Abstract
The chromosomal DNA of Streptomyces hygroscopicus 10-22, a derivative of strain 5102-6, was digested with several restriction endonucleases and analyzed by pulsed-field gel electrophoresis (PFGE). Digestions with AseI gave 11 fragments with a total length of ca. 7.36 Mb. The AseI sites were mapped by analysis of overlapping chromosomal deletions in different mutants and confirmed by Southern hybridizations using partially digested genome fragments and linking cosmids as probes. PFGE analysis of DNA with and without proteinase K treatment, together with the hybridization results, suggested a linear organization with terminal proteins and large terminal inverted repeats. Some deletion mutants had circular chromosomes.
Streptomyces species are gram-positive, mycelium-forming soil bacteria with a high G+C content (70 to 74%) in their DNA (5). They display a complex life cycle which involves the production of a large number of secondary metabolites, many of which are antibiotics. Pulsed-field gel electrophoresis (PFGE) allows the physical mapping of large bacterial chromosomes, and the physical maps of several Streptomyces chromosomes have been reported (11, 16, 17, 21). Different approaches were used for mapping bacterial chromosomes (8). For example, hybridization of “linking clones” to Southern blots of restriction fragments separated by PFGE was used to establish the physical map of Streptomyces coelicolor A3(2) (11), Streptomyces lividans 66 (16), Streptomyces griseus (17), Streptomyces rimosus (21), and Saccharopolyspora erythraea (26). Physical maps were constructed for Mycoplasma mobile (2) and Pseudomonas aeruginosa (27) by employing one- and two-dimensional PFGE. A high-resolution physical map was established by ordering a set of cosmids in S. coelicolor A3(2) (25).
One of the outstanding features of Streptomyces is its frequent chromosomal deletions (30, 31), which could occur spontaneously or be induced by exposure to physical or chemical factors, such as UV radiation (32) and antibiotics (33). This phenomenon is believed to be related to the linearity of the chromosomes, and this genetic instability has not been shown to occur in circular chromosomes (7, 20).
Streptomyces hygroscopicus 10-22 (37) can produce at least three useful antifungal antibiotics, one of which is used extensively and the others of which are used on a small scale in Chinese agriculture. Antibiotic 5102-I, an aminoglycoside similar to validamycin, is highly effective against Pellicularia sasakii, which causes rice sheath blight disease, Pellicularia filamentosa, which causes seedling blight of cotton, and Helminthosporium sigmoideum, which causes sclerotial disease of rice. Antibiotic II is a polypeptide useful for protection against leaf spot of corn caused by Cochlioobolus heterostrophus. Antibiotic III, whose structure is unknown, is useful for the control of cotton wilt caused by Fusarium oxysporum. Extensive efforts have therefore been devoted to identifying the compounds (35, 36), developing a gene cloning system (22), and improving the yield of the antibiotics (23).
Here, we report the physical mapping of the S. hygroscopicus 10-22 chromosome, not by the conventional methods of hybridization with linking plasmids or cosmids (11, 16, 17, 21) as a principal tool, but mainly by the use of overlapping chromosomal deletions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
S. hygroscopicus 10-22 was used throughout this study as a starting strain and as a control. It and its derivatives were grown at 30°C on HAUCM agar (23) for spores or in YEME medium (10) containing only 10.3% (wt/vol) sucrose for preparing the mycelium. Luria-Bertani liquid medium was used for growing Escherichia coli strains (28). All the strains and plasmids used are described in Table 1.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Origin and characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| S. hygroscopicus | ||
| 10-22 | Wild type | 37 |
| 25-85 | Acridine orange-induced mutant of 10-22 | 18 |
| N-103, N-1011 | NTG-induced mutant of 10-22 | 18 |
| Q1 | Obtained after transformation of 10-22 by pIJ702 | 23 |
| Q105 | Obtained through curing of pIJ702 in Q1 | 23 |
| Q1513 | Protoplast regenerant of Q105 | 22 |
| DX505 | Yellow isolate of Q1 grown on MM | This work (Fig. 4) |
| DX506 | White isolate of Q1 grown on MM | This work (Fig. 4) |
| Q303, Q304, Q305 | Obtained through curing of pIJ702 in N-103 transformant carrying pIJ702 | 22 |
| T100, T102, T103, T104, T106, T138, T140, T141, T150, T155, T157, T163, T167, T170, T171, T172, T177, T178 | Independent protoplast regenerants of 10-22 | 23 |
| WH1, WH1-6, WH-12, WH-14, WH-24, WH-26, WH-27 | 10-22 derivatives transformed with pIJ702 derivatives containing inserts of 10-22 DNA | 23 |
| S. coelicolor A3(2) | Wild type | 10 |
| E. coli K-12 | ||
| DH5α | supE44 hsdR17 recA1 | 9 |
| LE392 | supE supF hsdR | 4 |
| Plasmids | ||
| 2B3, 5H4, 6G1, 6G6, 11G12, 12E4, 15B8 | Linking cosmids carrying AseI site on each of their inserts | This work |
| pIJ4642 | ori (pACYC184) cat aadA | 12 |
| pHZ132 | ori (pSG5) tsr bla vph | 1 |
ori, origin of replication; cat, chloramphenicol resistance gene; tsr, thiostrepton resistance gene; bla, ampicillin resistance gene; vph, viomycin resistance gene; MM, minimal medium.
Preparation of linking plasmids.
