Abstract
Horizontal DNA transfer contributes significantly to the dissemination of antibiotic resistance genes in Bacteroides fragilis. To further our understanding of DNA transfer in B. fragilis, we isolated and characterized a new transfer factor, cLV25. cLV25 was isolated from B. fragilis LV25 by its capture on the nonmobilizable Escherichia coli-Bacteroides shuttle vector pGAT400ΔBglII. Similar to other Bacteroides sp. transfer factors, cLV25 was mobilized in E. coli by the conjugative plasmid R751. Using Tn1000 mutagenesis and deletion analysis of cLV25, two mobilization genes, bmgA and bmgB, were identified, whose predicted proteins have similarity to DNA relaxases and mobilization proteins, respectively. In particular, BmgA and BmgB were homologous to MocA and MocB, respectively, the two mobilization proteins of the B. fragilis mobilizable transposon Tn4399. A cis-acting origin of transfer (oriT) was localized to a 353-bp region that included nearly all of the intergenic region between bmgB and orf22 and overlapped with the 3′ end of orf22. This oriT contained a putative nic site sequence but showed no significant similarity to the oriT regions of other transfer factors, including Tn4399. Despite the lack of sequence similarity between the oriTs of cLV25 and Tn4399, a mutation in the cLV25 putative DNA relaxase, bmgA, was partially complemented by Tn4399. In addition to the functional cross-reaction with Tn4399, a second distinguishing feature of cLV25 is that predicted proteins have similarity to proteins encoded not only by Tn4399 but by several Bacteroides sp. transfer factors, including NBU1, NBU2, CTnDOT, Tn4555, and Tn5520.
Bacteroides fragilis is an obligate anaerobe of the colon and a significant opportunistic pathogen. Antibiotic resistance among Bacteroides spp. is rapidly increasing, largely due to the dissemination of DNA transfer factors (plasmids and transposons) harbored by members of this genus. Transfer factors can be divided into two classes, conjugative and mobilizable. Conjugative plasmids and transposons are self-transmissible, encoding both functions of DNA transfer, that is, DNA processing events for transfer initiation and mating apparatus formation, which brings donor and recipient bacteria into stable cell-cell contact. In contrast, mobilizable plasmids and transposons carry mobilization genes only for transfer initiation, utilizing a mating apparatus provided by a coresident conjugative transfer factor.
All transfer factors also contain a cis-acting origin of transfer (oriT), where transfer is initiated. DNA processing events at the oriT consist of specific protein-DNA and protein-protein interactions that result in the formation of a relaxed DNA-protein complex called the relaxosome (see references 7, 9, 14, and 25 for detailed descriptions). In brief, mobilization proteins assemble at the oriT, with one protein, the DNA relaxase, nicking a single DNA strand at the nic site. The nicked DNA strand is then transferred with 5′-to-3′ polarity to a recipient bacterium via a mating bridge that is formed between donor and recipient cells. DNA replication concomitantly synthesizes the complementary strand in both the donor and the recipient. The oriT is typically located adjacent to the mobilization genes (50), thus forming a compact mobilization region. A common feature of the oriT is the presence of inverted repeats near the nic site which are recognition sites for the binding of mobilization proteins (21, 35, 50).
Transfer of a mobilizable plasmid or transposon from Bacteroides spp. is thought to be dependent on a coresident conjugative transposon (CTn) to provide the mating apparatus. Nearly all identified Bacteroides conjugative transposons carry the tetracycline resistance gene, tetQ (28). Further, transfer of both plasmids and transposons from Bacteroides spp. is stimulated 1,000- to 10,000-fold upon exposure to a subinhibitory concentration of tetracycline (1 μg/ml) (28, 30, 39, 45). In addition, all Bacteroides spp. mobilizable plasmids and transposons tested to date are also mobilized in Escherichia coli when coresident with the conjugative IncP plasmid RP4 or R751 (15, 18, 20, 30, 44, 46). In E. coli, Bacteroides mobilization genes provide the DNA processing functions to form a relaxosome, while RP4 and R751 provide the mating apparatus.
Bacteroides spp. are the only gram-negative bacteria known to harbor mobilizable transposons. Five mobilizable transposons, Tn4399, Tn5520, Tn4555, NBU1, and NBU2, have been characterized to date, and both their mobilization and transposition properties have been studied (12, 31, 36, 46). Tn4399 (9.6 kb) requires two genes, mocA and mocB, for mobilization (18). MocA is the predicted DNA relaxase, having similarity to TraI from RP4 (7), MbpB from pLV22a (a B. fragilis mobilizable plasmid [20]), and MobA from CTnDOT (a Bacteroides thetaiotaomicron conjugative transposon [49]). During transposition, Tn4399 creates a 3-bp target site repeat. In addition, a novel feature of Tn4399 insertions is the presence of an extra 5 bp between the right inverted repeat and the target site repeat (13). A second mobilizable transposon, Tn5520, requires just one gene for mobilization, bmpH (46), whose predicted protein has homology with the single mobilization proteins from Tn4555 (MobA) (37), NBU1 (MobN1), and NBU2 (MobN2) (15). The single mobilization proteins encoded by Tn5520, Tn4555, NBU1, and NBU2 are predicted to be multifunctional, performing all site recognition, DNA binding, and DNA nicking functions. Tn5520 is the smallest mobilizable transposon described to date (4.69 kb) and encodes an integrase, bipH, in addition to its mobilization gene. Target site duplication is not seen at Tn5520 insertion sites. In addition to B. fragilis, Tn5520 also transposes in E. coli (46). A third mobilizable transposon, Tn4555 (12.1 kb), carries a cefoxitin resistance gene (36). Tn4555 integrates via a site-specific recombination mechanism without duplicating its target site, and its efficiency of integration is increased by two accessory genes, tnpA and tnpC (43). In addition to protein sequence identity with NBU1 and NBU2, the Tn4555 mobilization region has >75% nucleotide sequence homology with the mobilization regions of NBU1 and NBU2 (37). NBU1 (10.3 kb) and NBU2 (11.1 kb) integrate at the 3′ end of tRNA genes in Bacteroides spp. and also integrate in E. coli (32, 34, 48). In addition, NBU2 carries a lincosamide resistance gene, linA2N2, which confers a lincomycin resistance phenotype on B. thetaiotaomicron (48). Although Tn5520, Tn4555, NBU1, and NBU2 each encode a single homologous mobilization protein, their DNA sequences vary outside their mobilization regions. Since only the mobilization regions of these Bacteroides mobilizable transposons are highly similar, it has been suggested that these mobilization regions reside on cassettes (15, 18, 39, 48); that is, the mobilization gene and oriT are compactly organized within a discrete unit.
