture was used for analysis. The data were not corrected for losses. ZR, zeatin riboside.
FIG. 6.
RIA of the M. extorquens tRNA hydrolysate purified by anti-cis-zeatin immunoaffinity. The values are averages based on two separate determinations. Twenty percent of the total tRNA hydrolysate was used for analysis. The expected retention time of cis-zeatin (cZR) is indicated by a bar.
We found that a peak corresponding to iPA eluted from both the trans- and cis-isomer-specific column material, despite previously described evidence that the monoclonal antibodies do not cross-react with iPA (44). Since there was a large excess of binding capacity in the columns, it is possible that the residual affinity was sufficient to bind any iPA present in the tRNA. This was confirmed by testing the column material for the ability to bind iPA in the absence of zeatin (data not shown).
Isolation and disruption of the Methylobacterium miaA gene.
We generated a probe for the M. extorquens miaA gene by performing PCR with partially degenerate primers (four sense primers and four antisense primers) for conserved amino acid sequences in known miaA genes (Fig. 1). When the primers were used in pairwise combinations, only one primer pair produced a product of the appropriate size. This 745-bp fragment was used to probe a genomic library in order to isolate a 3.3-kb fragment containing the complete miaA sequence (see Materials and Methods). We sequenced 2.2 kb of this fragment, including the miaA gene (GenBank accession number AAF452713). miaA is flanked by homologs of serB (encoding phosphoserine phosphatase) and mmsB (encoding 3-hydroxybutyrate dehydrogenase).
We disrupted miaA by a single homologous crossover with a 600-bp internal gene fragment in suicide plasmid pACY61 (8). Three of 15 exconjugates had homologous insertions of the suicide plasmid into the miaA gene (see Materials and Methods). Since the serB and mmsB flanking genes are both transcribed in the orientation opposite that of miaA, we presumed that the insertion had no polar effects. All miaA mutants grew normally compared with the progenitor on AMS/methanol medium under standard growth conditions (see Materials and Methods). The mutants did exhibit the temperature-sensitive growth characteristic of miaA mutants (34) of E. coli K-12; the miaA mutant PPFMs did not grow at 37°C, while wild-type M. extorquens grew slowly at this temperature. One of these mutants was chosen for further study.
The tRNA hydrolysate of the M. extorquens miaA mutant lacked both trans-zeatin riboside and iPA, as shown by the absence of the HPLC absorbance peaks with retention times corresponding to those of these cytokinins in the progenitor (wild type) (Fig. 7). There was also a corresponding absence of active fractions associated with these peaks in specific RIA (data not shown).
FIG. 7.
HPLC analysis showing the absence of trans-zeatin riboside (tZR) in tRNA of miaA mutants of M. extorquens. The position of trans-zeatin riboside isolated from wild-type M. extorquens tRNA is indicated in panel A, while a corresponding peak is not present for the mutant in panel B. Ev, electron potential.
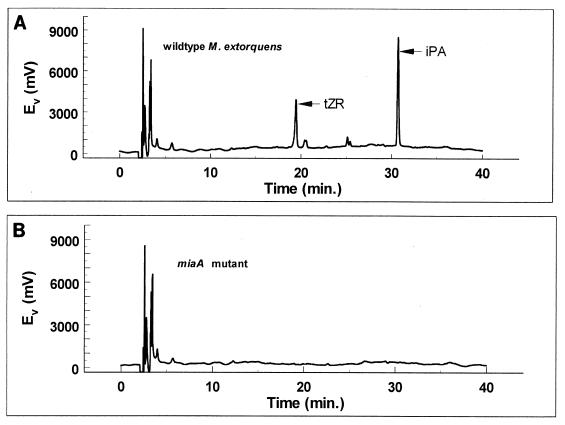
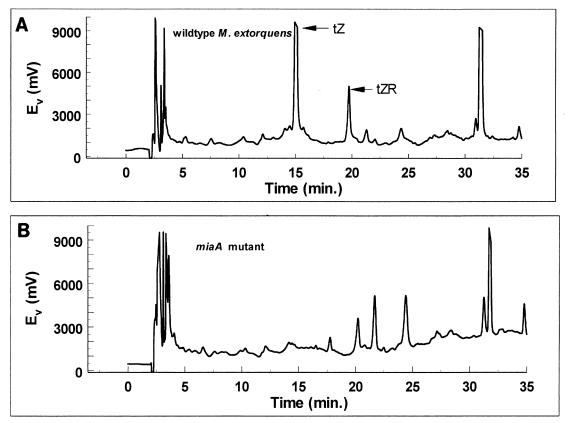
There was a similar absence of trans-zeatin in the culture supernatant of the miaA mutant, as shown by the absence of absorbance peaks corresponding to trans-zeatin and trans-zeatin riboside (Fig. 8). As determined by RIA, active fractions corresponding to trans-zeatin were present in the wild-type culture supernatant but not in the culture supernatant of the mutant (data not shown).