The random BamHI-AseI fragments of S. hygroscopicus 10-22 were ligated with the recovered AseI-BamHI fragment of pIJ4642 (12) carrying a chloramphenicol acetyltransferase gene (cat) originated from Tn9 (28) and a replication origin of pACYC184. After transformation of E. coli DH5α, chloramphenicol-resistant colonies were selected and confirmed by digestion with AseI plus BamHI. The inserts were used as probes to identify linking cosmids (i.e., containing AseI sites) in the cosmid library.
Construction of a cosmid library.
Total DNA of S. hygroscopicus 10-22 was partially digested with Sau3A1 to fragments with approximate sizes of 30 to 40 kb, treated with alkaline phosphatase, and ligated into the bifunctional cosmid vector pHZ132 (1). The ligated DNAs were packaged with in vitro packaging extracts and introduced into LE392. pHZ132 can accommodate a DNA fragment of 29 to 42 kb. Since pHZ132 has one AseI site, clones containing an insert with an AseI site were identified by digestion with AseI followed by conventional agarose gel electrophoresis. All the candidates were checked by hybridization to PFGE-separated fragments to identify their linking positions.
Preparation, digestion, and separation of DNA samples.
S. hygroscopicus 10-22 was grown in YEME medium in a 250-ml Erlenmeyer flask containing a coiled stainless steel spring for good aeration and cell dispersion. The resulting mycelium was harvested and was used to make plugs as described by Kieser et al. (11). For restriction analysis, slices of the agarose containing the intact chromosomal DNA were incubated (in the buffer recommended by the supplier [Promega]) with 20 to 30 U of restriction enzyme at 37°C for 4 to 10 h. All PFGE runs were performed in a contour-clamped homogeneous electric field system (Bio-Rad, Hercules, Calif.) (6). For all experiments, 1% agarose gels were run in an electrophoresis buffer of 0.5× TBE (50 mM Tris-borate buffer [pH 8.0], 0.1 mM EDTA). Pulse times were adjusted according to the sizes of the DNA fragments to be separated. Saccharomyces cerevisiae chromosomes (Bio-Rad) and lambda DNA concatemers were used as size standards. The proteinase-untreated samples of DNA were prepared as described by Lin et al. (20).
DNA labeling and hybridization.
After PFGE, DNA fragments were transferred to nylon membranes (Hybond-N; Amersham, Little Chalfont, England) by previously described methods (29) and cross-linked by exposure to 60 mJ of UV. DNA fragments were recovered from low-melting-point agarose gels for use as probes. Hybridization with cosmids digested with BamHI or chromosomal DNA labeled with α-32P was carried out as described previously (28). Hybridization using probes labeled with digoxigenin-labeled dUTP (Roche Diagnostics, Mannheim, Germany) was carried out at 68°C as specified by the manufacturer.
RESULTS
PFGE analysis demonstrates linearity of the S. hygroscopicus 10-22 chromosome with bounded proteins at both ends.
When intact chromosomal DNA of S. hygroscopicus 10-22 was electrophoresed in parallel with S. coelicolor A3(2), a band of ca. 8 Mb in size could be observed in both the S. coelicolor A3(2) and S. hygroscopicus 10-22 tracks, respectively (Fig. 1A). While the chromosomal DNA of S. hygroscopicus 10-22, together with that of E. coli DH5α, which is known to possess a circular chromosome (13), and S. coelicolor A3(2), which is known to have a linear chromosome (11), was electrophoresed under conditions suitable for the movement of linear structures (Fig. 1B), no obvious migrating band in the track of the E. coli DH5α was detected. The possibility of not having enough DNA in the E. coli DH5α sample was ruled out by NotI digestion before PFGE to generate several expected bands (Fig. 1B), indicating that the chromosome of S. hygroscopicus 10-22 is most likely linear.
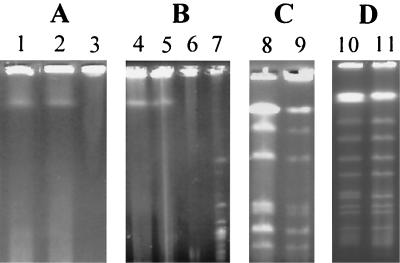
FIG. 1.
PFGE of S. hygroscopicus 10-22. Linear chromosomes of S. coelicolor M145, of ca. 8 Mb in size (panel A, lane 1), were run in parallel with the chromosome of S. hygroscopicus 10-22 (panel A, lane 2). A similar migrating property of the two samples could be observed. The two samples (panel B, lanes 4 and 5) were also run in parallel with the circular chromosome of E. coli DH5α without (panel B, lane 6) or with (panel B, lane 7) NotI digestion. The chromosome of S. hygroscopicus 10-22 treated with PK (panel A, lane 2) and SDS (panel A, lane 3) was digested with AseI before it was subjected to PFGE. Panel C, lane 8 (PK treated), and panel C, lane 9 (SDS treated), showed no obvious difference in banding pattern. While under the same conditions, obvious retardation of the AseI-J fragment of S. coelicolor M145 was observed for the SDS-treated sample (panel D, lane 11) but not for the PK-treated sample (panel D, lane 10). The run time was 30 h at 6 V/cm with an 80- to 200-s switch time at an included angle of 120° for panel B, 24 h at 6 V/cm with a 20- to 120-s switch time for panel C, and 22 h at 6 V/cm with a 20- to 80-s switch time for panel D in 0.5× TBE buffer.