We have isolated a new transfer factor, cLV25, from B. fragilis, and we report the requirements for cLV25 mobilization in E. coli, as well as the DNA sequence analysis of cLV25 and its relatedness to other known transfer factors. The results from this study demonstrate that the cLV25 mobilization proteins have similarity to the Tn4399 mobilization proteins and to mobilization proteins encoded by the CTnDOT ermF region. In addition, the genetic organization of the cLV25 mobilization genes and oriT is analogous to those of Tn4399 and the CTnDOT ermF region. Further, cLV25 can functionally cross-react with Tn4399. Also, we propose that cLV25 is a mobilizable transposon based on the presence of (i) imperfect inverted repeats at its termini, (ii) an apparent target site repeat, and (iii) an open reading frame whose predicted protein has similarity to integrases.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. B. fragilis strains were grown in supplemented brain heart infusion medium (3.7% brain heart infusion [Difco, Detroit, Mich.] supplemented with 0.0015% hemin and 5 g of yeast extract/liter) at 37°C in an anaerobic chamber (85% N2, 10% H2, and 5% CO2; Coy Laboratory Products, Inc., Ann Arbor, Mich.). E. coli strains were grown aerobically at 37°C in Luria-Bertani (LB) medium. E. coli strains containing R751 were grown in Mueller-Hinton medium (Difco). All media were solidified with 1.7% agar when required. E. coli HB101 and DH5α strains were used for routine cloning procedures. The antibiotic concentrations used for the selection of strains and plasmids were as follows: ampicillin, 200 μg/ml; chloramphenicol, 25 μg/ml; spectinomycin, 50 μg/ml; streptomycin, 50 μg/ml; tetracycline, 10 μg/ml; and trimethoprim, 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant phenotype and/or characteristicsa | Source or reference |
|---|---|---|
| B. fragilis LV25 | Tcr | Clinical isolate |
| E. coli | ||
| HB101 | Smr | 29 |
| DW1030 | Spr | 26 |
| DH5α | 2 | |
| Plasmids | ||
| R751 | IncPβ Tra+ Tmr | 17 |
| F′lac | IncF1 Tra+lac+ | Pasteur Institute |
| pGAT400 | Mob+ Clnr Apr Tcr (aerobic); E. coli-Bacteroides shuttle vector | 12 |
| pGAT400ΔBglII | Mob− Clnr Apr Tcr (aerobic); pGAT400 derivative | 12 |
| pBC SK(+) | Cmr; cloning vector | Stratagene |
| pGEM-T Easy | Apr; cloning vector | Promega |
| pCR2.1-TOPO | Apr Knr; cloning vector | Invitrogen |
| pACYC184 | Mob− Cmr Tcr; cloning vector | New England Biolabs |
| pBR328 | Mob− Apr Tcr Cmr; cloning vector | 6 |
| p25Δ.2 | Mob+ Clnr Apr; pGAT400ΔBglII::cLV25 (insertion in IS4400R) | This study |
| p25Δ.3 | Mob+ Clnr Apr; pGAT400ΔBglII::cLV25 (insertion in IS4400L) | This study |
| p21ΔB | Mob+ Clnr Apr; pGAT400ΔBglII::Tn4399 | 12 |
| pAC11 | Mob+ Cmr; 11.3-kb cLV25 ClaI/AvaI fragment ligated to the 2.8-kb ClaI/AvaI pACYC184 fragment | This study |
| pBR2N | Apr Tcr; 5.5-kb cLV25 EcoRI fragment ligated to EcoRI site of pBR328 | This study |
| pAC166 | Mob+ Cmr; Tn1000 insertion in orf25 of pAC11 | This study |
| pAC115 | Mob+ Cmr; Tn1000 insertion in orf25 of pAC11 | This study |
| pAC150 | Mob− Cmr; Tn1000 insertion in bmgA of pAC11 | This study |
| pAC114 | Mob− Cmr; Tn1000 insertion in bmgA of pAC11 | This study |
| pAC167 | Mob− Cmr; Tn1000 insertion in bmgB of pAC11 | This study |
| pAC148 | Mob+/− Cmr; Tn1000 insertion in orf22 of pAC11 | This study |
| pGEM-T4.1 | Apr; 4.1-kb cLV25 PCR product ligated to pGEM-T Easy | This study |
| pAC4.1 | Mob+ Cmr; 4.1-kb pGEM-T4.1 SalI/SphI fragment ligated to SalI/SphI sites of pACYC184 | This study |
| pGEM-T2.7 | Apr; 2.7-kb cLV25 PCR product ligated to pGEM-T Easy | This study |
| pBR2.5 | Mob+ Apr; 2.5-kb pGEM-T2.7 EcoRI fragment ligated to EcoRI site of pBR328 | This study |
| pCR23A | Apr; 798-bp PCR product containing cLV25 bmgB and 371-bp upstream sequence ligated to pCR2.1-TOPO | This study |
| pBR23A | Apr Tcr; 798-bp pCR23A EcoRI fragment ligated to EcoRI site of pBR328 | This study |
| pGEMoriT7 | Apr; 353-bp PCR product containing cLV25 oriT ligated to pGEM-T Easy | This study |
| pACoriT7 | Mob+ Tcr; pGEMoriT7 EcoRI insert fragment ligated to EcoRI site of pACYC184 | This study |
Tcr, Smr, Spr, Tmr, Clnr, Apr, Cmr, and Knr, resistance to tetracycline, streptomycin, spectinomycin, trimethoprim, clindamycin, ampicillin, chloramphenicol, and kanamycin, respectively; Tra+, transfer proficiency; Mob+, able to be mobilized; Mob−, unable to be mobilized; Mob+/−, reduced mobilization frequency.
Recombinant DNA techniques.
Plasmid DNA was prepared by alkaline lysis (29) and by affinity column purification (Qiagen Corp., Chatsworth, Calif.). Chromosomal DNA was prepared as previously described (27). Restriction endonucleases were purchased from New England Biolabs (Beverly, Mass.), and T4 DNA ligase was purchased from Promega (Madison, Wis.) and Epicentre Technologies (Madison, Wis.). All enzymes were used as recommended by the suppliers.
Plasmid construction.
To locate the cLV25 mobilization region, several cLV25 fragments were subcloned from p25Δ.2 into nonmobilizable vectors. p25Δ.2 contains an insertion of intact cLV25 in pGAT400ΔBglII. The 11.3-kb ClaI-AvaI fragment of p25Δ.2, containing the right two-thirds of cLV25, was ligated to the 2.8-kb AvaI-ClaI fragment of the cloning vector pACYC184 (New England Biolabs) to construct pAC11. The 5.5-kb EcoRI fragment of p25Δ.2 was ligated to the EcoRI site of the cloning vector pBR328 (6) to construct pBR2N. A 4.1- and a 2.7-kb region of cLV25 were obtained by PCR amplification and cloned into pGEM-T Easy (Promega) to generate pGEM-T4.1 and pGEM-T2.7, respectively. pGEM-T4.1 was digested with SphI/SalI, and the 4.1-kb fragment was ligated to the SphI/SalI sites of pACYC184 to generate pAC4.1. pGEM-T2.7 was digested with EcoRI, and the 2.5-kb insert fragment was ligated to the EcoRI site of pBR328 to generate pBR2.5. For complementation of the transposon insertions in bmgB and orf22, a 798-bp fragment containing cLV25 bmgB and upstream sequence was obtained by PCR amplification and cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.) to construct pCR23A. pCR23A was digested with EcoRI, and the 798-bp insert was ligated to the EcoRI site of pBR328 to generate pBR23A. The 353-bp fragment containing the cLV25 oriT was obtained by PCR amplification and cloned into pGEM-T Easy to generate pGEMoriT7. This 353-bp fragment was excised from pGEMoriT7 by an EcoRI digest and then ligated to the EcoRI site of pACYC184 to construct pACoriT7.
PCR.
The GeneAmp XL PCR kit (Roche Molecular Systems, Inc., Branchburg, N.J.) was used according to the manufacturer's instructions for DNA amplification of the cLV25 4.1-kb fragment. The 4.1-kb cLV25 fragment, bp 4182 to 8289, was amplified using primers oriTL3 (5′-CAATGACGAATTTGGTGAATGGATGC-3′) and MOB-R2 (5′-GATGACAACACAAGGGGACATATCGG-3′) with p25Δ.2 as the template. The following three cLV25 fragments were amplified using p25Δ.2 as the DNA template, native Pfu DNA polymerase (Stratagene, La Jolla, Calif.), and the GeneAmp XL PCR kit core reagents. The 2.7-kb cLV25 fragment, bp 4182 to 6927, was amplified using primers oriTL3 and MOB-R (5′-CGGCTCGGGTTGACAATGACAAGGC-3′). cLV25 bmgB and 371 bp of upstream sequence, bp 6130 to 6927, were amplified using primers orf23F (5′-CAAAGCCGCCTGTTCCTTGCCAAGC-3′) and MOB-R. A 353-bp fragment containing the cLV25 oriT, bp 6575 to 6927, was amplified using primers oriTL1 (5′-GGTATCTTAAAAATTGCGACCAACGG-3′) and MOB-R. All reactions were performed on an Applied Biosystems GeneAmp PCR System 2400 thermal cycler (Perkin-Elmer, Foster City, Calif.). For blunt-ended PCR products generated from native Pfu DNA polymerase, 3′ deoxyadenosine residues were added postamplification by incubating the PCR mixture with 1 U of AmpliTaq Gold DNA polymerase (Roche Molecular Systems, Inc.) for 10 min at 72°C.