FIG. 8.
HPLC analysis showing the absence of trans-zeatin (tZ) and trans-zeatin riboside (tZR) in the medium of an miaA mutant of M. extorquens. Peaks corresponding to trans isomers isolated from wild-type M. extorquens medium are indicated in panel A, while corresponding peaks are not present for the mutant in panel B. Ev, electron potential.
Cytokinin production and restoration of ability to germinate in soybean seeds.
Previously, a PPFM isolate recovered from soybean was reported to restore the ability of heat-treated soybean seeds to germinate (21). To determine whether stimulation of germination was due to cytokinin secretion, we tested the abilities of M. extorquens and the miaA mutant to restore the ability of heat-treated soybean seeds to germinate (Table 3). Both untreated and heat-treated (50°C, 48 h) soybean seeds were allowed to imbibe for 5 h in overnight cultures of wild-type M. extorquens or the miaA mutant, the spent supernatants of cultures of these bacteria, or fresh control AMS medium (with and without tetracycline). Both the mutant and the wild-type bacteria were found to have significant and consistent stimulatory effects on germination of the heat-treated seeds. The degrees of stimulation obtained with the mutant and the wild-type treatments were indistinguishable. Clarified spent media alone gave similar results, indicating that the effect was due to a diffusible substance secreted into the medium by the bacteria.
TABLE 3.
Stimulation of germination of heat-treated soybean seeds by PPFMs, expressed as percent increases over values for control treatments with fresh AMS medium (wild type) or fresh AMS medium containing tetracycline (15 μg/ml) (mutant)
| Treatmenta | % Increaseb
|
|||
|---|---|---|---|---|
| PPFMs | PPFM spent medium | miaA mutant | Mutant spent medium | |
| Nonheated | 4 ± 8 | −8c | 24 ± 8 | 8c |
| Heated (50°C, 48 h) | 36 ± 6 | 58c | 62 ± 22 | 60c |
The average rates of germination for control nonheated and heated seeds were 82 and 50%, respectively.
Unless indicated otherwise, the values are averages ± standard deviations based on three replicates, each consisting of 50 seeds.
Trials with spent medium were performed with only one replicate consisting of 50 seeds.
DISCUSSION
In contrast to the interactions of plants with pathogens and symbionts, the relationship between plants and commensal bacteria on their surfaces is little understood. Traditionally, the study of cytokinin production by plant-associated bacteria has been associated with microbes known to cause plant disease or to enter into an intimate symbiosis with a plant host. We sought to rectify the omission of plant commensal bacteria from this field of study by making a detailed examination of cytokinin production by the plant leaf bacteria belonging to the genus Methylobacterium, commonly called PPFMs. Circumstantial evidence indicated that plant hormones might be produced by these bacteria (12).
At first glance, the presence of the trans isomer of zeatin in spent medium was consistent with de novo cytokinin synthesis in PPFMs, despite the low levels recovered. All previous studies in which cytokinin biosynthesis was described indicated that tRNA-derived zeatin, both plant and bacterial, occurred solely as the cis isomer (7, 31, 32). However, in vitro assays for DMAT in M. extorquens and in a leaf PPFM isolate were consistently negative, and no signal was detected with anti-DMAT polyclonal antibodies in a total protein blot of either M. extorquens or a leaf isolate PPFM (from Arabidopsis) (unpublished data). Our inability to find evidence of DMAT activity is consistent with the results of the M. extorquens AM1 sequencing project (University of Washington, Seattle). No open reading frames with identity to ipt or tzs (the genes encoding isopentenyl transferase in Agrobacterium) have been identified yet, and an estimated 99% of the open reading frames have been examined by the project.