Two different procedures were used to prepare DNA samples for PFGE. First, intact DNA was prepared using a procedure including proteinase K (PK) treatment. Secondly, DNA samples were prepared without PK but with sodium dodecyl sulfate (SDS) (2% SDS in 0.5 M EDTA, pH 8.0) treatment to remove noncovalently bound proteins from DNA, as described by Lin et al. (20). Figure 1A shows the result of PK-treated and -untreated S. hygroscopicus 10-22 samples. There is one band in the 10-22 sample treated with PK whose size is indistinguishable from that of the undigested S. coelicolor A3(2) chromosome. The DNA in the SDS-treated sample, however, is trapped in the slot (Fig. 1A, lane 3), as seen for the SDS-treated S. coelicolor A3(2) chromosome, suggesting chromosomes with a circular conformation (34) which will be rendered linear when treated with PK due to the dissociation of covalently bound terminal proteins but will still keep a circular form when treated with SDS. Without PK treatment, the lysis might be incomplete or some binding proteins might still be present, thus retarding the mobility of the free chromosome and rendering it unable to enter the gel. This possibility was ruled out by the observation that the SDS-treated sample can be digested with AseI and resulting fragments can migrate into the gel after electrophoresis, as was the case for the PK-treated sample (Fig. 1C).
Macrorestriction analysis reveals a chromosome size of ca. 7.36 Mb.
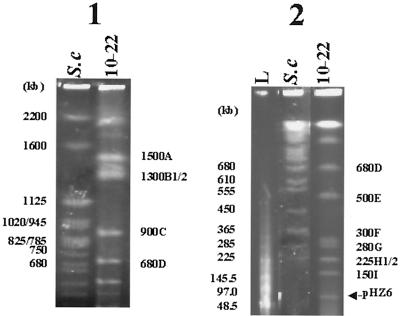
Because of the high G+C content of the Streptomyces DNA (70 to 74%), restriction enzymes with recognition sites containing only A and T nucleotides, such as AseI (ATTAAT), DraI (TTTAAA), and SspI (AATATT) will usually generate relatively few fragments and be suitable for PFGE experiments (11). DraI digestion resulted in smearing, but AseI gave nine bands (Fig. 2). Good resolution of all fragments in one track requires optimal adjustment of the pulse time program, but it would not necessarily satisfy the good separation of some very small or very large fragments. We thus adjusted the conditions as appropriate to separate the relatively small and large fragments separately. As can be seen in Fig. 2, panel 1, the intensity of the 1,300-kb band is obviously stronger than that of the 1,500-kb band, indicating that there could be two 1,300-kb comigrating fragments (designated 1300B1 and 1300B2, respectively). The same is true for the 225-kb band (Fig. 2, panel 2), where the twofold greater intensity of this band suggests that there could be two different 225-kb fragments (designated 225H1 and 225H2, respectively) in the same position. The smallest band in Fig. 2, panel 2, corresponds to a linear plasmid, pHZ6, which contains no AseI site (unpublished data). In conclusion, AseI digestion yielded 11 fragments, adding up to ca. 7.36 Mb. SspI digestion produced more than 27 bands, which were not studied further.
FIG. 2.
PFGE separation of the fragments of S. hygroscopicus 10-22 chromosomal DNA digested with AseI. Conditions were adjusted to separate large (panel 1; 60- to 150-s pulses, 130 V, 40 h) and small (panel 2; 20- to 80-s pulses, 130 V, 20 h) fragments. S.c, S. cerevisiae YNN295 chromosomes; L, lambda DNA concatemers used as size markers; 10-22, S. hygroscopicus 10-22 digested with AseI. Size markers are shown to the left and sizes of AseI fragments of S. hygroscopicus 10-22 are indicated at the right of gel. The arrow points to a linear plasmid detected in S. hygroscopicus 10-22.
Isolation of linking plasmids and cosmids.
The strategy used for constructing linking plasmids and obtaining linking cosmids was outlined in Materials and Methods. At least 16 colonies likely carrying BamHI-AseI inserts with different sizes were obtained, presumably corresponding to at least eight AseI fragments. Using these clones as probes to hybridize with the S. hygroscopicus 10-22 genomic library, six linking cosmids flanking six different AseI sites but eight different AseI fragments (see Fig. 6) were isolated. They were used as additional means to confirm the relationship of some of the neighboring fragments deduced by overlapping chromosomal deletions, as described below.
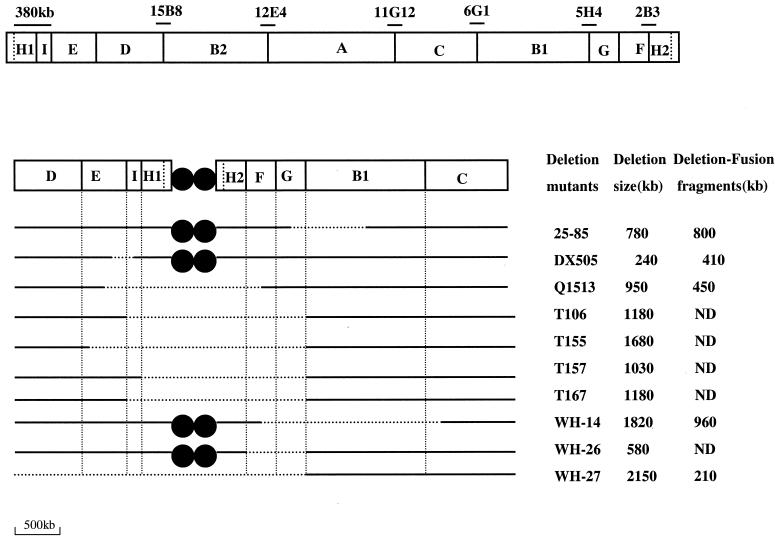
FIG. 6.