Plasmid mobilization experiments.
The E. coli-Bacteroides shuttle vector pGAT400ΔBglII was mobilized into B. fragilis strain LV25 by an RK2-mediated shuttle mating (8). The transfer of pGAT400ΔBglII from B. fragilis LV25 was measured by quantitative B. fragilis-to-E. coli filter matings as previously described (11). The mobilization of plasmids in E. coli was determined by mixing log-phase cultures of HB101 donors, containing R751 and the test plasmid, and DW1030 recipients in a 1:9 donor-to-recipient ratio (total volume, 1.5 ml). The mixed cells were collected and resuspended, placed on sterile 47-mm-diameter GN-6 Gelman membrane filters (Pall Corporation, Ann Arbor, Mich.) supported on LB agar, and incubated for 5 h at 37°C. The cells were then suspended and diluted in 1× modified phosphate-buffered saline (4.2 mM Na2HPO4, 1.7 mM NaH2PO4, 0.14 mM NaCl, pH 6.9), plated to selective media, and incubated overnight at 37°C. The frequency of mobilization was determined by normalizing the frequency of transfer of the test plasmid to that of R751 in the same experiment. All mobilization frequencies were reported as the mean ± standard error. Complementation studies were done similarly, with each donor strain containing three plasmids; R751, the mutant plasmid, and the complementing plasmid.
Transposon mutagenesis.
Tn1000 insertions were introduced into the target plasmid, pAC11, as previously described (10). Briefly, HB101 donors, containing F′lac and pAC11, and DW1030 recipients were mixed in a 1:2 donor-to-recipient ratio and incubated at 37°C with vigorous shaking for 1 h and then with gentle shaking for 2.25 h. The mating broths were diluted and plated to LB agar containing spectinomycin and chloramphenicol. For each Tn1000 insertion in pAC11, all internal BglII fragments were deleted to prevent further transposition events. Insertions were mapped initially by restriction enzyme analysis, and select insertions were mapped precisely by sequence analysis from the ends of Tn1000 with primers gamma (5′-AATCAGCTACAACATACG-3′) and delta (5′-CGAATTATCTCCTTAACG-3′).
Southern hybridization analysis.
DNA samples were digested with EcoRI and fractionated on a 0.7% agarose gel by electrophoresis using standard methods. The DNA was transferred to a neutral nylon membrane (S&S Nytran, Keene, N.H.) by downward capillary alkaline transfer using the Schleicher & Schuell (Keene, N.H.) TurboBlotter system. The 3.6-kb EcoRI cLV25 fragment was purified by agarose gel electrophoresis, labeled with psoralen biotin, and incubated with the DNA blot as specified by the Schleicher & Schuell Rad-Free probe labeling and hybridization kit protocol. DNA-DNA hybridizations were detected using a streptavidin-alkaline phosphatase conjugate and a chemiluminescent substrate.
DNA sequencing and analysis.
The seven cLV25 EcoRI fragments from transconjugant plasmid DNA p25Δ.2 (1.3, 3.6, 5.5, 2.4, 0.3, 1.0, and 7.5 kb) were subcloned into pBC SK(+) (Stratagene) to generate p25Δ.2H, p25Δ.2P, p25Δ.2N, p25Δ.2 M (containing 2.4- and 1.0-kb fragments), p25Δ.2C, and p25Δ.2L, respectively. These subclones were used as DNA templates to sequence one DNA strand of cLV25 by primer walking. Overlapping subclones and PCR products were used as templates to sequence across the EcoRI restriction sites. Double-stranded DNA sequence was obtained for 5.5 kb of cLV25 that encompasses the mobilization region and for 100 bp at each terminus. Plasmid DNA templates were purified by affinity column purification (Qiagen Corp.) for all sequencing reactions. Nucleotide sequences were determined by the Sanger method using Applied Biosystems DNA-sequencing systems at Loyola University Medical Center (model 373A) and Northwestern University Biotechnology Laboratory (model 377 XL). The nucleotide sequences and amino acid sequences of potential open reading frames were analyzed using available on-line programs (http://www.up.univ-mrs.fr/∼wabim/english/logligne.html and http://www.andrew.cmu.edu/user/yzhan/dnaoligo.html) and compared to sequences in the GenBank database using the BLAST programs (1).
Nucleotide sequence accession number.
The cLV25 sequence (15,343 bp) was submitted to the GenBank database and was given the accession number AY053505.
RESULTS
B. fragilis strain LV25 harbors a new 15.3-kb chromosomal transfer factor, designated cLV25.
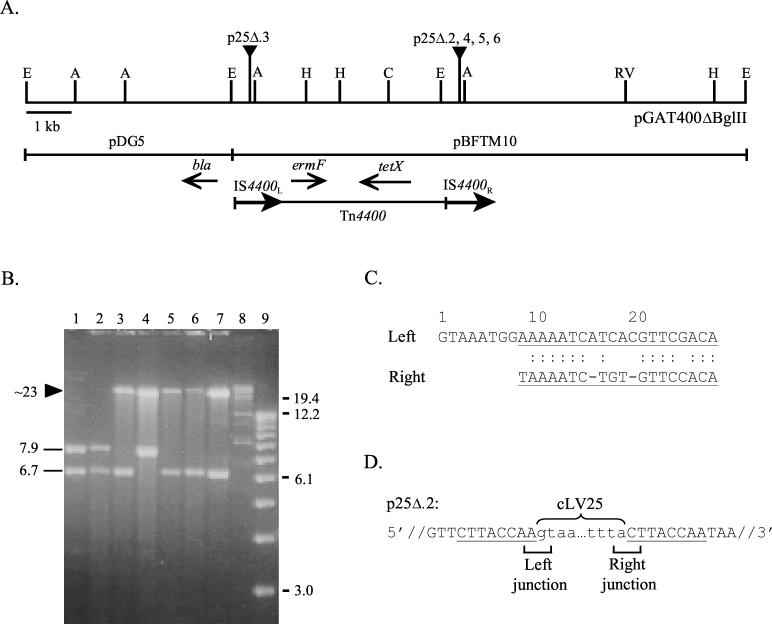
The E. coli-Bacteroides shuttle vector pGAT400ΔBglII was used to isolate a new transfer factor from a tetracycline-resistant B. fragilis clinical isolate, LV25. In E. coli, pGAT400ΔBglII can be mobilized by the conjugative plasmid pRK231. However, in Bacteroides spp., pGAT400ΔBglII is not mobilized by conjugative transposons unless it acquires, in cis, DNA that carries a mobilization region. pGAT400ΔBglII was mobilized by pRK231 from E. coli HB101 to LV25 by conjugation. LV25, containing pGAT400ΔBglII, was then used as a donor in a mating experiment with E. coli HB101 to determine if pGAT400ΔBglII “captured” a transfer factor. Six transconjugants, HB101 p25Δ.1 through -6, were obtained at a frequency of 2.6 × 10−8, but only when donor cells were pretreated with a subinhibitory concentration (1 μg/ml) of tetracycline. HindIII restriction enzyme analysis of plasmid DNA from five transconjugants, p25Δ.2 through -6, demonstrated that pGAT400ΔBglII had acquired approximately 15 kb of DNA in each case (Fig. 1B). This 15-kb DNA insert was designated cLV25. Each cLV25 insertion in pGAT400ΔBglII resulted in the disappearance of one pGAT400ΔBglII HindIII DNA fragment and the presence of a high-molecular-weight HindIII DNA fragment containing intact cLV25.