In the absence of de novo synthesis, the most logical alternative source of the secreted zeatin is tRNA. An examination of tRNA hydrolysates revealed the presence of zeatin and, surprisingly, the trans isomer instead of the cis isomer. This finding has only two previously described precedents. The trans isomer of zeatin was found in the tRNA of B. japonicum by Sturtevant and Taller (40), and the trans isomer of methylthioribosylzeatin was isolated from the tRNA of the rhizosphere bacterium Azotobacter vinelandii in 1985 (1). The evidence presented here for Methylobacterium is the first evidence that definitively links bacterial production of secreted trans-zeatin to a tRNA origin.
It should also be noted that no evidence for the presence of methylthiolated zeatin was found in the cytokinins isolated from Methylobacterium tRNA. Much of the work on modifications of the isopentenylated bases in tRNA has been performed with E. coli and Salmonella enterica serovar Typhimurium. E. coli does not hydroxylate the isopentenyl side chain of its tRNA. S. enterica serovar Typhimurium apparently requires that the adenine residue at position 37 first be methylthiolated on the purine ring by the miaB gene product (at position C-2) before hydroxylation of the isopentenyl side chain can occur (35). Only very small quantities of cis-zeatin riboside were found in Salmonella mutants lacking the ability to produce a methylthiolated adenine residue at position 37, suggesting that the cis-hydroxylase (miaE) requires methylthiolation to effectively recognize the tRNA substrate (36). No active peaks corresponding to the methythiolated derivative were obtained with PPFM tRNA, and thus far, no open reading frames with significant identity to any known miaE gene have been found in the M. extorquens AM1 genome.
The predominant form of zeatin secreted by PPFMs was the free base; the nucleoside was found irregularly and at low levels. B. japonicum, in contrast, secreted mainly the nucleoside (unpublished data). The opposite was the case for the nonhydroxylated counterparts; iPA was the only nonhydroxylated cytokinin isolated from Methylobacterium culture medium. No detectable iP was found. The reason for this remarkable difference is not known.
It is also difficult to determine if the levels of cytokinins produced by PPFMs are sufficient to have an effect in planta . Studies of cytokinin effects are often performed with cultured tissue and with cytokinin levels in the nanomolar to millimolar range. As cytokinin effects in the whole plant are in many cases the result of localized and transient changes in cytokinin levels, the possible effects of these low levels of production in the context of plant habitation by PPFMs are hard to project. The generation of a cytokinin-negative mutant should allow us to attempt to answer these questions.
The function of trans-zeatin in the tRNA of Methylobacterium in the absence of a relationship with plants is equally unclear. In Salmonella, hydroxylation-deficient mutants (miaE) are unable to utilize citric acid cycle intermediates as carbon sources (35, 36). Interestingly, a mutation in miaA is completely epistatic to this defect; miaA-miaE double mutants are able to utilize succinate, fumarate, and malate normally. E. coli, which naturally lacks a functional miaE gene but has a working miaA gene, is also able to utilize these intermediates, so the effect of hydroxylation of the isopentenyl side chain cannot be generalized among bacteria. It must be stressed that the detailed studies of the effects of the various modifications of the adenine residue at position 37 involved cis hydroxylations; so far, trans modifications have been found only in bacteria associated with plants either as nodule formers or as colonizers of the rhizhosphere or plant leaf surface. Considering the much higher activity of the trans isomer of zeatin than of the cis isomer of zeatin, it is attractive to speculate that there is a connection between this tRNA modification and a role in plant-microbe interactions.