AseI map of the linear chromosome of S. hygroscopicus 10-22. The vertical lines represent the AseI sites. Solid circles represent terminal proteins at both ends. The cosmids and the partially digested fragments, which confirm the AseI sites, were lined above the AseI junction region. The two assumed AseI sites are represented by two vertical dotted lines at the very ends of the chromosome. The deleted regions of the representative deletion mutants (especially those shown in Fig. 4) are indicated by dotted lines. ND, not detected.
Construction of the AseI restriction map of the chromosome.
The strategy used to deduce the location of chromosome fragments is shown in Fig. 3. In a preliminary experiment, we extensively analyzed mutants of S. hygroscopicus 10-22 (spontaneous mutations or those generated by protoplast regeneration, N-methyl-N′-nitro-N-nitrosoguanidine [NTG] mutagenesis, etc.) by PFGE. Thirty strains were found to carry independent genetic instability events that were mostly large deletions. Compared with the restriction pattern of the wild-type S. hygroscopicus 10-22, most of the mutant strains tested exhibited an altered AseI restriction profile with obvious chromosomal deletions of different extents. Based on the assumption that in each case a contiguous piece of DNA was deleted, those mutants were grouped according to their macrorestriction patterns on PFGE gels. New fragments could be detected in some, but not all, of the deletion mutants, as could be explained by the creation of fragments at deletion junctions, while in some other cases, no extra bands apart from the deleted AseI fragments could be observed. A junction fragment might nevertheless exist but could comigrate with one of the preexisting fragments or be small enough to have run out of the gel.
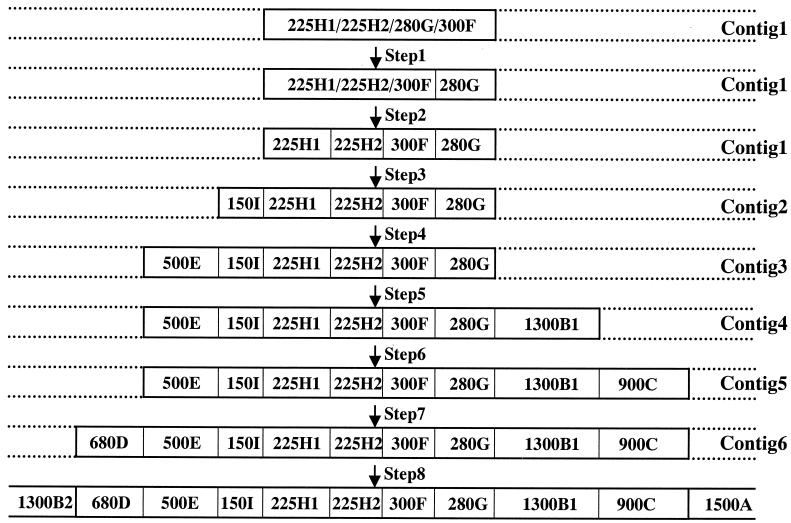
FIG. 3.
Schematic presentation of stepwise alignment of the relative positions of AseI fragments of S. hygroscopicus 10-22 by analysis of deletion mutants using PFGE. Chromosomal deletions occur most frequently in the terminal regions, and therefore Contig1 is likely to contain the two terminal fragments (which in turn contain TIR [terminal inverted repeats]). Contig2 to -6 covered less frequent deletion regions flanking both sides of Contig1, whose approximate extents of deletion are determined by the analysis of different deletion mutants by PFGE. See text for more details.
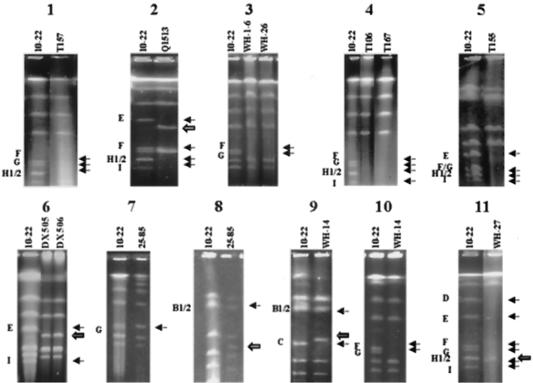
Mutant T157 (Fig. 4, panel 1) was obtained after protoplast regeneration. It lacked fragments 225H1, 225H2, 280G, and 300F. Similar banding patterns could also be observed in many other mutant strains, such as N103, Q303, Q304, Q305, T102, T103, T104, T140, T141, T150, T172, T177, and T178 (data not shown). Frequent codeletion of the above-mentioned four fragments in these strains suggested that this is a preferred area for deletion and that the deleted fragments are most likely contiguous (Fig. 3). For the convenience of description, this region was named “Contig1.”
FIG. 4.
Macrorestriction banding patterns of different S. hygroscopicus 10-22 mutants digested with AseI. The PFGE conditions for panels were as follows: panel 1, 20- to 80-s pulses, 130 V, 24 h; panel 2, 20- to 80-s pulses, 130 V, 20 h; panel 3, 20- to 80-s pulses, 130 V, 22 h; panel 4, 20- to 80-s pulses, 130 V, 26 h; panel 5, 60- to 140-s pulses, 130 V, 36 h; panel 6, 30- to 70-s pulses, 130 V, 23 h; panel 7, 60- to 160-s pulses, 130 V, 40 h; panel 8, 20- to 80-s pulses, 130 V, 24 h; panel 9, 70- to 140-s pulses, 130 V, 40 h; panel 10, 20- to 80-s pulses, 130 V, 25 h; panel 11, 20- to 80-s pulses, 130 V, 24 h. Black solid arrows indicate the missing fragments, and open arrows indicate the newly formed fragments.