FIG. 1.
New DNA in pGAT400ΔBglII acquired from B. fragilis LV25. (A) Restriction map of pGAT400ΔBglII. bla, ampicillin resistance; ermF, clindamycin resistance; tetX, aerobic tetracycline resistance; A, AvaI; C, ClaI; E, EcoRI; H, HindIII; RV, EcoRV. The solid triangles indicate the locations of cLV25 in pGAT400ΔBglII. (B) Agarose gel of HindIII restriction enzyme digestions of pGAT400ΔBglII and transconjugant plasmid DNA from a B. fragilis LV25-to-E. coli HB101 mating. Lanes: 1, pGAT400ΔBglII; 2, p25Δ.1; 3, p25Δ.2; 4, p25Δ.3; 5, p25Δ.4; 6, p25Δ.5; 7, p25Δ.6; 8, high-molecular-weight DNA markers; 9, 1-kb DNA ladder. Molecular size markers in kilobases are indicated on the right. The solid lines on the left indicate pGAT400ΔBglII HindIII fragments in kilobases. The arrowhead indicates the presence of 15 kb of new DNA in pGAT400ΔBglII. (C) Sequence analysis of the cLV25 termini. Shown is an alignment of the 20-bp imperfect inverted repeat (underlined) and the sequence of the additional 8 bp detected at the left end of cLV25 after insertion in pGAT400ΔBglII. (D) DNA sequence analysis of the left and right junctions of transconjugant plasmid DNA p25Δ.2. The pGAT400ΔBglII sequence is in uppercase letters, and the cLV25 sequence is in lowercase letters. The 8-bp target site repeat is underlined.
Restriction enzyme analysis of p25Δ.2 through -6 demonstrated that cLV25 was present at one of two locations in pGAT400ΔBglII, IS4400R and IS4400L, which are identical insertion sequences of Tn4400 (Fig. 1A). DNA sequence analysis of one cLV25 insertion in IS4400R, p25Δ.2, revealed the presence of a 20-bp imperfect inverted repeat at the ends of cLV25. Further analysis of the ends of cLV25 revealed that there was an additional 8 bp between the target site and the left end of cLV25 (Fig. 1C). Additionally, an 8-bp target site repeat was identified (Fig. 1D). The junctions of a second cLV25 insertion, p25Δ.3, were sequenced, showing that cLV25 was present in the identical site in IS4400L and in the same orientation as the cLV25 insertion in IS4400R. Sequence analysis of this second insertion confirmed the presence of the same 20-bp imperfect inverted repeat at the ends of cLV25, 8 bp between the target site and the left end of cLV25, and an 8-bp target site repeat identical to those of the cLV25 insertion in IS4400R (data not shown).
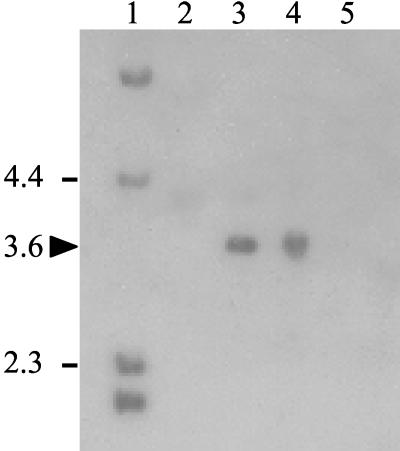
Southern hybridization analysis determined that cLV25 originated from the chromosome of B. fragilis LV25 (Fig. 2). This was demonstrated by hybridization of a cLV25 probe (an internal 3.6-kb EcoRI fragment) with LV25 chromosomal DNA. Lysis of strain LV25 revealed that it contained no plasmid DNA.
FIG. 2.
Southern hybridization analysis demonstrating that cLV25 originated from the chromosome of B. fragilis LV25. LV25 chromosomal and plasmid DNAs were digested with EcoRI and hybridized with a 3.6-kb internal cLV25 EcoRI fragment from p25Δ.2. Lanes: 1, biotinylated lambda DNA HindIII markers; 2, pGAT400ΔBglII; 3, p25Δ.2; 4, LV25 chromosomal DNA; 5, LV25 plasmid DNA. The solid lines indicate the positions of molecular size markers in kilobases. The arrowhead indicates the 3.6-kb cLV25 EcoRI fragment.
Sequence analysis of cLV25 reveals similarity with Tn4399, CTnDOT, and NBU2.
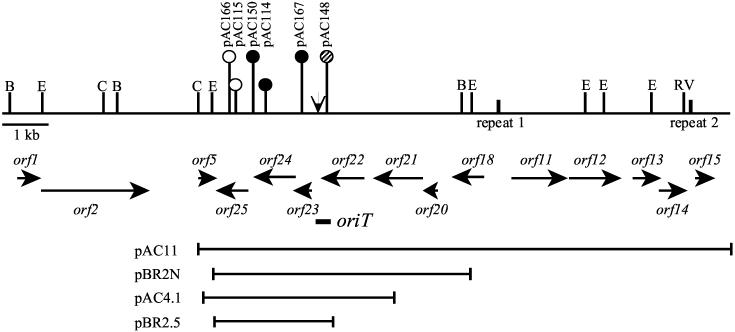
The complete nucleotide sequence of cLV25 (15,343 bp) was obtained by sequencing seven EcoRI restriction fragments, as well as subclones that overlapped the EcoRI restriction sites. DNA sequence analysis of cLV25 revealed 26 potential open reading frames, 15 of which encoded predicted proteins >11.0 kDa (Fig. 3). Predicted proteins that were homologous to Bacteroides proteins in the GenBank database are described in Table 2. The predicted protein for cLV25 orf24 has similarity to several DNA relaxases, including the Tn4399 DNA relaxase, MocA (42% identity over 278 amino acids) (18); MobA from the CTnDOT ermF region (42% identity over 228 amino acids) (49); and MbpB from pLV22a (22% identity over 192 amino acids) (20). Alignment of Orf24 with known DNA relaxases also revealed a 14-bp motif III sequence (amino acids 101 to 114) (data not shown) (22, 24). Motif III is part of a highly conserved DNA relaxase domain containing two invariant histidine residues involved in the cleaving-joining reaction (24). The predicted protein for orf23 has 31% identity (over 107 amino acids) with the second Tn4399 mobilization protein, MocB (18), and 30% identity (over 109 amino acids) with MobB from the CTnDOT ermF region (49).
FIG. 3.
Partial restriction map of cLV25 and locations of Tn1000 insertions (solid circles, Mob−; open circles, Mob+; hatched circles, reduced frequency of mobilization). The vertical arrow indicates the position of the putative nic site. Fifteen potential open reading frames (horizontal arrows), the location of the oriT, and the regions of cLV25 tested for mobilization are shown below the map. B, BstXI; C, ClaI; E, EcoRI; RV, EcoRV.
TABLE 2.