Unfortunately, the precise nature of this connection was not illuminated in this study. Initial experiments performed by Holland and Polacco demonstrated that PPFMs can stimulate the germination of heat-treated soybean seeds, an effect that could be mimicked by exogenous application of benzyl adenine (21). A role for cytokinins in germination is not unprecedented. It has been shown that cytokinin levels in germinating seeds exhibit transient spikes that are timed closely with the initiation of germination (4, 24, 47). Since the miaA mutant of M. extorquens, in whose medium there was no detectable cytokinin, stimulated germination at a level indistinguishable from the level stimulated by the wild type, a role for cytokinins in this process is unlikely. There are other substances, however, that can mimic cytokinin effects in plants, and there are precedents for bacterial production of these substances. Nod factors, produced by members of the genera Rhizobium and Bradyrhizobium, can stimulate cortical cell division in a manner identical to the stimulation observed with cytokinins (10). It is possible that PPFMs, which are phylogenetically related to these bacteria, are able to produce substances similar to Nod factors. While Nod factor biosynthesis has not yet been demonstrated in M. extorquens, genes that are similar to the nodB, -C, -D, -J, and -T genes and to other genes have been identified in the M. extorquens AM1 genome. Crucially, NodA, which is required for Nod factor production, is conspicuously absent. However, another member of the genus, Methylobacterium nodulans, does have the nodA gene and can nodulate legumes belonging to the genus Croatalaria (41). It is also possible that stimulation of germination by PPFMs occurs by mechanisms completely unrelated to the mechanisms that stimulate cortical division during nodulation. Gibberellins also play a large role in the release of seeds from dormancy, and the possibility that PPFMs produce or stimulate this class of hormones has not been examined yet.
Understanding the nature of the exchange between the bacteria described here and their plant hosts and how the bacteria manage to live apparently undetected in close association with the plants may shed light on plant-microbe interactions in general. In particular, this system may serve as a model system for studying the effects of cytokinin production through tRNA turnover by a plant-associated bacterium that is not a known phytopathogen or symbiont. In addition to elucidating the possible roles of cytokinins in plant-microbe relationships, an examination of PPFM-produced cytokinins may shed some light on the importance of bacterially produced cytokinins for normal plant growth and development. The contribution of plant hormones produced by nonphytopathogenic bacteria to the growth and development of plants has never been fully examined.
Acknowledgments
We thank Demosthenis Chronis, University of Missouri, for technical assistance with screening of the M. extorquens genomic library. We thank Maren Klich, Terrence J. Evens, and Mark A. Holland for critically reading the manuscript.
This work was supported by a National Science Foundation graduate research fellowship (to R. Koenig).
Footnotes
Contribution 13,196 of the Missouri Agricultural Experiment Station Journal Series.
For a commentary on this article, see page 1818 in this issue.
REFERENCES
- 1.Ajitkumar, P., and J. D. Cherayil. 1985. Presence of 2-methylthioribosyl-trans-zeatin in Azotobacter vinelandii tRNA. J. Bacteriol. 162:752-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.) 1989. Short protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 4.Biddington, N. L., and T. H. Thomas. 1976. Influence of different cytokinins on the germination of lettuce (Latuca sativa) and celery (Apium graveolens) seeds. Physiol. Plant. 37:12. [Google Scholar]
- 5.Bousfield, I. J., and P. N. Green. 1985. Reclassification of bacteria of the genus Protomonas Urakami and Komagata 1984 in the genus Methylobacterium (Patt, Cole, and Hanson) emend. Green and Bousfield 1983. Int. J. Syst. Bacteriol. 35:209. [Google Scholar]
- 6.Chapman, R. W., and R. O. Morris. 1976. Occurrence of trans-ribosylzeatin in Agrobacterium tumefaciens tRNA. Nature 262:153-154. [DOI] [PubMed] [Google Scholar]
- 7.Cherayil, J. D., and M. N. Lipsett. 1977. Zeatin ribonucleosides in the transfer ribonucleic acid of Rhizobium leguminosarum, Agrobacterium tumefaciens, Corynebacterium fascians, and Erwinia amylovora. J. Bacteriol. 131:741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chistoserdov, A. Y., L. V. Chistoserdova, W. S. McIntire, and M. E. Lidstrom. 1994. Genetic organization of the mau cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characterization of mau mutants. J. Bacteriol. 176:4052-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, K. D. 1991. Purification of plasmid and high molecular mass DNA using PEG-salt two phase extraction. BioTechniques 11:18-24. [PubMed] [Google Scholar]
- 10.Cooper, J. B., and S. R. Long. 1994. Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. Plant Cell 6:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corpe, W. A. 1985. A method for detecting methylotrophic bacteria on solid surfaces. J. Microbiol. Methods 3:215-221. [Google Scholar]
- 12.Corpe, W. A., and D. V. Basile. 1982. Methanol-utilizing bacteria associated with green plants. Dev. Ind. Microbiol. 23:483-493. [Google Scholar]
- 13.Corpe, W. A., and S. Rheem. 1989. Ecology of the methylotrophic bacteria on living leaf surfaces. FEMS Microbiol. Ecol. 62:243-250. [Google Scholar]
- 14.DeShazer, D., and D. E. Woods. 1996. Broad-host-range cloning and cassette vectors based on the R388 trimethoprim resistance gene. BioTechniques 20:762-764. [DOI] [PubMed] [Google Scholar]
- 15.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freyermuth, S. K., R. L. G. Long, S. Mathur, M. A. Holland, T. P. Holtsford, N. E. Stebbins, R. O. Morris, and J. C. Polacco. 1996. Metabolic aspects of plant interaction with commensal methylotrophs, p. 277-284. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishers, Dodrecht, The Netherlands.