The relative positions of the four codeleted AseI fragments in Contig1 were deduced by the analysis of some other deletion mutants. Figure 4, panel 2, shows the AseI digestion pattern of Q1513, in which three fragments (225H1, 225H2, and 300F), but not 280G of Contig1, were missing (Fig. 4). The 280G band must therefore be at one side of Contig1, arbitrarily placed on the right (Fig. 3, step 1).
Analysis of the restriction banding patterns of strains WH-1-6 and WH-26 (Fig. 4, panel 3) and WH1 and WH-24 (data not shown) provided additional information that 300F is immediately adjacent to 280G. These strains have codeleted 300F and 280G, while keeping 225H1 and 225H2 intact (as could be judged by the relative band intensities). Therefore, Contig1 is most likely to be present in the order of 225H1, 225H2, 300F, and 280G, from left to right (Fig. 3, step 2). The placing of 225H1 and 225H2 is arbitrary.
The AseI map of wild-type S. hygroscopicus 10-22 could be further extended by analysis of other overlapping deletions. Compared with S. hygroscopicus 10-22, strains T106 and T167 (Fig. 4, panel 4) have not only deleted the entire Contig1 (as in the case of T157; Fig. 4, panel 1), but have also deleted an extra 150I band. (N-1011, an NTG-mutagenized strain, and T100, T138, T163, and T171, four independent strains generated by protoplast regeneration, had a similar deletion profile to that of T106 and T167 [data not shown].) The codeletion of 150I with Contig1 led us to think that 150I could be located at either side of Contig1. With the help of the restriction banding pattern of Q1513, in which 150I was codeleted with 225H1, 225H2, and 300F, but which retained an intact 280G fragment from Contig1 (Fig. 4, panel 2), 150I was shown to be next to, and to the left of, the 225H1 band, forming Contig2 (Fig. 3, step 3).
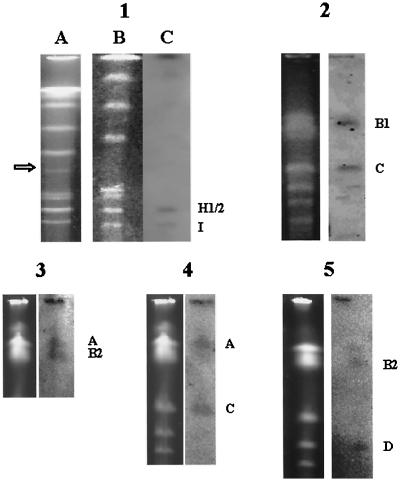
When the chromosomal DNA of S. hygroscopicus 10-22 was digested with AseI, one partially digested fragment, of ca. 380 kb, occasionally appeared (Fig. 5, panel 1, lane A). This partially digested fragment was shown to consist of 150I and the adjacent 225H1 by its recovery from a PFGE gel and use as a probe against the PFGE-separated S. hygroscopicus 10-22 genome fragments (Fig. 5, panel 1, lanes B and C). This experiment confirmed the above order of the AseI fragments.
FIG. 5.
Southern hybridization to detect AseI fragments of S. hygroscopicus 10-22 genomic DNA using linking cosmids or partially digested PFGE fragments as probes. All samples were digested with AseI (e.g., panel 1, lane B) and are shown with their respective autoradiographs (e.g., panel 1, lane C). The probes for panels were as follows: panel 1, partially digested fragment of ca. 380 kb, indicated by an open arrow in lane A; panel 2, linking cosmid 6G1; panel 3, linking cosmid 12E4; panel 4, linking cosmid 11G12; panel 5, linking cosmid 15B8. Corresponding positive signals are indicated at the right of the gels.
T155 (Fig. 4, panel 5) and WH-12 (data not shown) possess another type of altered restriction pattern in which several large-molecular-weight bands, including 500E and previously determined Contig2 (150I, 225H1, 225H2, 300F, and 280G), were codeleted, implying that 500E abutted with Contig2. With the help of Q1513 (Fig. 4, panel 2), in which 500E was codeleted with 225H1, 225H2, 300F, and 150I, but not with 280G, we reasoned that 500E is next to 150I in Contig2, forming Contig3 (Fig. 3, step 3). As further evidence, a newly formed 450-kb fragment in Q1513 (as indicated by an open arrow in Fig. 4, panel 2) showed homology in a Southern hybridization experiment with 300F (as probe) of 10-22 (data not shown), suggesting that this 450-kb fragment originated from the deletion-fusion of a partial 500E and a partial 300F.
Analysis of the AseI banding patterns of DX505 and DX506 (Fig. 4, panel 6), two naturally occurring mutants of S. hygroscopicus 10-22, supported the relative alignment of 500E and 150I in Contig4. The chromosome of the two strains has codeletions of 500E and 150I, with the appearance of an additional fragment (ca. 410 kb). It is likely, but not proven, that this new fragment is a deletion-fusion of 500E and 150I in Contig3 (Fig. 3, step 4). Owing to the high DNA concentration, the bands in the 10-22 control moved a little slower than the corresponding fragments in DX505 and DX506.