Features of cLV25 potential open reading frames
| ORFa | Coding position (bp) | ORF size (bp) | Molecular mass (kDa) | Homology |
|---|---|---|---|---|
| 11 | 10731-11954 | 1,224 | 47.2 | 23% identity, 44% similarity over 332 amino acids with Tn4555 Int (42); 30% identity, 50% similarity over 165 amino acids with C terminus of Tn5520 BipH (46) |
| 12 | 11980-13116 | 1,137 | 44.2 | 26% identity, 42% similarity over 391 amino acids with N terminus of NBU2 Orf2 (48) |
| 21 | 7833-8906 | 1,074 | 40.8 | 55% identity, 72% similarity over 346 amino acids with NBU2 Orf3 (48); 45% identity, 63% similarity over 288 amino acids with CTnDOT ermF region Orf3F (49) |
| 22 | 6704-7669 | 966 | 37.1 | 30-62% identity, 50-72% similarity over 318, 318, 288, and 316 amino acids with NBU1 PrmN1 (34), NBU2 PrmN2 (48), CTnDOT OrfP (3), and CTnDOT ermF region PrmF (49) |
| 23 | 6173-6556 | 384 | 14.3 | 31% identity, 56% similarity over 107 amino acids with Tn4399 MocB (18); 30% identity, 51% similarity over 109 amino acids with CTnDOT MobB (49) |
| 24 | 5281-6207 | 927 | 34.7 | 42% identity, 63% similarity over 278 amino acids with Tn4399 MocA (18); 42% identity, 64% similarity over 228 amino acids with CTnDOT MobA (49); 22% identity, 48% similarity over 192 amino acids with pLV22a MbpB (20) |
ORF, open reading frame.
orf22 encodes a predicted protein that is similar to the amino termini of several DNA primases. Orf22 also has similarity over its entire length to proteins of unknown function encoded by other Bacteroides sp. transfer factors. These proteins include PrmN1 from NBU1 (62% identity), PrmN2 from NBU2 (57% identity), OrfP from the CTnDOT transfer region (41% identity), and PrmF (30% identity) from the CTnDOT ermF region (3, 34, 48, 49). In addition, the N terminus of Orf22 (126 amino acids) has 68% identity with Tn4399 Orf1, while the C terminus (98 amino acids) has 53% identity with Tn4399 Orf3 (18).
The carboxyl terminus of Orf11 has similarity to numerous integrase and recombinase proteins, including integrases encoded by the Bacteroides mobilizable transposons Tn4555 (Int) (23% identity over the length of the protein) (42) and Tn5520 (BipH) (30% identity over 165 amino acids in the C terminus) (46). The predicted protein for Orf20 has similarity to a putative excisionase (Xis) from Salmonella enterica (32% identity over the length of the protein) (4).
The genetic organization of the region comprising orf21, orf22, orf23, and orf24 was highly similar to the organization of the genes on Tn4399 (18), CTnDOT (49), and NBU2 (48) that encode homologous proteins. Also, cLV25 orf11 and orf12 were organized similarly to NBU2 intN2 and orf2 (48) but located on the opposite DNA strand and were flanked by two sets of perfect inverted repeats (Fig. 3).
cLV25 is mobilized in E. coli by R751.
The Bacteroides transfer factors pBFTM10, pLV22a, Tn4399, and Tn5520 are mobilized in E. coli when coresident with the E. coli conjugative plasmid R751 (11, 18, 20, 46). During mobilization, the Bacteroides transfer factor provides the DNA processing functions for transfer initiation at its own oriT, while R751 provides only the mating apparatus. cLV25 was also tested for mobilization (as part of the E. coli-Bacteroides shuttle vector), to determine if the DNA processing functions encoded by cLV25 would allow it to be mobilized in E. coli. When HB101 containing R751 and p25Δ.2 was mated with DW1030, p25Δ.2 was mobilized at a frequency of (1.7 ± 0.44) × 10−4, 2 orders of magnitude lower than a control plasmid, pGAT400 (containing B. fragilis pBFTM10 [12]) (Table 3). Analysis of transconjugant plasmid DNA by restriction enzyme digestion showed that both R751 and p25Δ.2 were unaltered (data not shown). pGAT400ΔBglII, the negative control, either was not mobilized or formed a cointegrate with R751.
TABLE 3.
R751 mobilization of cLV25 and subclones in E. coli
| Plasmida | Frequency of mobilization (mean ± SE)b |
|---|---|
| pGAT400 | (1.1 ± 0.18) × 10−2 |
| pGAT400ΔBglII | ND |
| p25Δ.2 | (1.7 ± 0.44) × 10−4 |
| pACYC184 | ND |
| pBR328 | ND |
| pAC11 | (1.2 ± 0.22) × 10−4 |
| pAC4.1 | (1.7 ± 0.20) × 10−4 |
| pBR2.5 | (9.6 ± 3.8) × 10−4 |
| pACoriT7 | (2.1 ± 0.53) × 10−3 |
Plasmids were mobilized from E. coli HB101 R751 to E. coli DW1030.
The frequency of mobilization was determined by dividing the number of transconjugants that received the mobilized plasmid by the number of transconjugants that received R751. The mean was obtained from a minimum of four independent matings. SE, standard error; ND, mobilization not detectable.
A 4.1-kb region contains the mobilization genes and oriT.
To determine the minimum region of cLV25 required for mobilization in E. coli by R751, several subclones of cLV25 were tested for mobilization (data not shown). Of the subclones tested, only pAC11 (Fig. 3), containing an 11.3-kb ClaI/AvaI cLV25 restriction fragment, was mobilized in E. coli by R751 at a frequency comparable to that of wild-type p25Δ.2 (Table 3). To further define the region required for mobilization, transposon mutagenesis of pAC11 was performed. A total of 61 Tn1000 insertions in pAC11 were mapped by restriction enzyme analysis. Select insertions were mapped precisely by sequencing outward from the ends of Tn1000. Each insertion mutation of cLV25 was tested separately for mobilization in E. coli. Three insertion mutations, pAC150, pAC114, and pAC167, resulted in a complete loss of mobilization, and one insertion mutation, pAC148, resulted in a reduced frequency of mobilization (Fig. 3). These insertions were located within a 1.9-kb region of cLV25. To test this region for mobilization, a 4.1-kb fragment of cLV25, encompassing this 1.9-kb region, was cloned into pACYC184 to construct pAC4.1 (Fig. 3). pAC4.1 was mobilized in E. coli by R751 at a wild-type frequency (Table 3), thus further delimiting the cLV25 mobilization region.
The gene product of orf24, BmgA, is required for mobilization and functions in trans.
Insertions in pAC150 and pAC114 disrupted orf24, the putative DNA relaxase gene, and resulted in a loss of mobilization in E. coli. Complementation experiments were performed in E. coli using p25Δ.2 (intact cLV25) and pBR2N (5.5-kb cLV25 subclone) to provide wild-type genes in trans. pAC150 and pAC114, coresident with either p25Δ.2 or pBR2N, were tested for mobilization in E. coli by R751. To detect the mobilization of mutant plasmids, transconjugants were selected for resistance to chloramphenicol for all complementation experiments. In the presence of p25Δ.2 or pBR2N, both pAC114 and pAC150 were mobilized in E. coli at wild-type frequencies (Table 4). Restriction enzyme analysis of transconjugant plasmid DNA verified that the mutant plasmids were mobilized and that there were no alterations to the plasmids (data not shown). Further, the insertions in orf24 were not polar on orf25, since two insertions in orf25, pAC166 and pAC115, did not affect mobilization of pAC11 (Fig. 3). These results demonstrated that orf24 encodes a trans-acting factor that is essential for the mobilization of cLV25. We designated this gene bmgA (Bacteroides mobilization gene).
TABLE 4.
R751 mobilization of pAC11 containing Tn1000 insertions in the presence and absence of cLV25 in trans
| Tn1000 location | Plasmida | cLV25 in transc
|
Frequency of mobilization (mean ± SE)b | |
|---|---|---|---|---|
| p25Δ.2 | pBR2N | |||
| orf24 | pAC150 | − | − | ND |
| + | − | (4.1 ± 0.92) × 10−4 | ||
| − | + | (4.7 ± 0.87) × 10−5 | ||
| pAC114 | − | − | ND | |
| + | − | (4.3 ± 0.78) × 10−5 | ||
| − | + | (3.1 ± 0.72) × 10−5 | ||
| orf23 | pAC167 | − | − | ND |
| + | − | (1.8 ± 1.3) × 10−4 | ||
| − | + | (7.5 ± 4.0) × 10−5 | ||
| orf22 | pAC148 | − | − | (7.0 ± 4.4) × 10−7 |
| + | − | (1.6 ± 0.76) × 10−4 | ||
| − | + | (9.4 ± 1.7) × 10−5 | ||
Plasmids were mobilized from E. coli HB101 to E. coli DW1030.