- 17.Golberg, S. B., J. S. Flick, and S. G. Rogers. 1984. Nucleotide sequence of the tmr locus of Agrobacterium tumefaciens pTiT37 T-DNA. Nucleic Acids Res. 12:4665-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray, J., S. B. Gelvin, R. Meilan, and R. O. Morris. 1996. Transfer RNA is the source of extracellular isopentenyladenine in a Ti-plasmidless strain of Agrobacterium tumefaciens. Plant Physiol. 110:431-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidekamp, F., W. G. Dirkse, J. Hille, and H. van Ormondt. 1983. Nucleotide sequence of the Agrobacterium tumefaciens octopine Ti plasmid-encoded tmr gene. Nucleic Acids Res. 11:6211-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland, M. A., and J. C. Polacco. 1992. Urease-null and hydrogenase-null phenotypes of a phylloplane bacterium reveal altered nickel metabolism in two soybean mutants. Plant Physiol. 98:942-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holland, M. A., and J. C. Polacco. 1994. PPFMs and other covert contaminants: is there more to plant physiology than just plant? Annu. Rev. Plant Physiol. Plant Mol. Biol. 45:197-209. [Google Scholar]
- 22.Kamínek, M., V. Paces, J. Corse, and J. S. Challice. 1979. Effect of stereospecific hydroxylation of N6-(Δ2-isopentenyl)adenosine on cytokinin activity. Planta 145:239-243. [DOI] [PubMed] [Google Scholar]
- 23.Kelmers, A. D., C. W. Hancher, E. F. Phares, and G. D. Novelli. 1974. Large-scale fermentation of Escherichia coli and recovery of transfer ribonucleic acids. Methods Enzymol. 20:3-8. [Google Scholar]
- 24.Khan, A. A. 1971. Cytokinins: permissive role in seed germination. Science 171:853.. [DOI] [PubMed] [Google Scholar]
- 25.Li, M., and H. P. Schweizer. 1992. Resolution of common DNA sequencing ambiguities of GC-rich DNA templates by terminal deoxynucleotidyl transferase without dGTP analogues. Focus 15:19-20. [Google Scholar]
- 26.Libbenga, K. R., F. Van Iren, R. J. Bogers, and M. F. Schraag-Lamers. 1973. The role of hormones and gradients in the initiation of cortex proliferation and nodule formation in Pisum sativum L. Planta 114:29-39. [DOI] [PubMed] [Google Scholar]
- 27.Lichtenstein, C., H. Klee, A. Montoya, D. Garfinkel, S. Fuller, C. Flores, E. Nester, and M. Gordon. 1984. Nucleotide sequence and transcript mapping of the tmr gene of the pTiA6NC octopine Ti plasmid: a bacterial gene involved in plant tumorigenesis. J. Mol. Appl. Genet. 2:354-362. [PubMed] [Google Scholar]
- 28.Long, R. L. 2000. Ph.D. thesis. University of Missouri, Columbia.