Some mutants contained large new bands. As shown in Fig. 4, panels 7 and 8, strain 25-85 has a new fragment (ca. 800 kb), which must have originated from a fragment of more than 900 kb because the natural 900C, 680D, and 500E fragments, which might form a joint fragment of more than 800 kb, are still present in strain 25-85. The apparent doublet of 1,300-kb fragments seemed to have a much lower band intensity than in the control, while 280G was also absent from 25-85. It is thus likely that the new, ca. 800-kb fragment is a deletion-fusion fragment of one of the 1,300-kb fragments (1300B1) with 280G, which was confirmed by the Southern hybridization using the 800-kb deletion-fusion fragment as a probe (not shown). Another strain, T170, with a deletion-fusion involving 1300B1 and 280G, which form a new fragment of ca. 950 kb, also seemed to support the abuttal of the two fragments.
Another strain, WH-14 (Fig. 4, panels 9 and 10), had a new band of ca. 960 kb and lacked 300F, 280G, one 1,300-kb fragment (1300B1), and 900C. It appears that the 960-kb new fragment is a deletion-fusion of 300F and 900C, with complete loss of 280G and 1300B1. A Southern hybridization using a linking cosmid (6G1) as a probe (Fig. 5, panel 2) confirmed this hypothesis. Positive signals were seen in the bands corresponding to 1300B1 and 900C, as expected. Thus, 1300B1 abuts 900C (Fig. 3, step 6, Contig5).
In all of the deletion mutants analyzed, only one deletion event involved 680D (Fig. 4, panel 11). This mutant strain (WH-27) had a deletion of all of the fragments smaller than 900 kb, with formation of a new fragment (of 200 kb). Because 900C was not lost, 680D is unlikely to be adjacent to the right side of 900C in Contig5, but rather to the left of 500E, as represented in Contig6 (Fig. 3, step 6), to constitute codeletions with adjacent relatively smaller fragments. This reasoning is further supported by partial digestion of the wild-type S. hygroscopicus 10-22 genome: occasionally, a 1,200-kb fragment, about equivalent to 680 kb plus 500 kb under the PFGE conditions favoring separation of large fragments, could be observed. No pair of fragments other than those ordered in Contig6, however, would form a ca. 1,200-kb fragment. It is thus likely that 680D is to the left of 500E, as outlined for Contig6. The ca. 200-kb new fragment in WH-27 could possibly be a 680D-280G (or [unlikely] 680D-1300B1) deletion-fusion fragment.
Since only two AseI fragments, 1300B2 and 1500A, remain unassigned, they must be adjacent and located between 680D and 900C. Hybridization with linking cosmid 12E4 as a probe confirmed the adjacency of 1300B2 and 1500A (Fig. 5, panel 3). In Fig. 5, panel 3, the 1300B1 and 1300B2 doublet was slightly separated to appear as a thick band, and the probe (12E4) hybridized to a band corresponding to the 1300B2 position as well as to 1500A at about equal intensity. Which of the two undefined fragments is next to 900C and which one will be next to 680D? This question was answered by hybridization with linking cosmids 11G12 (Fig. 5, panel 4) and 15B8 (Fig. 5, panel 5) as probes. The fact that 11G12 hybridized to 1500A and 900C (Fig. 5, panel 4) is a good indication that 1500A is immediately adjacent to 900C as outlined in Fig. 3. Similarly, the hybridization of 15B8 to 680D and 1300B2 (1300B1 was already assigned earlier) (Fig. 5, panel 5) indicates that 1300B2 is next to 680D, as shown in Fig. 3. These results are all in good agreement with the early deduction based on the analysis by overlapping deletions, supported the earlier reasoning about the location of 900C and 680D fragments, and proved again the feasibility of alignment of chromosome fragments by the overlapping deletion approach.
Localization of the terminal fragments.
We tried to detect the terminal fragments by comparing the relative movement of the PK-treated and SDS-treated chromosomal DNAs of S. hygroscopicus after digestion with AseI. Consistently, no obvious difference of the banding pattern was observed on PFGE (Fig. 1), while obvious retardation of the AseI-J fragment of the PK-untreated sample but not the PK-treated S. coelicolor A3(2) sample can be observed under the same conditions (Fig. 1D), making direct localization of the terminal fragments by conventional methods rather difficult.
A second attempt was made to localize the terminal fragments by analyzing the deletion mutants again, assuming that deletions involving both terminal fragments would circularize the chromosome. Indeed, T155 (Fig. 4, panel 5), a mutant with a large deletion involving the entire Contig3 (covering 500E, 150I, 225H1, 225H2, 300F, and 280G), has a circular chromosome whose DNA was not trapped after AseI digestion but was retarded in the gel slot even when the plug sample for PFGE was treated with PK. The same was also true for strains T106 (Fig. 4, panel 4) and T157 (Fig. 4, panel 1), whose chromosomal DNA deletions covered only Contig2 (150I, 225H1, 225H2, 300F, and 280G) and Contig1 (225H1, 225H2, 300F, 280G), respectively. Therefore, the two end fragments would seem to be confined in Contig1. However, demonstration of the linear topology of the strain WH-14 chromosome (Fig. 4, panel 9), whose deletion was confined to two of the four fragments inside Contig1 (300F and 280G), excluded the possibility that 300F and 280G could be the two terminal fragments, leaving 225H1 and 225H2 as the only options.
DISCUSSION
Many laboratory strains of Streptomyces species suffer chromosomal deletions of genomic DNA in an area which has been called the unstable or deletion-prone region of the chromosome. Chromosomal deletions in Streptomyces glaucescens can be of 800 kb or more (3); in S. lividans, the deletion-prone region encompasses at least 1 Mb (24); and in Streptomyces ambofaciens the region can reach 2 Mb of contiguous DNA (7, 14, 15). In S. hygroscopicus 10-22, frequent deletions ranging from 240 to 2,150 kb, encompassing several AseI fragments of the genome, were observed, making it possible to order the relative positions of the eleven AseI fragments on the chromosome and to finally draw a complete chromosomal physical map. The deletion-prone region contains a relatively high density of AseI sites, suggesting that there are patches of DNA with low G+C content on the chromosome.