The frequency of mobilization was determined by dividing the number of transconjugants that received the mobilized plasmid by the number of transconjugants that received R751. The mean was obtained from a minimum of four independent matings. SE, standard error; ND, mobilization not detectable.
+, present; −, absent.
The gene product of orf23, BmgB, is also required for mobilization and functions in trans.
An insertion in pAC167 disrupted orf23, a putative mobilization gene, and abolished mobilization of pAC11. To determine whether pAC167 could be complemented in trans, pAC167, coresident with either p25Δ.2 or pBR2N, was tested for mobilization in E. coli by R751. In the presence of p25Δ.2 or pBR2N, pAC167 was mobilized at a wild-type frequency (Table 4). Restriction enzyme analysis of transconjugant plasmid DNA confirmed that pAC167 was mobilized and that there were no alterations (data not shown). To determine whether the insertion in orf23 was polar on orf24, a 798-bp region containing orf23 and upstream sequence was cloned into pCR2.1-TOPO to generate pCR23A. In the presence of pCR23A, mobilization of pAC167 in E. coli was restored to wild-type frequency. Complementation of pAC167 by orf23 alone indicated that the transposon insertion in orf23 was not polar on orf24. The transposon insertion in pAC148, located at the 3′ end of orf22, reduced the frequency of cLV25 mobilization 100- to 1,000-fold compared to wild-type pAC11. When coresident with p25Δ.2 or pBR2N, pAC148 was mobilized at a wild-type frequency (Table 4). Restriction enzyme analysis of transconjugant plasmid DNA demonstrated that pAC148 was unaltered (data not shown). To determine if the transposon insertion in orf22 was polar on orf23, pAC148 was tested for complementation by orf23 alone (pBR23A) and by orf23 and orf24 (pBR2.5). pAC148 was mobilized at a wild-type frequency when coresident with pBR23A or pBR2.5, indicating that the mutation in orf22 was polar on orf23. Further, pBR2.5 was mobilized by R751 at a frequency of (9.6 ± 3.8) × 10−4 and is the smallest subclone of cLV25 thus far mobilized (Fig. 3). Therefore, the gene product of orf23 is a trans-acting factor also required for mobilization of cLV25. This gene was designated bmgB.
Tn4399 partially complements cLV25 bmgA.
Given the high degree of protein sequence similarity between cLV25 BmgA and BmgB and Tn4399 MocA and MocB, respectively, we tested whether the insertion mutations in bmgA and bmgB could be complemented by Tn4399 in trans. pAC150, pAC114, and pAC167 were tested for mobilization by R751 in E. coli when coresident with p21ΔB, which contains Tn4399. p21ΔB was previously obtained at the same time as other Tn4399 insertions in pGAT400ΔBglII (12). In the presence of p21ΔB, pAC150 was mobilized at a frequency of (3.6 ± 1.3) × 10−7, 3 orders of magnitude lower than wild-type cLV25, while the frequency of mobilization of p21ΔB was (1.5 ± 0.53) × 10−5. There was no complementation of pAC114 or pAC167 by p21ΔB. Thus, Tn4399 partially complements one of the mutations in cLV25 bmgA, the gene for the MocA homolog, but not the mutation in cLV25 bmgB, the gene for the MocB homolog.
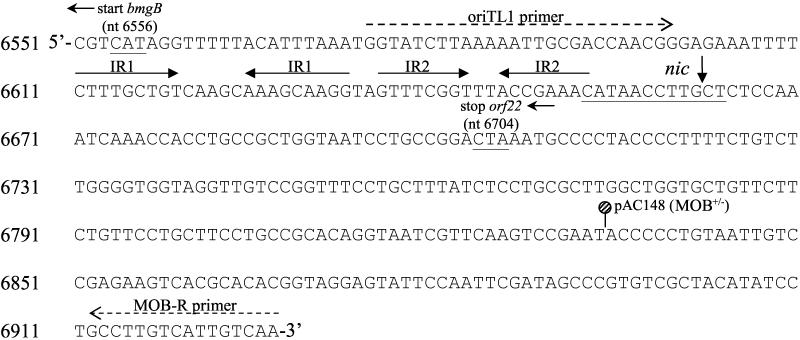
Cloning of the cLV25 origin of transfer.
Examination of the cLV25 DNA sequence revealed the presence of a potential 12-bp nic site sequence, containing the sequence CTTGC, 106 bp upstream of the bmgB start codon on the cDNA strand (Fig. 4) (24). Two sets of inverted repeats (9 and 8 bp) were identified immediately upstream of this putative nic site. To determine whether the functional cLV25 oriT was located within this region, a 353-bp fragment that encompassed nearly all of the intergenic region between bmgB and orf22 and overlapped with the 3′ end of orf22 was cloned into pACYC184 to generate pACoriT7 (Fig. 4). When cLV25 was provided on p25Δ.2 in trans, pACoriT7 was mobilized in E. coli by R751 at a frequency of (2.1 ± 0.53) × 10−3, a 10-fold-greater frequency than p25Δ.2 (Table 3). Restriction enzyme analysis confirmed that there were no alterations to the mobilized plasmid (data not shown). When the DNA sequence of the cLV25 oriT was compared directly with those of other Bacteroides sp. transfer factors, the putative 12-bp cLV25 nic site sequence was found to be nearly identical (10 of 12 bp) to the nic site sequence of Tn4399 (19). Other than the cLV25 putative nic site, there was no significant homology with the Tn4399 oriT sequence or other Bacteroides transfer factor oriT sequences.
FIG. 4.
Nucleotide sequence of the 353-bp cLV25 oriT. The vertical arrow indicates the position of the putative nic site. The putative nic site sequence, the proposed start codon for bmgB (orf23), and the proposed stop codon for orf22 are underlined. bmgB and orf22 are coded by the cDNA strand. The solid arrows above the sequence indicate two sets of inverted repeats (IR). The positions of the oriTL1 and MOB-R primers used to amplify the oriT are indicated above the sequence by the dashed arrows. The location of the Tn1000 insertion in pAC148 is indicated.
DISCUSSION
We isolated, sequenced, and characterized a new 15.3-kb chromosomal transfer factor, cLV25, from B. fragilis clinical isolate LV25. cLV25 was isolated by its insertion into the E. coli-Bacteroides shuttle vector, pGAT400ΔBglII. cLV25 was presumably mobilized (as part of pGAT400ΔBglII) from B. fragilis LV25 to E. coli HB101 by a conjugative transposon and does not appear to encode a transfer apparatus to permit its self-transfer. cLV25 was also mobilized in E. coli by R751, requiring two mobilization genes, bmgA and bmgB, and a 353-bp functional oriT.
Features of cLV25 suggest that it is a mobilizable transposon, and its potential transposition properties are most similar to those of Tn4555, a Bacteroides vulgatus mobilizable transposon. cLV25 insertions in pGAT400ΔBglII were site specific and resulted in an 8-bp target site repeat. Additionally, there was an extra 8 bp associated with the left end of cLV25, which may represent a coupling sequence similar to that required for transposition of Tn4555 (41). The ends of Tn4555 consist of a 12-bp imperfect inverted repeat, and a 6-bp variable coupling sequence is utilized as a crossover region for recombination. In contrast to cLV25, transposition of Tn4555 does not result in a target site repeat (41). Data to support probable transposition activity of cLV25 are the presence of a 20-bp imperfect inverted repeat at the termini and putative integrase- and excisionase-encoding genes. The predicted protein for cLV25 orf11 has the most similarity to the entire Tn4555 integrase, Int (42). Further, carboxyl-terminus regions of Orf11 and Int, as well as BipH (Tn5520), IntN1 (NBU1), and IntN2 (NBU2), have similarity to the lambda family of site-specific integrases (33, 42, 46, 48). The putative excisionase encoded by cLV25 orf20 has similarity to a putative excisionase from S. enterica (4) as well as limited similarity to a second putative excisionase, Xis, from Tn4555 (40% identity over an internal stretch of 55 amino acids) (42). cLV25 was tested for indirect transposition using a standard assay in E. coli for cointegrate formation with pOX38-Km, a derivative of the F factor (5). No cointegrate formation between pOX38-Km and cLV25 was observed. However, this result does not rule out this mode of transposition, since pOX38-Km may not be the ideal test system for cLV25, or direct transposition. Alternatively, cLV25 may not transpose in E. coli, especially since transposition in E. coli has only been demonstrated for one Bacteroides mobilizable transposon, Tn5520 (46).