- 29.MacDonald, E. M. S., and R. O. Morris. 1985. Isolation of cytokinins by immunoaffinity chromatography and analysis by HPLC-radioimmunoassay. Methods Enymol. 110:347-358. [Google Scholar]
- 30.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.McGaw, B. A., and L. R. Burch. 1995. Cytokinin biosynthesis and metabolism, p. 98-117. In P. J. Davies (ed.), Plant hormones: physiology, biochemistry, and molecular biology, 2nd ed. Kluwer Academic Publishers, Dodrecht, The Netherlands.
- 32.Morris, R. O., D. A. Regier, R. M. Olson, Jr., L. A. Struxness, and D. J. Armstrong. 1981. Distribution of cytokinin-active nucleosides in isoaccepting transfer ribonucleic acids from Agrobacterium tumefaciens. Biochemistry 20:6012-6017. [DOI] [PubMed] [Google Scholar]
- 33.Murea, N. 1994. Cytokinin biosynthesis in tRNA and cytokinin incorporation into plant RNA, p. 87-100. In D. W. S. Mok and M. C. Mok (ed.), Cytokinins: chemistry, activity and function. CRC Press, Boca Raton, Fla.
- 34.Nakayashiki, T., and H. Inokuchi. 1998. Novel temperature-sensitive mutants of Escherichia coli that are unable to grow in the absence of wild-type tRNA6Leu. J. Bacteriol. 180:2931-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Persson, B. C., and G. R. Björk. 1993. Isolation of the gene (miaE) encoding the hydroxylase involved in the synthesis of 2-methylthio-cis-ribosylzeatin in the tRNA of Salmonella typhimurium and the characterization of mutants. J. Bacteriol. 175:7776-7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Persson, B. C., O. Olafsson, H. K. Lundgren, L. Lederstedt, and G. R. Björk. 1998. The ms2io6A37 modification of tRNA in Salmonella typhimurium regulates growth on citric acid cycle intermediates. J. Bacteriol. 180:3144-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips, D. A., and J. G. Torrey. 1972. Studies on cytokinin production by Rhizobium. Plant Physiol. 49:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 39.Schmidt, J., R. Wingender, M. John, U. Wieneke, and J. Schnell. 1988. Rhizobium meliloti nodA and nodB genes are involved in generating compounds that stimulate mitosis of plant cells. Proc. Natl. Acad. Sci. USA 85:8578-8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturtevant, D. B., and B. J. Taller. 1989. Cytokinin production by Bradyrhizobium japonicum. Plant Physiol. 89:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sy, A., E. Giraud, P. Jourand, N. Garcia, A. Willems, P. de Lajudie, Y. Prin, M. Neyra, M. Gillis, C. Boivin-Masson, and B. Dreyfus. 2001. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J. Bacteriol. 183:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taller, B. J. 1994. Distribution, biosynthesis and function of cytokinins in tRNA, p. 101-112. In D. W. S. Mok and M. C. Mok (ed.), Cytokinins: chemistry, activity and function. CRC Press, Boca Raton, Fla.
- 43.Taller, B. J., and T. Y. Wong. 1989. Cytokinins in Azotobacter vinelandii culture medium. Appl. Environ. Microbiol. 55:266-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trione, E. J., B. B. Krygier, G. M. Banowetz, and J. M. Kathrein. 1985. The development of monoclonal antibodies against the cytokinin zeatin riboside. J. Plant Growth Regul. 4:101-109. [Google Scholar]
- 45.Urakami, T., and K. Komagata. 1984. Protomonas, a new genus of facultatively methylotrophic bacteria. Int. J. Syst. Bacteriol. 34:188-201. [Google Scholar]
- 46.Wang, T. L., E. A. Wood, and N. J. Brewin. 1982. Growth regulators, Rhizobium, and nodulation in peas. Planta 155:350-355. [DOI] [PubMed] [Google Scholar]
- 47.Webb, D. P., J. van Staden, and P. F. Wareing. 1973. Changes in endogenous cytokinins, gibberellins and germination inhibitors during the breaking of dormancy in Acer saccharum Marsh. J. Exp. Bot. 24:105. [Google Scholar]