The two chromosomal terminal fragments for S. hygroscopicus 10-22 could not be determined conveniently by comparing the PK-treated and SDS-treated chromosomal DNAs embedded in plugs after digestion with AseI and looking for dissociated fragments, but again the analysis of the topology of a series of deletion mutants was helpful and led to the conclusion that 225H1 and 225H2 are the two detectable terminal fragments. Could 225H1 and 225H2 fragments have identical sequences present as terminal inverted repeats? In the experiment for the determination of the relative locations of 150I and 225H1 using a ca. 380-kb partially digested genome fragment as a probe (Fig. 5, panel 1), two positive signals corresponding to 150I and 225H1 or 225H2 were detected and the hybridizing band at the 225H1 and 225H2 position seemed to be at least twice intensified compared with the 150I band, which could be explained as a result of the hybridization to two fragments (225H1 plus 225H2) running at the same position. If 225H1 and 225H2 were indeed identical terminal sequences, the relative hybridization signals of 225H1 or 225H2 and 150I to the 380-kb partial fragments would be 4.5:1.5 or 3:1. This is close to the measured ratio of 2.5:1 by scanning the hybridizing bands at the 225- and 150-kb band positions. Therefore, the 225H2 fragment also seemed to be involved in hybridization. It is highly possible that only the 225H1 fragment contributes to the formation of the 380-kb partial fragment, but 225H1 contains a complete or partial direct or inverted repeat sequence(s) with that of 225H2, and hence the 225H2 fragment also hybridized with the 380-kb probe. We also checked the possible hybridizations using linking clone 2B3 as a probe to 225H1 and/or 225H2, 300F, and 150I but only observed the obvious hybridization with bands corresponding to 225H1 and/or 225H2 and 300F but not to 150I. It was assumed that the region flanking the 300F side of the AseI site of the linking clone 2B3 has very little homology with 150I, suggesting that the AseI site is very close to one end of the terminal inverted repeats. Terminal inverted sequences ranging from 44 bp to 550 kb (5, 19) were reported in invertron structures of Streptomyces species. The above hybridization result implies that 225H1 and 225H2 are likely to be the terminal fragments of the S. hygroscopicus 10-22 chromosome with identical sequences, but this has not been proven. The reason for the failure could be explained if there was another AseI site flanking each of the two inverted repeats (225H1 and 225H2) proximal to the ends bound by terminal proteins. In such a case, 225H1 and 225H2 would not be considered the terminal fragments but the second fragments next to both ends. An effort to look for small AseI DNA fragments by running normal agarose gel electrophoresis and PFGE (at a switch time of 5 to 30 s for ca. 10 h) after the plug was digested with AseI was also unsuccessful. The real end fragments might be either too small to run out of the gel or too small to detect by ethidium bromide staining. We tentatively add two symmetrical AseI sites as conceivably an extension of long inverted repeats (more than 225 kb) on the physical map of strain 10-22 in Fig. 6.
Acknowledgments
This work received support from the National Science Foundation of China, the 863 Fund from the Ministry of Science and Technology, the Shanghai Municipal Council of Science and Technology, the International Foundation for Science, and the Rockefeller Foundation.
We thank T. Kieser and D. A. Hopwood for the gifts of plasmid, phage vectors, probes, and strains. We thank D. A. Hopwood and K. F. Chater for the critical reading of the manuscript.
REFERENCES
- 1.Bao, K. 1997. Ph.D. thesis. Huazhong Agricultural University, Wuhan, China.
- 2.Bautsch, W. 1988. Rapid physical mapping of the Mycoplasma mobile genome by two-dimensional field inversion gel electrophoresis techniques. Nucleic Acids Res. 16:11461-11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birch, A., A. Häusler, M. Vögtli, W. Krek, and R. Hütter. 1989. Extremely large chromosomal deletions are intimately involved in genetic instability and genomic rearrangements in Streptomyces glaucescens. Mol. Gen. Genet. 217:447-458. [DOI] [PubMed] [Google Scholar]
- 4.Borck, K., J. D. Beggs, W. J. Brammar, A. S. Hopkins, and N. E. Murray. 1976. The construction in vitro of transducing derivatives of phage lambda. Mol. Gen. Genet. 146:199-207. [DOI] [PubMed] [Google Scholar]
- 5.Chater, K. F., and D. A. Hopwood. 1993. Streptomyces genetics. American Society for Microbiology, Washington, D.C.
- 6.Chu, G., D. Vollrath, and R. W. Davis. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582-1585. [DOI] [PubMed] [Google Scholar]
- 7.Fischer, G., B. Decaris, and P. Leblond. 1997. Occurrence of deletions associated with genetic instability in Streptomyces ambofaciens is independent of the linearity of the chromosomal DNA. J. Bacteriol. 179:4553-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fonstein, M., and R. Haselkorn. 1995. Physical mapping of bacterial genome. J. Bacteriol. 177:3361-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 10.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces—a laboratory manual. The John Innes Foundation, Norwich, Conn.
- 11.Kieser, H. M., T. Kieser, and D. A. Hopwood. 1992. A combined genetic and physical map of the Streptomyces coelicolor A3(2) chromosome. J. Bacteriol. 174:5496-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, Conn.