Two features of cLV25 differentiate it from previously characterized Bacteroides transfer factors. The first distinguishing feature is the in vivo functional cross-reaction between cLV25 and Tn4399, whose mobilization proteins, MocA and MocB, have similarity to BmgA and BmgB, respectively. An insertion mutation in cLV25 bmgA, the putative DNA relaxase, was complemented by Tn4399 in trans, although the frequency of mobilization was considerably lower than that of wild-type cLV25. This suggests that the Tn4399 DNA relaxase, MocA, can nick the cLV25 oriT, albeit inefficiently. A previous study reported a functional cross-reaction between Bacteroides mobilization proteins of NBU1 and NBU2 and their noncognate, but virtually identical, oriTs (15). However, the NBU1 and NBU2 mobilization proteins, MobN1 and MobN2, have a significantly higher degree of similarity (86% identity) than BmgA and MocA (42% identity). Likewise, MobN1 can also complement Tn4555 MobA (37). These two mobilization proteins also have a very high degree of similarity (76% identity).
The cLV25-Tn4399 cross-reaction also differs from the MobN1-MobN2 and MobN1-MobA cross-reactions in the extent of oriT DNA sequence identity shared between complementing transfer factors. The NBU1 and NBU2 oriT DNA sequences are nearly identical (89%) (48), while 55 bp of the Tn4555 nic site region has 90% sequence identity with that of NBU1 (38). In contrast, the cLV25 oriT has no significant sequence identity with the Tn4399 oriT except for its putative nic site, which includes the sequence CTTGC. An identical 5-bp sequence is also present in the predicted nic sites of other Bacteroides transfer factors, namely, pBFTM10, pLV22a, and Tn5520 (35, 20, 46). It is likely that this 5-bp sequence plus a few adjacent nucleotides is important and sufficient for recognition and/or DNA nicking by a homologous DNA relaxase. Thus, the results of our complementation study are an example of a cross-reaction between two mobilizable transfer factors that have related DNA relaxases but distinct oriTs. Such a functional cross-reaction has been previously demonstrated for pSC101 and R1162. These two E. coli mobilizable plasmids have homologous DNA nickase proteins, but their oriT DNA sequences are unrelated except for the 12-bp cleavage sites (16). cLV25 bmgB was not complemented by its Tn4399 homolog, MocB. This may indicate that there are specific interactions between the BmgB mobilization protein and the oriT and/or BmgA not complemented by Tn4399. The function of BmgB in the mobilization of cLV25 is unknown.
The second distinguishing feature of cLV25 is that it has protein sequence similarity to multiple Bacteroides transfer factors, including Tn4399 (MocA, MocB, Orf1, and Orf3), CTnDOT (MobA, MobB, PrmF, Orf3F, and OrfP), NBU1 (PrmN1), NBU2 (PrmN2, Orf3, and Orf2), Tn4555 (Int), and Tn5520 (BipH). The mobilization region of cLV25 is similar in both protein sequence and organization to two different Bacteroides transfer factors, a mobilizable transposon (Tn4399) and a conjugative transposon (CTnDOT). This high degree of protein sequence similarity and analogous genetic organization suggests that cLV25, Tn4399, and a portion of CTnDOT (speculated to be a mobilizable transposon [49]) may have diverged from a common mobilization region cassette.
Other significant homology was seen between cLV25 Orf22 and Orf1 and Orf3 of Tn4399, PrmN1 of NBU1, PrmN2 of NBU2, OrfP and PrmF of CTnDOT, and the amino termini of several bacterial DNA primases. Based upon the homology between Orf22 and both Orf1 and Orf3 of Tn4399, we suggest that their encoding genes, orf1 and orf3, are part of a single open reading frame that would encode a putative full-length DNA primase. This single open reading frame would include Tn4399 orf2, which appears to be out of frame with orf1 and orf3. Sequence analysis of cLV25 Orf22 also revealed that it contains a putative CHCC-type zinc finger, a domain primarily involved in DNA binding in bacterial and bacteriophage DNA primases (47), but the conserved zinc binding cysteine and histidine residues are not present. Similarly, these conserved residues are absent from the putative zinc finger motif in Orf1 of Tn4399, PrmN1, PrmN2, and OrfP, and no DNA primase function has been described for any of these proteins. A second sequence motif, EGFMD(Y/F/D) (3), derived from an alignment of putative DNA primases from Bacteroides conjugative transposons and Lactococcus lactis DnaG was present in Orf22 (amino acids 195 to 200). This Bacteroides sequence is similar to the consensus motif (EGYATA) identified in DNA primases encoded by bacterial chromosomes and bacteriophages, as well as conjugative plasmids. Substitutions of specific amino acids in the latter motif result in a loss of priming activity (23, 40). If Orf22 functions as a DNA primase, it may play an important role in the recipient to ensure cDNA strand synthesis after transfer of cLV25.
The predicted cLV25 proteins Orf12 and Orf21 have similarity to NBU2 Orf2 and both NBU2 Orf3 and CTnDOT Orf3F, respectively. In addition, cLV25 may also encode a number of different proteins, including those which have similarity to CRP/FNR-type transcriptional regulators and putative heme receptors. These putative proteins are encoded in the terminal regions of cLV25. Interestingly, a percent G+C content profile of cLV25 reveals that both its 5′- and 3′-terminal regions (4.1 and 2.2 kb, respectively) have an average G+C content of 35%, similar to those of gram-positive bacteria; the central 9 kb of cLV25 has an average G+C content of 44%, consistent with the average G+C content reported for Bacteroides spp. (40 to 48%) (42). Thus, cLV25 may comprise both Bacteroides and gram-positive DNA. Further analyses of these potential open reading frames will be helpful in understanding the divergence of cLV25 from the Bacteroides transfer factors to which it is similar.
In summary, we have characterized a unique transfer factor from B. fragilis, cLV25, that resembles multiple Bacteroides transfer factors and is putatively a mobilizable transposon. Further, both cLV25 and Tn4399 mobilization proteins can functionally nick the cLV25 oriT, despite the lack of extended DNA sequence identity in this region.
Acknowledgments
We thank Gayatri Vedantam for helpful discussions and critical reading of the manuscript.
This work was supported by VA Merit Review grant 006 to D.W.H.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 3.Bonheyo, G., D. Graham, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon, CTnDOT. Plasmid 45:41-51. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, D. A., G. A. Peters, L. Ng, and M. R. Mulvey. 2000. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 189:285-291. [DOI] [PubMed] [Google Scholar]
- 5.Chandler, M., and D. J. Galas. 1983. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J. Mol. Biol. 170:61-91. [DOI] [PubMed] [Google Scholar]
- 6.Covarrubias, L., L. Cervantes, A. Covarrubias, X. Soberon, I. Vichido, A. Blanco, Y. M. Kupersztoch-Portnoy, and F. Bolivar. 1981. Construction and characterization of new cloning vehicles. V. Mobilization and coding properties of pBR322 and several deletion derivatives including pBR327 and pBR328. Gene 13:25-35. [DOI] [PubMed] [Google Scholar]
- 7.Furste, J. P., W. Pansegrau, G. Ziegelin, M. Kroger, and E. Lanka. 1989. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc. Natl. Acad. Sci. USA 86:1771-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guiney, D. G. 1984. Promiscuous transfer of drug resistance in gram-negative bacteria. J. Infect. Dis. 149:320-329. [DOI] [PubMed] [Google Scholar]
- 9.Guiney, D. G., and E. Lanka. 1989. Conjugative transfer of IncP plasmids, p. 27-56. In C. M. Thomas (ed.), Promiscuous plasmids of gram negative bacteria. Academic Press Ltd., London, United Kingdom.
- 10.Guyer, M. S. 1978. The gamma delta sequence of F is an insertion sequence. J. Mol. Biol. 126:347-365. [DOI] [PubMed] [Google Scholar]
- 11.Hecht, D. W., T. J. Jagielo, and M. H. Malamy. 1991. Conjugal transfer of antibiotic resistance factors in Bacteroides fragilis: the btgA and btgB genes of plasmid pBFTM10 are required for its transfer from Bacteroides fragilis and for its mobilization by IncPβ plasmid R751 in Escherichia coli. J. Bacteriol. 173:7471-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hecht, D. W., and M. H. Malamy. 1989. Tn4399, a conjugal mobilizing transposon of Bacteroides fragilis. J. Bacteriol. 171:3603-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecht, D. W., J. S. Thompson, and M. H. Malamy. 1989. Characterization of the termini and transposition products of Tn4399, a conjugal mobilizing transposon of Bacteroides fragilis. Proc. Natl. Acad. Sci. USA 86:5340-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 15.Li, L. Y., N. B. Shoemaker, G. R. Wang, S. P. Cole, M. K. Hashimoto, J. Wang, and A. A. Salyers. 1995. The mobilization regions of two integrated Bacteroides elements, NBU1 and NBU2, have only a single mobilization protein and may be on a cassette. J. Bacteriol. 177:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer, R. 2000. Identification of the mob genes of plasmid pSC101 and characterization of a hybrid pSC101-R1162 system for conjugal mobilization. J. Bacteriol. 182:4875-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, R. J., and J. A. Shapiro. 1980. Genetic organization of the broad-host-range IncP-1 plasmid R751. J. Bacteriol. 143:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy, C. G., and M. H. Malamy. 1993. Characterization of a “mobilization cassette” in transposon Tn4399 from Bacteroides fragilis. J. Bacteriol. 175:5814-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy, C. G., and M. H. Malamy. 1995. Requirements for strand- and site-specific cleavage within the oriT region of Tn4399, a mobilizing transposon from Bacteroides fragilis. J. Bacteriol. 177:3158-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novicki, T. J., and D. W. Hecht. 1995. Characterization and DNA sequence of the mobilization region of pLV22a from Bacteroides fragilis. J. Bacteriol. 177:4466-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pansegrau, W., D. Balzer, V. Kruft, R. Lurz, and E. Lanka. 1990. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc. Natl. Acad. Sci. USA 87:6555-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pansegrau, W., and E. Lanka. 1991. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 19:3455.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pansegrau, W., and E. Lanka. 1992. A common sequence motif among prokaryotic DNA primases. Nucleic Acids Res. 20:4931.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pansegrau, W., and E. Lanka. 1996. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 54:197-251. [DOI] [PubMed] [Google Scholar]
- 25.Pansegrau, W., and E. Lanka. 1996. Mechanisms of initiation and termination reactions in conjugative DNA processing. J. Biol. Chem. 271:13068-13076. [DOI] [PubMed] [Google Scholar]
- 26.Robillard, N. J., F. P. Tally, and M. H. Malamy. 1985. Tn4400, a compound transposon isolated from Bacteroides fragilis, functions in Escherichia coli. J. Bacteriol. 164:1248-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito, H., and K. I. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 28.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Shoemaker, N. B., C. Getty, E. P. Guthrie, and A. A. Salyers. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 166:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoemaker, N. B., and A. A. Salyers. 1988. Tetracycline-dependent appearance of plasmidlike forms in Bacteroides uniformis 0061 mediated by conjugal Bacteroides tetracycline resistance elements. J. Bacteriol. 170:1651-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 1996. NBU1, a mobilizable site-specific integrated element from Bacteroides spp., can integrate nonspecifically in Escherichia coli. J. Bacteriol. 178:3601-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 1996. The Bacteroides mobilizable insertion element, NBU1, integrates into the 3′ end of a Leu-tRNA gene and has an integrase that is a member of the lambda integrase family. J. Bacteriol. 178:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 2000. Multiple gene products and sequences required for excision of the mobilizable integrated Bacteroides element NBU1. J. Bacteriol. 182:928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sitailo, L. A., A. M. Zagariya, P. J. Arnold, G. Vedantam, and D. W. Hecht. 1998. The Bacteroides fragilis BtgA mobilization protein binds to the oriT region of pBFTM10. J. Bacteriol. 180:4922-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, C. J., and A. C. Parker. 1993. Identification of a circular intermediate in the transfer and transposition of Tn4555, a mobilizable transposon from Bacteroides spp. J. Bacteriol. 175:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, C. J., and A. C. Parker. 1996. A gene product related to TraI is required for the mobilization of Bacteroides mobilizable transposons and plasmids. Mol. Microbiol. 20:741-750. [DOI] [PubMed] [Google Scholar]
- 38.Smith, C. J., and A. C. Parker. 1998. The transfer origin for Bacteroides mobilizable transposon Tn4555 is related to a plasmid family from gram-positive bacteria. J. Bacteriol. 180:435-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, C. J., G. D. Tribble, and D. P. Bayley. 1998. Genetic elements of Bacteroides species: a moving story. Plasmid 40:12-29. [DOI] [PubMed] [Google Scholar]
- 40.Strack, B., M. Lessl, R. Calendar, and E. Lanka. 1992. A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the alpha protein of the Escherichia coli satellite phage P4. J. Biol. Chem. 267:13062-13072. [PubMed] [Google Scholar]
- 41.Tribble, G. D., A. C. Parker, and C. J. Smith. 1997. The Bacteroides mobilizable transposon Tn4555 integrates by a site-specific recombination mechanism similar to that of the gram-positive bacterial element Tn916. J. Bacteriol. 179:2731-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tribble, G. D., A. C. Parker, and C. J. Smith. 1999. Genetic structure and transcriptional analysis of a mobilizable, antibiotic resistance transposon from Bacteroides. Plasmid 42:1-12. [DOI] [PubMed] [Google Scholar]
- 43.Tribble, G. D., A. C. Parker, and C. J. Smith. 1999. Transposition genes of the Bacteroides mobilizable transposon Tn4555: role of a novel targeting gene. Mol. Microbiol. 34:385-394. [DOI] [PubMed] [Google Scholar]
- 44.Trinh, S., and G. Reysset. 1997. Identification and DNA sequence of the mobilization region of the 5-nitroimidazole resistance plasmid pIP421 from Bacteroides fragilis. J. Bacteriol. 179:4071-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valentine, P. J., N. B. Shoemaker, and A. A. Salyers. 1988. Mobilization of Bacteroides plasmids by Bacteroides conjugal elements. J. Bacteriol. 170:1319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vedantam, G., T. J. Novicki, and D. W. Hecht. 1999. Bacteroides fragilis transfer factor Tn5520: the smallest bacterial mobilizable transposon containing single integrase and mobilization genes that function in Escherichia coli. J. Bacteriol. 181:2564-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Versalovic, J., and J. R. Lupski. 1993. The Haemophilus influenzae dnaG sequence and conserved bacterial primase motifs. Gene 136:281-286. [DOI] [PubMed] [Google Scholar]
- 48.Wang, J., N. B. Shoemaker, G. R. Wang, and A. A. Salyers. 2000. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J. Bacteriol. 182:3559-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whittle, G., B. D. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT. Appl. Environ. Microbiol. 67:3488-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkins, B., and E. Lanka. 1993. DNA processing and replication during plasmid transfer between gram-negative bacteria, p. 105-136. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.