- 13.Kohara, Y., K. Akiyama, and K. Isono. 1987. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50:495-508. [DOI] [PubMed] [Google Scholar]
- 14.Leblond, P., and B. Decaris. 1999. “Unstable” linear chromosomes: the case of Streptomyces, p.235-261. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. American Society for Microbiology, Washington, D.C.
- 15.Leblond, P., P. Demuyter, J. M. Simonet, and B. Decaris. 1991. Genetic instability and associated genome plasticity in Streptomyces ambofaciens: pulsed-field gel electrophoresis evidence for large DNA alterations in a limited genomic region. J. Bacteriol. 173:4229-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leblond, P., M. Redenbach, and J. Cullum. 1993. Physical map of the Streptomyces lividans 66 genome and comparison with that of the related strain Streptomyces coelicolor A3(2). J. Bacteriol. 175:3422-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lezhava, A., T. Mizukami, T. Kajitani, D. Kameoka, M. Redenbach, H. Shinkawa, O. Nimi, and H. Kinashi. 1995. Physical map of the linear chromosome of Streptomyces griseus. J. Bacteriol. 177:6492-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang, R. F., and Q. Zhou. 1991. Blocked mutants of antibiotic 5102-III obtained by acridine orange treatment of S. hygroscopicus 10-22. J. Huazhong Agric. Univ. 10:362-366. [Google Scholar]
- 19.Lin, Y. S., and C. W. Chen. 1997. Instability of artificially circularized chromosomes of Streptomyces lividans. Mol. Microbiol. 26:709-719. [DOI] [PubMed] [Google Scholar]
- 20.Lin, Y. S., H. M. Kieser, D. A. Hopwood, and C. W. Chen. 1993. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 10:923-933. [DOI] [PubMed] [Google Scholar]
- 21.Pandza, K., G. Pfalzer, J. Cullum, and D. Hranueli. 1997. Physical mapping shows that the unstable oxytetracycline gene cluster of Streptomyces rimosus lies close to the ends of the linear chromosome. Microbiology 143:1493-1501. [DOI] [PubMed] [Google Scholar]
- 22.Qin, Z., K. Peng, X. Zhou, R. Liang, Q. Zhou, H. Chen, D. A. Hopwood, T. Kieser, and Z. Deng. 1994. Development of a gene cloning system for Streptomyces hygroscopicus subsp. yingchengensis, a producer of three useful antifungal compounds, by elimination of three barriers to DNA transfer. J. Bacteriol. 176:2090-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin, Z. 1992. Ph.D. thesis. Huazhong Agricultural University, Wuhan, China.
- 24.Redenbach, M., F. Flett, W. Piendl, I. Glocker, U. Rauland, O. Wafzig, R. Kliem, P. Leblond, and J. Cullum. 1993. The Streptomyces lividans 66 chromosome contains a 1 MB deletogenic region flanked by two amplifiable regions. Mol. Gen. Genet. 241:255-262. [DOI] [PubMed] [Google Scholar]
- 25.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 26.Reeves, A. R., D. A. Post, and T. J. Vanden Boom. 1998. Physical-genetic map of the erythromycin-producing organism Saccharopolyspora erythraea. Microbiology 144:2151-2159. [DOI] [PubMed] [Google Scholar]
- 27.Romling, U., D. Grothues, W. Bautsch, and B. Tummler. 1989. A physical genome map of Pseudomonas aeruginosa PAO. EMBO J. 8:4081-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Smith, G. E., and M. D. Summers. 1980. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal. Biochem. 109:123-129. [DOI] [PubMed] [Google Scholar]
- 30.Volff, J. N., and J. Altenbuchner. 1998. Genetic instability of the Streptomyces chromosome. Mol. Microbiol. 27:239-246. [DOI] [PubMed] [Google Scholar]
- 31.Volff, J. N., and J. Altenbuchner. 2000. A new beginning with new ends: linearisation of circular chromosomes during bacterial evolution. FEMS Microbiol. Lett. 186:143-150. [DOI] [PubMed] [Google Scholar]
- 32.Volff, J. N., D. Vandewiele, J. M. Simonet, and B. Decaris. 1993. Ultraviolet light, mitomycin C and nitrous acid induce genetic instability in Streptomyces ambofaciens ATCC23877. Mutat. Res. 287:141-156. [DOI] [PubMed] [Google Scholar]
- 33.Volff, J. N., D. Vandewiele, J.-M. Simonet, and B. Decaris. 1993. Stimulation of genetic instability in Streptomyces ambofaciens ATCC23877 by antibiotics that interact with DNA gyrase. J. Gen. Microbiol. 139:2551-2558. [DOI] [PubMed] [Google Scholar]
- 34.Wang, S. J., H. M. Chang, Y. S. Lin, C. H. Huang, and C. W. Chen. 1999. Streptomyces genomes: circular genetic maps from the linear chromosomes. Microbiology 145:2209-2220. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, S. H., H. Z. Zhao, and J. L. Liu. 1982. Studies on the agricultural antibiotic 5102-III. Isolation and characterization of antibiotic 5102-III. J. Huazhong Agric. Univ. 1:39-43. [Google Scholar]
- 36.Zhang, S. H., S. Z. Zhao, and J. L. Liu. 1981. Studies on the agricultural antibiotic 5102-II. Isolation and characterization of antibiotic 5102-II. Acta Microbiol. Sin. 22:145-150. [Google Scholar]
- 37.Zhou, Q., and J. L. Liu. 1981. Studies on antifungal activities of the agricultural antibiotic 5102. Chin. J. Antibiot. 6:1-6. [Google Scholar]